Abstract
In the present study the expression of LRIG1 (leucine rich repeats and immunoglobin-like domains 1) and its relation to EGFR (epidermal growth factor receptor) was examined in tumour samples and adjacent non-neoplastic tissues from 30 patients with colorectal cancer. The LRIG1 gene, at chromosome 3p14, encodes an intergral membrane protein, which counteracts signalling by receptor tyrosine kinases belonging to the ERBB (epidermal growth factor receptor) family. LRIG1 is expressed in all tissues and organs analysed to date, including breast, brain, skin, kidney, spleen and colon. Overexpression of EGFR is seen in 70–90% of colorectal cancers, and is associated with a poor survival. Western blot analysis showed LRIG1 upregulation in 43% and downregulation in 43% of the colorectal cancers compared to adjacent non-neoplastic tissue. No correlation was evident between LRIG1, analysed by Western Blot and the expression of EGFR analysed by immunohistochemistry. FISH (fluoroscence in situ hybridisation) analysis showed increased LRIG1 copy number in one of nine tumours. Four colorectal cancer cell lines demonstrated two LRIG1 gene copies. In conclusion, there was a great heterogeneity in the expression of the LRIG1 protein in colorectal cancer, which was not related to gene dosage of the LRIG1 gene. Further studies can be of interest to evaluate whether alteration in LRIG1 expression in colorectal cancer is of biological or clinical significance.
The human gene LRIG1 at chromosome 3p14 encodes a protein with extracellular or lumenal leucine-rich repeats and immunoglobulin-like domains, a transmembrane part, and a cytoplasmic tail Citation[1–3]. LRIG1 antagonises EGFR-mediated signalling by enhancing ubiquitinylation and degradation of the receptor Citation[4], Citation[5]. EGFR (ERBB1) is expressed in many colon carcinomas and is associated with poor survival Citation[6–8]. Previous studies have shown that response to EGFR targeted treatments does not correlate to immunohistochemical EGFR expression Citation[9] and therefore other EGFR regulating proteins are important to study. In squamous cell carcinoma of the skin, patients with tumours underexpressing LRIG1 show worse survival Citation[10]. Since LRIG1 is a potential suppressor of EGFR driven colon cancer growth, we found it of interest to explore LRIG1 gene copy number and the expression of LRIG1 protein in sporadic colon cancer. The LRIG1 locus was analysed by fluorescence in situ hybridisation (FISH) and the protein expression was analysed by Western blot analysis.
Material and methods
Patients and sample preparation
Tumour samples and non-neoplastic colon tissues were collected during 2002 from ten patients with colorectal cancer (group A). In group A, samples of the tumour and adjacent non-neoplastic colon tissue were frozen in liquid nitrogen immediately after excision and stored at −80°C. The other adjacent parts of the tissue samples were fixed in formalin, paraffin embedded and used for routine morphological examination and tumour grading.
Fresh frozen and formalin fixed paraffin embedded cancer tissue and surrounding normal colorectal tissue from patients undergoing surgery for colorectal cancer has been collected at the Department of Pathology since 1987. From this collection we randomly sampled 20 tumors diagnosed from 1989 to 1998 (Group B). The colorectal tumours were of different grading and had a lot of fresh frozen tumour available for the western blot analysis. Group B was used to expand the analysis using a larger number of samples for Western blot analysis of LRIG1 in fresh frozen samples and immunohistochemical analysis of the EGFR in formalin fixed tissue. Clinical characteristics of the patients are presented in . The ethical committee of Umeå University approved the study. None of the patients had received any treatment prior to specimen collection.
Table I. Clinical data of the 30 colorectal cancer patients in the study.
Cell lines
To avoid the bias of interference by stroma and/or healthy colon cells, LRIG1 gene copy number was evaluated in four cell lines derived from colon cancer tumours. The colorectal cancer cell lines LOVO, SW480, HT29 and DLD-1 were obtained from American Type Culture Collection (Manassas, VA, USA). The cell lines were cultivated in Dulbecco's modified Eagle′s medium, supplemented with 10% v/v fetal bovine serum and 50 µg/ml gentamicin from Invitrogen AB (Täby, Sweden). The FISH slides were prepared according to standard procedures.
FISH
FISH was performed according to standard techniques as described in a previous study Citation[11]. In each sample, two independent investigators scored LRIG1 signals. At least 100, and if possible 200 nuclei were counted, depending on the quality of the sample. The presence of three or more signals in more than 20% of the nuclei was the criteria for scoring an increased copy number of LRIG1.
Western blot analysis
Western blot was performed as previously described Citation[11]. The primary antibodies used were LRIG1-151 Citation[2] and rabbit anti-actin (Sigma-Aldrich, St. Louis, MO, USA). The samples were stepwise diluted by 50% in three steps. The results were analysed visually and an apparent change between tumour and non-neoplastic tissue of at least 50% was considered as a significant difference (). Attempts to establish a reliable EGFR western blot method did not result in a method stable enough for analysis.
Immunohistochemistry
A considerable effort has been made to achieve an immunohistochemical analysis of LRIG1. In colorectal cancer, unlike other tumours, a persistent background staining made the analysis unreliable, and therefore LRIG1 Western blot was performed instead. Immunohistochemical analysis of the EGFR was performed on formalin fixed and paraffin embedded colorectal cancer specimens according to routine procedures. For this study, 4 µm sections were placed into a completely automated immunostaining machine, Benchmark LT, Ventana Medical Systems Inc., Tucson, AZ, USA). The processes in the machine include deparaffinization, protease treatment (Protease 1), EGFR antibody reaction (mouse monoclonal 3C6, prediluted 1:200), and avidin-biotin blockage. Antibody reaction was demonstrated using iVEIW kit with DAB (diaminobenzidine) as chromogen and hematoxylin for counterstaining according the manufacturer's protocol. All reagents were from Ventana Medical System, Inc. All slides were independently evaluated by two investigators and intraobserver disagreements (<10%) were reviewed a third time followed by a conclusive judgement. One of the investigators was an experienced pathologist (R.P), Staining was scored as follows: 0, no membranous staining in any tumor cells; 1+, membranous staining in more than 10% of the tumor cells with any intensity.
Results
LRIG1 gene copy number in fresh frozen colorectal cancer tissues
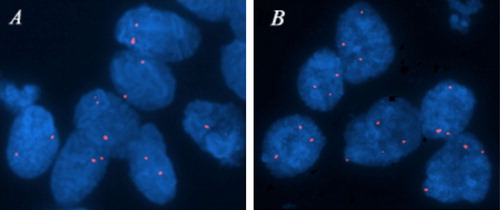
To evaluate the number of LRIG1 gene copies, FISH was performed on cell nuclei from colorectal cancer samples (Group A, n = 10; ). Nine tissue samples were successfully evaluated; a tenth sample was of poor quality and analysis failed. An increased copy number of LRIG1 in more than 20% of the nuclei was found in one tumour, showing three to five LRIG1 signals per nucleus. Normal signal pattern corresponding to two copies per nucleus was detected in 8 of 9 tumours. The colorectal cancer cell lines LOVO, SW480, HT29, and DLD-1 were also examined by FISH and demonstrated two LRIG1 signals per nucleus corresponding to normal gene copy numbers.
Figure 1. FISH analyses of LRIG1 locus in colorectal cancer cells from two patients. (A) Nuclei showing normal gene copy number (two copies per nucleus) of the LRIG1 gene. (B) Nuclei showing increased gene copy number (more than two copies per nucleus) of the LRIG1 gene at 3p14. Analysis was performed with a specific LRIG1 probe (red).
Western blot analysis of LRIG1 expression in colorectal cancer samples and matched non-neoplastic tissue
Western blot analysis was performed on samples from both group A and B (, , n = 30). In 13/30 (43%) tumours the LRIG1 protein was downregulated in the tumour compared to matched non-neoplastic tissues. In the same number of tumours (13/30) the LRIG1 protein was upregulated. In 4/30 (13%), no significant difference in LRIG1 expression was evident.
Figure 2. Example of Western blot analyses of LRIG1 in samples from two colorectal cancer patients. For comparison, the samples were stepwise diluted by 50% in three steps. Tumours (T) versus matched non-neoplastic tissue (N) from the same patient. Pat 6 was considered underexpressing the LRIG1 protein in the tumour compared to adjacent non-neoplastic tissue and Pat 12 was considered as overexpressing the LRIG1 protein in the tumour compared to non-neoplastic tissue.
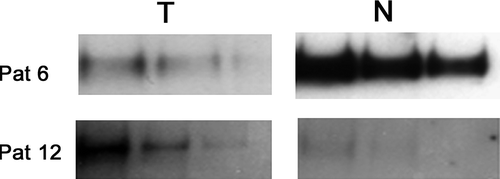
Table II. Crosstable of LRIG1 Western blot (WB) results and immunohistochemical (IHC) EGFR scores in 30 colorectal cancer samples and matching adjacent non-neoplastic tissue.
Immunohistochemical EGFR expression in colorectal cancer samples
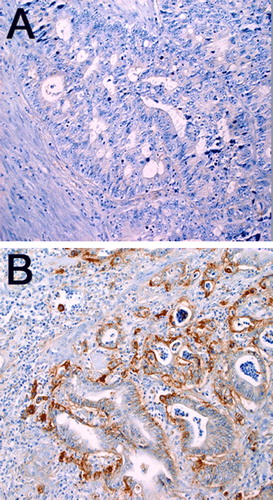
Immunohistochemical analysis was performed on the 20 available samples of formalin fixed and paraffin embedded colorectal cancer tissue (, ). In 13/20 samples (65%) a positive staining for the EGFR was found. Negative staining was found in 7/20 (35%) samples. No significant correlation between LRIG1 expression determined by Western blotting and the immunohistochemical EGFR expression was seen.
Figure 3. Immunohistochemical analyses of EGFR in primary colorectal tumors. (A) Negative staining in colorectal cancer cells and surrounding tissues. (B) Positive membranous staining in more than 10% of colorectal cancer cells (original magnification ×20), and mostly negative staining in surrounding tissues.
Discussion
The present study revealed a great heterogeneity in the expression of LRIG1 in colorectal cancer. FISH analysis showed chromosomal abnormalities at the 3p14 locus in only one tumour, with slightly increased copy number. This was in contrast to a recent report concerning LRIG1 in breast cancer, where a significant (40%) proportion of the tumours showed increased copy number of the LRIG1 locus Citation[11]. In this study, chromosomal aberrations at 3p14 do not appear to be common events in colorectal carcinoma. This is also supported by the analysis of four colon cancer cell lines, which did not show any changes in LRIG1 copy number.
Of the 30 tumours examined by Western blotting, up- or downregulation of LRIG1 was evident in 26 cases, with equal numbers of tumours over- and underexpressing the protein, respectively. The differences in LRIG1 protein expression was not due to chromosomal loss or gain and is therefore due to other mechanisms. In a study of LRIG1 in squamous cell carcinoma of the skin Citation[10], a correlation between increased LRIG1 expression and a more favourable prognosis has been shown. In the present study, the small number of tumours investigated precluded analysis of clinical outcome.
LRIG1 has recently been shown to downregulate the ERBB receptors Citation[4], Citation[5], including the EGFR, a receptor of major clinical importance in the pathogenesis of colorectal cancer Citation[7]. The EGFR is a well-established factor of importance in colorectal cancer progression both as a predictor of outcome and as the target of new drugs Citation[12]. Moreover, the observation that response to anti-EGFR treatment is independent of immunohistochemical EGFR expression levels makes EGFR regulation studies of current importance Citation[9]. As LRIG1 downregulates the ERBB receptors by receptor ubiquitinylation, immunohistochemical analysis of EGFR was performed to see if there was a correlation between LRIG1 Western blot levels and immunohistochemical EGFR levels. In this report, LRIG1 expression did not correlate to EGFR protein expression. This is in contrast to our previous report of LRIG1 in breast cancer, where a correlation between LRIG1 and EGFR RNA levels was evident Citation[11]. From this perspective the relationship between LRIG1 and EGFR could be suggested to be tissue specific and not evident for all tumour types. However, determination of the actual impact of LRIG1 on EGFR levels would require more detailed biochemical analyses and therefore this study cannot exclude a correlation that could be detected by other methods. Moreover, LRIG1 also has regulatory effects on ERBB2, 3 and 4, and such potential interactions were not investigated in this study Citation[5]. In conclusion, the LRIG1 protein was up- or downregulated in colorectal cancer without evident impact on EGFR protein levels.
Acknowledgements
We would like to thank Kerstin Bergh for help with the immunohistochemistry and Charlotte Andersson for help with the FISH analysis. This study was supported by grants from The Swedish Cancer Society and the Cancer Research Foundation, Northern Sweden.
References
- Suzuki Y, Sato N, Tohyama M, Wanaka A, Takagi T. cDNA cloning of a novel membrane glycoprotein that is expressed specifically in glial cells in the mouse brain. LIG-1, a protein with leucine-rich repeats and immunoglobulin-like domains. J Biol Chem 1996; 271: 22522–7
- Nilsson J, Vallbo C, Guo D, Golovleva I, Hallberg B, Henriksson R, et al. Cloning, characterization, and expression of human LIG1. Biochem Biophys Res Commun 2001; 284: 1155–61
- Nilsson J, Starefeldt A, Henriksson R, Hedman H, et al. LRIG1 protein in human cells and tissues. Cell Tissue Res 2003; 312: 65–71
- Gur G, Rubin C, Katz M, Amit I, Citri A, Nilsson J, et al. LRIG1 restricts growth factor signaling by enhancing receptor ubiquitylation and degradation. Embo J 2004; 23: 3270–81
- Laederich MB, Funes-Duran M, Yen L, Ingalla E, Wu X, Carraway KL, 3rd, et al. The leucine-rich repeat protein LRIG1 is a negativeregulator of ErbB family receptor tyrosine kinasesregulator of ErbB family receptor tyrosine kinases. J Biol Chem 2004; 279: 47050–6
- Goldstein NS, Armin M. Epidermal growth factor receptor immunohistochemical reactivity in patients with American Joint Committee on Cancer Stage IV colon adenocarcinoma: Implications for a standardized scoring system. Cancer 2001; 92: 1331–46
- Mayer A, Takimoto M, Fritz E, Schellander G, Kofler K, Ludwig H, et al. The prognostic significance of proliferating cell nuclear antigen, epidermal growth factor receptor, and mdr gene expression in colorectal cancer. Cancer 1993; 71: 2454–60
- Koretz K, Schlag P, Moller P. Expression of epidermal growth factor receptor in normal colorectal mucosa, adenoma, and carcinoma. Virchows Arch A Pathol Anat Histopathol 1990; 416: 343–9
- Chung KY, Shia J, Kemeny NE, Shah M, Schwartz GK, Tse A, et al. Cetuximab shows activity in colorectal cancer patients with tumors that do not express the epidermal growth factor receptor by immunohistochemistry. J Clin Oncol 2005; 23: 1803–10
- Tanemura A, Nagasawa T, Inui S, Itami S. LRIG-1 provides a novel prognostic predictor in squamous cell carcinoma of the skin: Immunohistochemical analysis for 38 cases. Dermatol Surg 2005; 31: 423–30
- Ljuslinder I, Malmer B, Golovleva I, Thomasson M, Grankvist K, Hockenstrom T, et al. Increased copy number at 3p14 in breast cancer. Breast Cancer Res 2005; 5: 719–727
- Tabernero J, Salazar R, Casado E, Martinelli E, Gomez P, Baselga J, et al. Targeted therapy in advanced colon cancer: The role of new therapies. Ann Oncol 2004; 15: 55–62
