Abstract
Introduction. EGF/EGFR interactions are important mechanisms behind colorectal tumour development and growth. Recently a single nucleotide polymorphism in the EGF gene has been identified (EGF A61G). It may be a potential predictor for survival of patients receiving EGFR-inhibitor cetuximab treatment, but the clinical importance and the functional influence on EGF gene expression levels in colorectal cancer (CRC) patients have not yet been further assessed. The aim of the present study was to investigate the relationship between EGF A61G genotype and EGF gene expression levels in colorectal adenocarcinomas and normal colon tissue. Material and methods. Eighty-one CRC patients were included in the study. Tissue samples from normal colon, adenocacinomas and corresponding blood samples were analysed by real-time PCR for EGF gene expression and EGF A61G genotype, respectively. Results. Thirty-three percent were AA, 48% and 19% A/G and G/G respectively. We found a significantly lower median age in the A/A group compared to the G/G group, suggesting a later time of diagnosis in the G/G patients. There was a significant difference between the median EGF gene expression among the three genotypes in normal colon (p < 0.001) but not in adenocarcinomas. Furthermore, the median EGF gene expression was lower in CRC tissue than in normal colon samples, (0.13 (range 0.01–6.4) vs. 0.76, (range 0.013–5.55)). Conclusion. We suggest that EGF A61G genotype has a functional influence on EGF gene expression in normal colon in CRC patients. The clinical implications warrant further investigations in prospective trials.
The epidermal growth factor (EGF) is one of the natural ligands of the epidermal growth factor receptor (EGFR), which is a transmembrane tyrosine kinase receptor critical to normal cell proliferation and differentiation. Dysregulation of the EGFR signalling system occurs frequently in colorectal cancer (CRC) by different mechanisms e.g. receptor overexpression, gene amplification and/or mutations, thereby contributing to tumour development and growth Citation[1], Citation[2]. The EGF/EGFR interaction is suggested to function as an important autocrine loop Citation[3], which promotes tumour development by stimulating the intracellular tyrosine kinase activity.
EGFR has become a promising target for anti cancer treatment, and clinical trials of both monoclonal antibodies (cetuximab, panitumumab) and small molecule tyrosine kinase inhibitors (erlotinib, gefitinib) have reached larger phase III studies in a variety of different solid tumours including CRC Citation[4–7]. Prediction of response to these new targeted therapies has become a critical issue since none of the apparent targets seem to contain predictive information. The EGFR staining intensity by immunohistochemistry (IHC) is not a reliable marker for selection of patients to cetuximab containing regiments, as shown in the trials by Cunningham et al. and Saltz et al. Citation[6], Citation[8] Furthermore, patients with EGFR IHC negative tumours are potential responders as reported by Chung et al. Citation[9]. On the other hand, a study on NSCLC demonstrated a correlation between response to EGFR TKI gefitinib and mutational status of the EGFR gene located in exon 18, 19 and 21, but these mutations are rare in CRC Citation[10], Citation[11]. These data suggest that individual alterations of genes involved in the complex EGF/EGFR signalling pathway might influence the efficacy of EGFR inhibitors as well as provide prognostic and predictive information in CRC. Thus we have previously evaluated the importance of a functional single nucleotide polymorphism (SNP) in the EGFR promoter region (EGFR Sp1-216) in patients with rectal cancer and we suggest that it may be an important predictor of response to preoperative chemoradiation Citation[12].
Recently a SNP in the EGF gene has been identified. The EGF A61G polymorphism is located in the 5′-untranslated region at position 61 and reported to imply functional influence on EGF production. Mononuclear cells from individuals with the A/A genotype have been reported to produce significantly lower levels of EGF than cells from G containing variants Citation[13]. Furthermore Bhowmick and colleagues showed that intra-tumoural EGF gene expression levels varied according to the different genotypes in glioblastoma patients Citation[14]. A single study has evaluated the clinical value of EGF polymorphism in CRC patients receiving EGFR inhibitor cetuximab and found a trend for association between EGF genotype and survival Citation[15], but the functional influence of EGF A61G has not yet been investigated in CRC. Thus, the literature on EGF A61G in CRC is sparse and calls for further investigation.
The present study aims to provide a basic knowledge of the functional influence of EGF A61G genotype on EGF- and EGFR gene expression levels in colorectal adenocarcinomas and normal colon from CRC patients.
Materials and methods
The study included 81 patients with histologically confirmed colorectal adenocarcinomas, who were operated at Vejle Hospital during the period of December 2003 to June 2005. Blood samples were drawn at surgery and tissue samples were collected from surgical specimens including both colorectal adenocarcinomas and normal colon. Fresh tissue was stored in RNA later (Qiagen, CA, USA). The amount of adenocarcinoma cells in the tumour specimens was estimated by an experienced G-I pathologist by visual macroscopical examination and the specimens from tumours and normal colon was stored in RNA later. The study was approved by the Regional Ethics Committee of Vejle and Funen Counties according to Danish law.
EGF A61G gene polymorphism analysis
Genomic DNA was extracted from whole blood as previously described Citation[16]. PCR analysis of the EGF A61G polymorphisms was performed using the ABI PRISM 7900 HT Sequence Detection System. For detection of the EGF A61G polymorphism a pre-developed assay (PDAR, Applied Biosystems, no c __27031637 _10) was used. EGFR-Sp1 polymorphism analysis was performed as previously described Citation[12].
Gene expression analysis
Total RNA was isolated using RNeasy kit from Qiagen according to manufacturer's instructions. The concentration and purity of the samples was determined by optical density at wavelengths 260 nm and 280 nm (The isolated RNA was quantitated by Spectophotometry) (Eppendorf, Hamburg, Germany). cDNA synthesis was performed using M-MLV RT (Invitrogene) as previously described Citation[17], and subsequently, the mRNA was quantified by Real-time PCR (ABI PRISM 7900 HT FAST Sequence Detection System). The PCR amplification was conducted using the TaqMan® Fast Universal PCR Master mix (2x) (Applied Biosytems) according to manufactures instructions. For EGF gene expression analysis the following primers and probe were used: forward primer (exon 22-24): 5′-TGT CCT CTT GCC CTC AAC CTT-3′, reverse primer (exon 22-24) 5′-TGC TGC CTG GCC ATC CT-3′ and probe: 5′-FAM-TGT GGT TAT AAA AGA ACA CCA AGA CCT CAA GAA TGG-TAMRA-3′. The housekeeping gene ß-actin (BA) was measured by the assay; 4310881E, (Applied Biosystems) and used for normalization of EGF (qEGF/qBA). Quantification of the PCR reactions was based on a standard curve composed of total RNA from the HRT cell line (Stratagene). A serial dilution of six concentrations was performed. Positive and negative controls were included in each run, and all analyses were carried out in triplicates. A pre-developed assay (PDAR, Hs01076088_m1, Applied Biosystems) was used for detection of EGFR mRNA as previously published Citation[18].
Statistical methods
Fisher's exact test was used for comparison of groups and Students t-test or Wilcoxon statistics were used for analysis of continues variables when appropriate. χ2 test was used to evaluate the Hardy-Weinberg equilibrium (HWE) of allelic frequencies. P-values were considered significant if p ≤ 0.05. All statistics were carried out using the NCSS statistical software (NCSS Statistical Software, Utah 84037, USA).
Results
Patient characteristics are shown in . Disease extension was evaluated by Dukes classification. A total of 81 patients had available blood samples for EGF A61G polymorphism analysis and corresponding EGF gene expression analysis in tissue samples.
Table I. Patient characteristics.
Thirty-three percent of patients were A/A genotype, 48% and 19% A/G and G/G respectively. χ2 test was compatible with Hardy-Weinberg equilibrium (p > 0.10). There was no correlation between the different genotypes and clinicopathological parameters (gender, classification and topography) except for a difference in median age between the A/A and G/G group (69 and 79 years respectively, p = 0.026), as shown in . The mean and median EGF gene expression levels did not differ according to the clinocopathological parameters.
Figure 1. Distribution of age and EGF gene expression levels in normal colon tissue according to EGF A61G genotype. The left side boxplots (orange) represent the EGF gene expression levels in normal colon and the right side box plots (blue) the age in years.
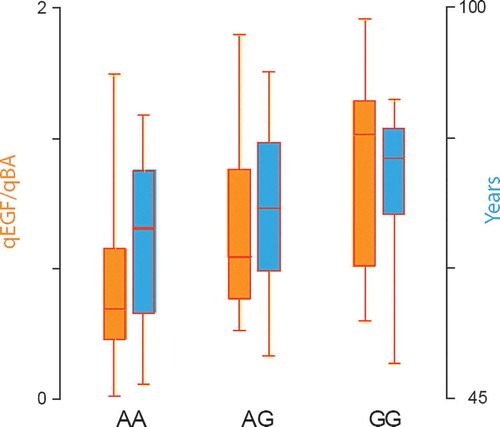
The median gene expression levels according to EGF A61G genotype are shown in . No difference was found in the CRC tissue according to genotype. There was a significant different median qEGF/qBA level in normal colon tissue between the three groups (). The differences between A/A and A/G (p = 0.0003), A/G and G/G (p = 0.0007) and A/A and G/G (p = 0.02) genotypes were all significant.
Table II. Median gene expression levels according to EGF A61G genotype (*p < 0.05).
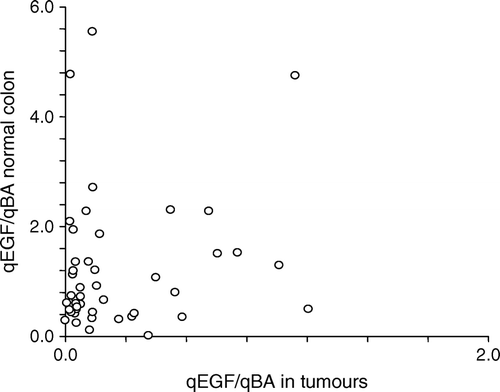
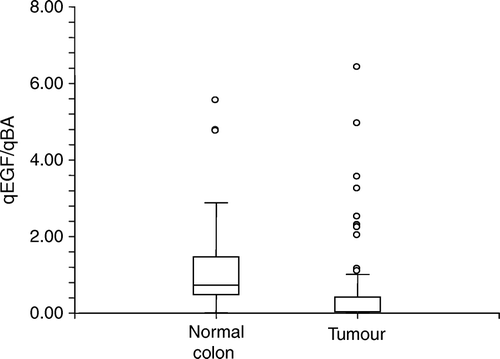
There was no correlation between EGF gene expression levels in colon and corresponding normal colon tissue as illustrated by scatter plot in . However, the median gene expression level of EGF was significantly lower (p < 0.0001) in the CRC tissue than in the normal colon samples (, 0.13 (0.01–6.4) and 0.76 (0.013–5.55), respectively (Median (range)).
The median EGFR gene expression levels did not differ with respect to EGF A61G genotypes and there was no relation between the EGF A61G and EGFR Sp1-216 genotype. Data presenting the EGFR gene expression levels according to EGFR Sp1-216 genotype are previously published Citation[18]. Furthermore Sp1-216 genotypes were not correlated to EGF gene expression levels (data not shown).
Discussion
The clinical importance of EGF A61G polymorphism has been evaluated in several malignant diseases e.g. malignant melanoma and glioblastomas Citation[14], Citation[19–23]. Recently, Zhang and colleagues investigated the predictive value of EGF A61G polymorphism in addition to several potential SNPs in CRC patients receiving EGFR inhibitor cetuximab. EGF A61G genotype showed a trend for association with survival and the authors suggest that a combined analysis of EGF A61G and cyclin D1 A870G polymorphism may be useful markers for clinical outcome of patients treated with single agent cetuximab Citation[15]. Only a few studies have assessed the functional importance of the EGF genotype, and no studies have addressed the issue in colorectal cancer.
The overall genotype frequencies in our population of CRC patients are comparable to the frequencies in previously reported healthy control groups. The present study found 33% of patients with A/A, 48% A/G and 19% with the G/G genotype as compared to 32–39% A/A, 42–50% A/G and 12.9–20.2% G/G in previous studies Citation[13], Citation[19–24]. Furthermore the test for Hardy-Weinberg equilibrium showed frequencies within the probability limits for HWE. On the other hand we found a significant difference of age between two different genotypes. This finding might suggest that patients with A/A variant are younger at the time of diagnosis than patients with the G/G genotype (median 69 versus 79 years, respectively). Furthermore the median EGF gene expression in normal colon was higher in the high-age genotype group (G/G), and low in the A/A patients. These observations could support the hypothesis that EGF G/G genotype might be a potential indicator for late CRC diagnosis, but is not necessarily an indicator for a lower risk of disease. Obviously, no firm conclusion on these aspects based on this rather small subset of patients can be drawn.
The present data indicates that EGF A61G polymorphism has a functional influence on EGF gene expression levels in normal colon tissue as shown in . These findings support the observations by Shahbazi and colleagues, who demonstrated a significantly increased EGF expression in primary cultures of peripheral mononuclear cells from normal individuals with G containing variants Citation[13]. Furthermore Bhowmick and colleagues showed a similar difference in EGF gene expression levels between the different genotypes in tissue from 42 glioblastomas Citation[14]. However the gene expression level of EGF in normal tissue was not assessed. The mechanism by which the EGF A61G genotype contributes to regulation of the EGF expression is unknown. However, this SNP is located in the EGF 5′UTR and might therefore be of critical importance to EGF gene expression.
The EGF A61G genetic variant did not seem to influence the EGFR gene expression in neither tumour nor normal colon tissue, which is also in agreement with the data from Bhowmich et al.
In addition to the findings discussed above, the present study provides us with two significant observations. Firstly, we found that the median EGF gene expression was significantly lower in tumours than in normal colon tissue. Secondly the EGF A61G genotype seems to influence the gene expression levels in normal colon tissue, but not in tumour samples.
The present observations are supported by a recent study by Schneider et al., who investigated different candidate genes (including EGFR) in tumour-adjacent and intra-tumoural colon tissue and reported a significant lower mean gene expression levels in tumours compared to “normal colon” tissue. Furthermore they found that tumour-adjacent normal tissue gene expression of key candidate genes were better overall predictors for early recurrence than intra-tumoural gene-expression Citation[24]. Additionally, Odin et al. previously reported that the gene expression alterations of folate enzymes in tumour-adjacent mucosa, but not in the adenocarcinomas, were associated with survival in CRC Citation[25]. These studies emphasis the potential value of determining “normal” tissue gene expression levels as markers for clinical outcome in CRC patients.
The present study has the obvious limitations of a rather small sample size, and furthermore the possible heterogeneous nature of tumour tissue compared to normal colon should also be taken into consideration when interpreting these data. However, this is the first report of the possible functional influence of the EGF A61G polymorphism in CRC patients, which could serve as a basis for further investigations.
In conclusion, the present study indicates a functional influence of the EGF polymorphism in normal colon in contrast to carcinomas. It also indicates a low EGF gene expression level in colorectal adenocarcinomas compared to normal colon. The potential clinical implications need to be investigated.
References
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001; 2: 127–37
- Lockhart C, Berlin JD. The epidermal growth factor receptor as a target for colorectal cancer therapy. Semin Oncol 2005; 32: 52–60
- Gleave ME, Hsieh JT, Wu HC, Hong SJ, Zhau HE, Guthrie PD, et al. Epidermal growth factor receptor-mediated autocrine and paracrine stimulation of human transitional cell carcinoma. Cancer Res 1993; 53: 5300–7
- Tyagi P. Recent results and ongoing trials with panitumumab (ABX-EGF), a fully human anti-epidermal growth factor receptor antibody, in metastatic colorectal cancer. Clin Colorectal Cancer 2005; 5: 21–3
- Shepherd FA, Rodrigues PJ, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005; 353: 123–32
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 2004; 351: 337–45
- Calvo E, Rowinsky EK. Clinical experience with monoclonal antibodies to epidermal growth factor receptor. Curr Oncol Rep 2005; 7: 96–103
- Saltz LB, Meropol NJ, Loehrer PJ, Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol 2004; 22: 1201–8
- Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005; 23: 1803–10
- Nagahara H, Mimori K, Ohta M, Utsunomiya T, Inoue H, Barnard GF, et al. Somatic mutations of epidermal growth factor receptor in colorectal carcinoma. Clin Cancer Res 2005; 11: 1368–71
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–39
- Spindler KL, Nielsen JN, Lindebjerg J, Brandslund I, Jakobsen A. Prediction of response to chemoradiation in rectal cancer by a gene polymorphism in the epidermal growth factor receptor promoter region. Int J Radiat Oncol Biol Phys 2006; 66: 500–4
- Shahbazi M, Pravica V, Nasreen N, Fakhoury H, Fryer AA, Strange RC, et al. Association between functional polymorphism in EGF gene and malignant melanoma. Lancet 2002; 359(9304)397–401
- Bhowmick DA, Zhuang Z, Wait SD, Weil RJ. A functional polymorphism in the EGF gene is found with increased frequency in glioblastoma multiforme patients and is associated with more aggressive disease. Cancer Res 2004; 64: 1220–3
- Zhang W, Gordon M, Press OA, Rhodes K, Vallbohmer D, Yang DY, et al. Cyclin D1 and epidermal growth factor polymorphisms associated with survival in patients with advanced colorectal cancer treated with Cetuximab. Pharmacogenet Genomics 2006; 16: 475–83
- Jakobsen A, Nielsen JN, Gyldenkerne N, Lindeberg J. Thymidylate synthase and methylenetetrahydrofolate reductase gene polymorphism in normal tissue as predictors of fluorouracil sensitivity. J Clin Oncol 2005; 23: 1365–9
- Lindebjerg J, Nielsen JN, Hoeffding LD, Bisgaard C, Brandslund I, Jakobsen A. Expression of thymidylate synthase in primary colorectal adenocarcinoma. Appl Immunohistochem Mol Morphol 2006; 14: 37–41
- Spindler KL. Epidermal growth factor receptor analyses in colorectal cancer. A comparison of methods. Int J Oncol 2006; 29: 1159–65
- Amend KL, Elder JT, Tomsho LP, Bonner JD, Johnson TM, Schwartz J, et al. EGF gene polymorphism and the risk of incident primary melanoma. Cancer Res 2004; 64: 2668–72
- James MR, Hayward NK, Dumenil T, Montgomery GW, Martin NG, Duffy DL. Epidermal growth factor gene (EGF) polymorphism and risk of melanocytic neoplasia. J Invest Dermatol 2004; 123: 760–2
- McCarron SL, Bateman AC, Theaker JM, Howell WM. EGF +61 gene polymorphism and susceptibility to and prognostic markers in cutaneous malignant melanoma. Int J Cancer 2003; 107: 673–5
- Okamoto I, Roka F, Krogler J, Endler G, Kaufmann S, Tockner S, et al. The EGF A61G polymorphism is associated with disease-free period and survival in malignant melanoma. J Invest Dermatol 2006.
- Randerson-Moor JA, Gaut R, Turner F, Whitaker L, Barrett JH, Silva IS, et al. The relationship between the epidermal growth factor (EGF) 5′UTR variant A61G and melanoma/nevus susceptibility. J Invest Dermatol 2004; 123: 755–9
- Schneider S, Park DJ, Yang D, El Khoueiry A, Sherrod A, Groshen S, et al. Gene expression in tumor-adjacent normal tissue is associated with recurrence in patients with rectal cancer treated with adjuvant chemoradiation. Pharmacogenet Genomics 2006; 16: 555–63
- Odin E, Wettergren Y, Nilsson S, Willen R, Carlsson G, Spears CP, et al. Altered gene expression of folate enzymes in adjacent mucosa is associated with outcome of colorectal cancer patients. Clin Cancer Res 2003; 9(16 Pt 1): 6012–9