Introduction
The diagnosis of acute promyelocytic leukemia (APL) is defined by the detection of a reciprocal translocation between chromosomes 15 and 17 [t(15;17)(q24;q21)], resulting in fusion of the promyelocytic leukemia (PML) gene on chromosome 15 and the retinoic acid receptor α (RARA) gene on chromosome 17. PML is a tumor suppressor gene implicated in cell proliferation and regulation of apoptosis [Citation1]. The RARA gene codes for a nuclear receptor that regulates transcription of genes involved in differentiation, apoptosis, and granulopoiesis [Citation2]. The PML-RARA gene product acts as a transcription factor increasing cell proliferation and blocking differentiation of myeloid cells [Citation3].
Genetically, the PML‑RARA translocation forms as the PML gene breaks at three breakpoint cluster regions (bcrs): intron 6 (bcr1), exon 6 (bcr2) and intron 3 (bcr3). RARA breakpoints occur in intron 2, preserving five of its six functional domains in the three PML-RARA isoforms: long (bcr1), variant (bcr2) and short (bcr3). Detection of PML‑RARA formation by means of conventional karyotyping, fluorescence in situ hybridization (FISH) or reverse transcription‑polymerase chain reaction (RT‑PCR) is required for the diagnosis of APL [Citation4]. Studies have demonstrated that this abnormality can also be detected by microarray and next-generation sequencing (NGS) of DNA or RNA [Citation5,Citation6].
In rare cases, FISH or molecular methods demonstrate the presence of the PML‑RARA fusion gene without the reciprocal RARA‑PML [Citation7]. This may result from a submicroscopic insertion of RARA into PML or PML movement to a chromosome other than 15 or 17. Since such cryptic insertions are not frequently reported, no prognostic significance has been clearly established [Citation8]. A prompt diagnosis is essential for administration of targeted therapies, including all‑trans retinoic acid (ATRA) to improve outcomes in patients with APL.
Here, we present a unique case of a patient with a normal karyotype and molecular evidence of the PML‑RARA short isoform (bcr-3)‑fusion transcript, with FISH analysis revealing two fusion signals on the two copies of chromosome 15, but absence of the reciprocal translocation on the two copies of chromosome 17. The clinical, cytogenetic and molecular characteristics of this case are presented and discussed in the current case report.
Case
A 68-year-old female with history of hypertension, diabetes mellitus (type 2) and hyperlipidemia initially presented to an outside hospital with complaints of dizziness and fatigue with imaging findings of acute chronic pulmonary embolism and laboratory findings of pancytopenia. Given concern for acute leukemia, the patient was transferred to our institution, Rush University Medical Center (RUMC), for escalation of care.
Bone marrow biopsy/aspirate at RUMC revealed a hypercellular bone marrow packed with acute promyelocytic leukemia. On aspirate smear, there were 70% blasts/promyelocytes with indented or bilobed nuclei containing fine to clumped chromatin and moderate amounts of cytoplasm with many azurophilic granules and some with prominent Auer rods. About 10% of the cells had a predominately promonocytic morphology. Given clinical suspicion for APL, ATRA was started pending results of diagnostic studies.
Preliminary studies at the prior hospital did not show a typical PML-RARA translocation, and the karyotype was normal (). FISH analysis was performed at RUMC using a break-apart probe (17q21) SpectrumGreen® covering (Tel) RARA, TOP2A, CCR7 and SpectrumOrange® covering the entire RARA gene (Cen) (Abbott Molecular, Inc.). Interphase nuclei contained a separation of the RARA locus on chromosome 17 into another site, presumably the PML gene (). Of 500 interphase nuclei hybridized by the RARA break-apart probe, 267 (53.4%) contained this abnormal RARA signal pattern with an extra dim fusion signal, suggesting a possible RARA gene rearrangement, and 233 (46.6%) contained a normal RARA signal pattern. The RARA gene rearrangement was also observed on metaphase FISH ().
Figure 1. Molecular analysis of white blood cell DNA performed at RUMC. The karyotype did not show the expected t(15;17) rearrangement (a), but interphase and metaphase FISH revealed separation of the green and orange RARA break apart (BA) probe signals from their location on chromosome 17 (white arrows, b and c). Interphase and metaphase FISH with single fusion (SF) PML and RARA probes revealed the movement of RARA to chromosome 15 at the location of PML as a small yellow fusion signal (white arrows, d and e). The PML-RARA fusion gene was detected by reverse transcriptase PCR using primers to exon 3 and exon 6 of the PML gene and reverse primer to exon 3 of RARA (f: Lane 1, molecular weight standard, lane 2 PML exon 6 forward primer, lane 3 PML exon 3 forward primer lane 4, RARA exon 2 forward primer, lane 5 reagent blank). The major PCR product detected at about 500 bp (starred, lane 3) contained exons 3–6 of the PML gene.
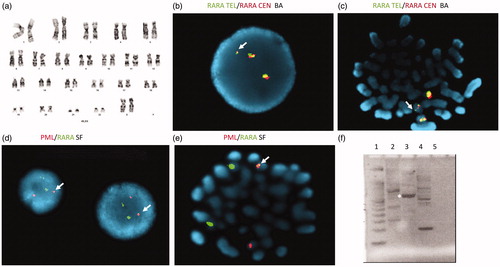
Further analysis was performed using the SpectrumOrange® single fusion probe (15q24) ∼239 kb, covering PML exons 1-7 and RARA SpectrumGreen® single fusion probe covering RARA, TOP2A and CCR7. This PML-RARA dual color single fusion translocation analysis revealed an atypical signal pattern in 265 of 500 nuclei (53%) examined. These nuclei contained one fusion with dim yellow intensity, suggesting a possible PML-RARA gene rearrangement. The remaining 235 nuclei counted (47%) contained a normal signal pattern (). Metaphase FISH analysis using the PML-RARA dual color single fusion probes supported the insertion of a segment of the RARA gene into chromosome 15 at the location of the PML gene ().
Molecular studies using quantitative reverse transcriptase PCR (qRT-PCR) were performed to detect/confirm a PML-RARA fusion gene [Citation7,Citation9]. These studies detected the PML-RARA transcript (PML-RARA/RARA = 62%). Agarose gel electrophoresis revealed a major 500 bp product from PML exon 3 – RARA exon 3 primers and a minor 700 bp product from PML exon 6 – RARA exon 3 primers (). Sanger sequencing of the 500 bp PCR amplicon produced 278 b of PML followed by 119 bp of unknown sequence followed by 144 bp of PML with no RARA detected.
Whole genome sequencing was performed to further describe the gene rearrangement structure. DNA for whole genome sequencing was isolated from a diagnostic sample of the patient’s blood including 57% tumor cells. The sequencing library was constructed from sheared DNA (250 bp fragments) using the KAPA Hyper library kit (Roche/Sigma, St. Louis, MO). The resulting library was sequenced at Novogene (Sacramento, CA) on the HiSeqX Sequencer (Illumina, San Diego, CA). Test and reference sequences were aligned with Bowtie 2. After indexing and sorting with samtools, structural variant calling was performed with the Breakdancer package algorithm. Secondary analysis of sequence data was performed using the Illumina DRAGEN (Dynamic Read Analysis for GENomics) platform.
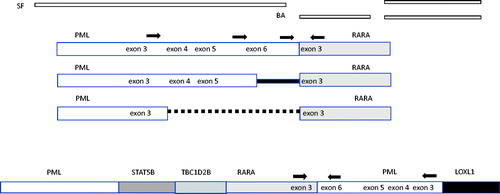
The results revealed multiple structural variants including a rearrangement of chromosome 15 at position 73,783,638 (Homo sapiens GRCh38.p13) in the region of PML (73,994,673–74,047,827) partnered with a translocation at position 40,802,877 in chromosome 17 located near the RARA gene (40,309,180–40,357,643). The results suggest a complex translocation with a possible intrachromosomal rearrangement of chromosome 15 ().
Figure 2. Structure and suggested structure of classical and cryptic translocations. Binding location of the single fusion (SF), break apart (BA) probes and RT-qPCR primers (arrows) are shown. In the classical APL t(15;17) translocations, the PML gene breaks at three breakpoint cluster regions (bcrs): intron 6 (bcr1), exon 6 (bcr2) and intron 3 (bcr3). RARA breakpoints occur in intron 2. A proposed structure based molecular analysis includes intra-chromosomal rearrangement of PML in addition to the insertion of RARA.
The patient received induction chemotherapy with ATRA and arsenic trioxide (standard of care based on the amplifiable translocation). Treatment course was complicated by differentiation syndrome (treated with dexamethasone), and hyperleukocytosis requiring hydroxyurea and one dose of idarubicin (9 mg/m2). Restaging bone marrow biopsy/aspirate demonstrated a moderately hypercellular marrow with trilineage maturation, and no residual APL by morphology nor by flow cytometry. Cytogenetics revealed a normal female karyotype but FISH results were inconclusive. Of all interphase nuclei analyzed, a low percentage (∼10%) was suspicious for the previously detected abnormal RARA and PML/RARA gene rearrangement signal patterns. Because the previous noted gene rearrangement signal patterns were quite subtle, however, we were unable to unequivocally determine the presence or absence of residual leukemia cells in this analysis. PCR remained positive for the PML-RARA translocation (PML-RARA/RARA = 10.2%). The patient went on to receive consolidation with ATO and ATRA.
A repeat bone marrow biopsy/aspirate performed after first consolidation chemotherapy revealed a mildly hypercellular marrow with trilineage hematopoiesis with erythroid predominance. Erythroid precursors showed increased dyspoiesis, including nuclear budding/blebs, which was thought to be reactive in etiology. There was no overt morphologic nor immunophenotypic evidence of acute promyelocytic leukemia. PCR for the PML-RARA translocation was negative.
Post-treatment bone marrow biopsy/aspirate showed no overt morphologic or immunophenotypic evidence of involvement by acute promyelocytic leukemia. FISH analysis was performed using both RARA break-apart and PML-RARA fusion probes. A low percentage (2.8–6.2%) of interphase nuclei analyzed were suspicious for the previously detected abnormal RARA and PML-RARA gene rearrangement signal patterns. Again, the presence or absence of the residual leukemia cells could not unequivocally be determined. PCR for the PML-RARA translocation remained negative. To date, the patient remains in complete remission more than 12 months from completing treatment.
Discussion
Cryptic APL is characterized by absence of the characteristic translocation t(15:17) either due to submicroscopic insertion of RARA into PML or complex rearrangements. Cryptic APL may have variable cytogenetic abnormalities or completely normal cytogenetics (i.e. normal karyotype, and/or no detection of fusion signal by FISH). In cases of cryptic APL, the diagnosis is almost always confirmed by PCR. Abnormal karyotype has been seen in patients with FISH negative cases of APL [Citation10]. Some of the notable abnormal karyotype findings that have been reported include isochromosome 17q, trisomy 8, or complex karyotypes [Citation11,Citation12]. FISH may show the PML‑RARA without the reciprocal RARA‑PML due to a submicroscopic insertion of RARA into PML. The 15qter region may be deleted in some cases. A rearrangement was reported with two PML‑RARA FISH fusion signals present on the two copies of chromosome 15. This was interpreted as a cryptic insertion of RARA into PML and chromosome 15 uniparental isodisomy, likely due to loss of the normal chromosome 15 and duplication of the rearranged one [Citation13].
Not all variants produce an actionable rearrangement. PML-RARA translocations with long “spacer” regions including intron 2 of RARA have aberrant protein binding leading to clinical resistance to ATRA [Citation14]. A minor percentage of APL cases have a partner gene different from PML fused to the RARA gene, such as the TBL1XR1-RARA fusion [Citation15]. Cases of FISH-negative APL have also been reported with rearrangements involving chromosomes 3, 7, 9, 11, 17, or 22 [Citation12].
The current case is among several reported with cryptic PML‑RARA fusions in cytogenetically normal samples. FISH analysis revealed a cryptic insertion of the RARA gene into the PML gene on chromosome 15. There was a normal karyotype and molecular evidence of the PML‑RARA short isoform 3‑fusion transcript by qRT-PCR. RARA break-apart FISH analysis revealed one separated RARA signal in addition to the two intact RARA signals on the two copies of chromosome 17. PML and RARA single fusion probes showed one tiny fusion signal on one of the copies of PML. This finding supports the hypothesis of an insertion of RARA into chromosome 15. Metaphase FISH break-apart probes confirmed two normal copies of RARA along with a RARA separation. Metaphase single fusion probes produced a yellow signal on chromosome 15. Genome sequencing was consistent with an intrachromosomal rearrangement of chromosome 15 with insertion of at least part of the RARA gene similar to a recently reported complex rearrangement [Citation16].
Genome sequencing has previously been used to detect masked genetic alterations in another case of APL, demonstrating that genome sequencing may be a useful tool to confirm diagnosis in equivocal cases, especially if there is high clinical suspicion of APL [Citation5,Citation17]. APL may be associated with diverse PML-RARA variants and unusual translocations/insertions. The diagnostic strength of whole-genome sequencing is that all classes of mutations, even a rare or unusual mutation; can be detected and reported in an effective manner. Most cases of cryptic APL are reported in adults as is the current patient (age 68), however pediatric cases of cryptic APL have been noted [Citation18]. Patient outcomes are reported in only a limited number of cases [Citation10,Citation19]. There are sporadic cases of patients with cryptic APL responding poorly to ATRA treatment [Citation7,Citation14]. A long-term follow-up of patients with cryptic APL compared with a series of consecutive classical APL cases found that the clinical courses were comparable when managed with the same approach [Citation10,Citation12]. This is consistent with the clinical outcome in the present case.
Conclusion
In this karyotypically normal case, the PML-RARA fusion gene is present as indicated by the positive qRT-PCR results and FISH results. The rearrangement is complex, with intrachromosomal rearrangement of chromosome 15. Assessment of PML‑RARA formation, or variant RARA gene rearrangements by means of conventional karyotyping, FISH or qRT‑PCR, is required for the diagnosis of APL, however, diagnosis can be challenging in APL cases with cryptic PML-RARA rearrangements. While this cryptic insertion has rarely been reported and no prognostic significance has been clearly established, this patient responded well to APL-directed therapy including ATRA treatment. A prompt diagnosis and the administration of targeted therapies are essential to improve the outcome in patients with APL.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Reineke EL, Kao HY. PML: an emerging tumor suppressor and a target with therapeutic potential. Cancer Ther. 2009;7(A):219–226.
- Melnick A, Licht JD. Deconstructing a disease: RAR-alpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93(10):3167–3215.
- De Braekeleer E, Douet-Guilbert N, De Braekeleer M. RARA fusion genes in acute promyelocytic leukemia: a review. Expert Rev Hematol. 2014;7(3):347–357.
- Choppa PC, Gomez J, Vall HG, et al. A novel method for the detection, quantitation, and breakpoint cluster region determination of t(15;17) fusion transcripts using a one-step real-time multiplex RT-PCR. Am J Clin Pathol. 2003;119(1):137–144.
- Abel HJ, Al-Kateb H, Cottrell CE, et al. Detection of gene rearrangements in targeted clinical next-generation sequencing. J Mol Diagn. 2014;16(4):405–417.
- Koshy J, Qian YW, Bhagwath G, et al. Microarray, gene sequencing, and reverse transcriptase-polymerase chain reaction analyses of a cryptic PML-RARA translocation. Cancer Genet. 2012;205(10):537–540.
- Iaccarino L, Divona M, Ottone T, et al. Identification and monitoring of atypical PML/RARA fusion transcripts in acute promyelocytic leukemia. Genes Chromosomes Cancer. 2019;58(1):60–65.
- Rashidi A, Fisher S. FISH-negative, cytogenetically cryptic acute promyelocytic leukemia. Blood Cancer J. 2015;5:e320.
- Slack JL, Bi W, Livak KJ, et al. Pre-clinical validation of a novel, highly sensitive assay to detect PML-RARalpha mRNA using real-time reverse-transcription polymerase chain reaction. J Mol Diagn. 2001;3(4):141–149.
- Greenfield G, Michail O, Merron B, et al. Acute promyelocytic leukaemia (APML) with cryptic PML-RARA fusion has a clinical course comparable to classical APML with t(15;17)(q24.1;q21.2) translocation. Br J Haematol. 2019;186(1):155–157.
- Zaccaria A, Valenti A, Toschi M, et al. Cryptic translocation of PML/RARA on 17q. A rare event in acute promyelocytic leukemia. Cancer Genet Cytogenet. 2002;138(2):169–173.
- Tang Y, Wang Y, Hu L, et al. Acute promyelocytic leukemia with cryptic t(15;17) on isochromosome 17: a case report and review of literature. Int J Clin Exp Pathol. 2015;8(11):15294–15300.
- Venci A, Mazza R, Spinelli O, et al. Acute promyelocytic leukemia with a cryptic insertion of RARA into PML on chromosome 15 due to uniparental isodisomy: a case report. Oncol Lett. 2017;13(6):4180–4184.
- Gu BW, Xiong H, Zhou Y, et al. Variant-type PML-RAR(alpha) fusion transcript in acute promyelocytic leukemia: use of a cryptic coding sequence from intron 2 of the RAR(alpha) gene and identification of a new clinical subtype resistant to retinoic acid therapy. Proc Natl Acad Sci USA. 2002;99(11):7640–7645.
- Osumi T, Watanabe A, Okamura K, et al. Acute promyelocytic leukemia with a cryptic insertion of RARA into TBL1XR1. Genes Chromosomes Cancer. 2019;58(11):820–823.
- Cao Y, Yao L, Liu Y, et al. An atypical PML-RARA rearrangement resulting from submicroscopic insertion of the RARA gene at the PML locus with novel breakpoints within PML exon 7b and RARA exon 3. Acta Haematol. 2019;142(2):98–104.
- Welch JS, Westervelt P, Ding L, et al. Use of whole-genome sequencing to diagnose a cryptic fusion oncogene. JAMA. 2011;305(15):1577–1584.
- Mai B, Liang C, Nguyen A, et al. Cryptic acute promyelocytic leukemia (APL) presenting as seizures in an adolescent. Ann Clin Lab Sci. 2020;50(2):270–273.
- Shepshelovich D, Oniashvili N, Parnes D, et al. Acute promyelocytic leukemia with isochromosome 17q and cryptic PML-RARA successfully treated with all-trans retinoic acid and arsenic trioxide. Cancer Genet. 2015;208(11):575–579.

