Abstract
Background
As a new kind of diallelic genetic marker, insertion/deletion (InDel) polymorphisms have recently been used in forensic science. However, there are relatively few studies on the forensic evaluation of InDel genetic polymorphisms from different populations.
Aim
The aim of the present work is to assess the genetic polymorphism and forensic applicability of 57 InDels from the Yi ethnic group and explore the genetic background of this group.
Subjects and methods
A total sample of 122 unrelated individuals of Yi group from the Yunnan province were genotyped by the AGCU indel 60 Kit. Multiplex population genetic analyses on the same 57 InDels were carried out among the Yunnan Yi group and 29 reference populations.
Results
The average allele frequency of these loci in the Yi ethnic group was 0.485. Heterozygosity, polymorphism information content, and the power of discrimination were 0.477, 0.362, and 0.612, respectively. The combined power of discrimination and the combined power of exclusion reached to 0.99999999999999999669 and 0.999962965, respectively. The results showed that 57 InDels polymorphisms have high genetic polymorphisms in the Yi ethnic group.
Conclusions
The 57 InDels could be used for forensic individual identification, paternity testing, and intercontinental population discrimination, with the potential for use in biogeographic ancestry inference.
1. Introduction
Genetic markers with high identification ability are widely used in forensic science for individual identification and paternity testing. Currently, the commonly used genetic markers in forensic genetics laboratories include short tandem repeats (STR) (Zhang Y et al. Citation2021; Zhou et al. Citation2022), single nucleotide polymorphisms (SNP) (Tillmar et al. Citation2021; Zhao et al. Citation2021) as well as insertions/deletions (InDel) (Chen L et al. Citation2021; Zhang X et al. Citation2021). However, using STRs is limited to a relatively high mutation rate, long amplicon, and limited number, and many genotyping techniques and platforms have restrained the application of SNPs in forensic practice. InDels, the biallelic genetic markers, are widely distributed in the human genome, just like SNPs. They are not only characterised by short amplicons and low mutation rates, but are also suitable for polychromatic fluorescent multiplex polymerase chain reaction (PCR) combined with capillary electrophoresis (CE) typing platform. With the disadvantages of STRs and SNPs, the use of InDels has attracted intensive attention in recent years.
In the past decades, several InDel typing systems suitable for forensic application have been established by scholars at home and abroad, proving the importance of InDels in forensic research. Pereira et al. in Citation2009 constructed a multiplex PCR genotyping system for individual identification based on 38 autosomal InDels, and demonstrated its potential value for individual identification, paternity testing, and degradation material identification. In the following year, Pimenta and Pena (Citation2010) designed a multiplex amplification system containing 40 InDels for paternity testing. The forensic parameter efficacy of this system was comparable with that of 13 STR loci in the Combined DNA Index System (CODIS), indicating the ability for paternity testing (Budowle et al. Citation1999). However, some loci mentioned above were not entirely applicable to the Chinese populations. Due to China’s large population, vast territory, and different nationalities, personalised loci panels applicable to different populations are required for better individual identification and paternity testing. In 2011, Li et al. (Li C et al. Citation2011; Li CT et al. Citation2011) established two multiplex amplification systems including 29 InDels and 30 InDels, respectively, for human individual identification based on the genomic features of the Chinese Han population. Recently, InDels have also been applied to forensic anthropology, in which they can clearly distinguish population structure and establish ancestry information (Santos, Fondevila, et al. Citation2015; Santos, Phillips, et al. Citation2015). Investigator® DIP plex kit is the first commercialised kit, which enables the amplification of 30 autosomal InDels (LaRue et al. Citation2012), and forensic experts have proved the value of this kit as an auxiliary detection tool in forensic personal identification and paternity testing.
The AGCU InDel 60 kit is a newly developed kit with six colour fluorescent-labeled multiplex amplification, including 57 autosome-InDels, 2 Y-chromosome InDels, and the sex identification locus Amelogenin. However, its efficacy in identifying different populations has not yet been well assessed. The Yi ethnic minority represents the sixth largest of 55 ethnic minority groups in China with a population of 9,830,327 individuals according to the China Statistical Yearbook 2021 (https://www.stats.gov.cn/sj/ndsj/2021/indexeh.htm). People of the Yi ethnic group mainly reside in Yunnan, Sichuan and other provinces. It is documented that the Yi originated from the ancient Qiang people in southeast China and merged with many indigenous people in the southwest (Song et al. Citation2022). Additionally, during the 14–17th centuries, a large cohort of the Han Chinese population entered the Yi settlement area due to cantonment, and there were variable degrees of genetic combination between the Yi ethnic group and Han population. During these processes, the Yi ethnic group acquired a unique genetic background, and therefore it is an ideal population for genetic studies. In the present study, the genetic polymorphisms of 122 Yi samples from the Honghe Hani Yi autonomous prefecture of Yunnan province were analysed by the AGCU InDel 60 kit, aiming to evaluate its efficacy in forensic DNA identification and provide basic data for the application to individual identification and paternity testing. Furthermore, the genetic relationships between the Yi ethnic group in Yunnan and 29 reference populations were analysed using principal component analysis (PCA), multidimensional scaling (MDS), and phylogenetic trees.
2. Subjects and methods
2.1. Sample collection, DNA extraction and quantification
Venous blood samples were collected from 122 unrelated individuals of Yi ethnic group, living in the Honghe Hani Yi autonomous prefecture of Yunnan province. There were no consanguineous relationships among these volunteers, and all were from families that had resided in the same locality for at least three generations. Written informed consents were obtained from all volunteers, which was approved by the ethics committee of Xi’an Jiaotong University (no. 2019-1039). Genomic DNA was extracted using chelex-100 resin method (Walsh et al. Citation1991). DNA concentrations were measured spectrophotometrically, and all samples were diluted to a 2 ng/μL final concentration.
2.2. Reference population
The 57 A-InDels genotyping data of 26 populations from different countries or regions were acquired from the 1000 Genomes Project Phase 3, and the data of three previously published Chinese populations (Fan H et al. Citation2021; Fang et al. Citation2022; Wang et al. Citation2022) (Han Chinese from Hunan, Li from Hainan, and Sharba) were collected.
2.3. Polymerase chain reaction and InDels genotyping
The multiplex PCR was carried out according to the manufacturer’s instructions of the AGCU indel 60 Kit (AGCU ScienTech Incorporation, Wuxi, China). The PCR reaction volume was 25 μl, containing 1 μL DNA (approximately 2 ng), 10 μL reaction mix, 5 μL InDel 60 primers, 1 μL HotStart U-Taq DNA Polymerase, as well as 8 μL sdH2O.
The PCR conditions were 95 °C for 5 min, 28 cycles at 94 °C for 30 s, 60 °C for 1 min, and 62 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR products were analysed by capillary electrophoresis on a 3500 × L Genetic Analyser (Applied Biosystems, Foster City, USA). The products were first centrifuged at 3000 rpm for 5 min in case of PCR product loss during opening the lid. The loading samples were prepared by mixing 1 μL of PCR products with 12 μL deionised formamide and 0.5 μL AGCU Marker SIZ-500. The loading samples were denatured at 95 °C for 3 min and immediately placed on ice. The results of InDels genotyping were analysed using the GeneMapper ID v3.2 software (Applied Biosystems, Foster City, USA). DNA 9948 and ultra-pure water were used as positive and negative controls, respectively.
2.4. Statistical analysis
The online website STRAF 2.0.8 (http://www.straf.fr/) was used to calculate the allele frequencies, observed heterozygosity (Ho), expected heterozygosity (He), matching probability (PM), power of discrimination (PD), power of exclusion (PE), polymorphism information content (PIC), the typical paternity index (TPI), and other forensic parameters. The Hardy-Weinberg equilibrium (HWE) test and linkage disequilibrium (LD) analyses were also performed using this online tool. The genetic distance (DA), the genetic differentiation index (FST) matrix and corresponding p values between different populations were calculated based on allele frequencies using DISPAN (Nei Citation1973; Nei et al. Citation1983) and Arlequin v3.5 (Excoffier and Lischer Citation2010). Phylogenetic trees were constructed using MEGA v11.0 (Tamura et al. Citation2011) based on the neighbor-joining (N-J) method. Genetic population structuring between the Yunnan Yi group and the other 29 populations was calculated using STRUCTURE v2.3.4 software (Pritchard et al. Citation2000). Meanwhile, the STRUCTURE results were uploaded to the online web Structure Harvester (http://taylor0.biology.ucla.edu/structure Harvester/) to determine the optional K value. The STRUCTURE results with K values from 2 to 7 were further permuted with program CLUMPP v1.1 (Jakobsson and Rosenberg Citation2007), and then visualised using DISTRUCT v1.1 (Rosenberg et al. Citation2002). The Snipper App suite version 3.0 (http://mathgene.usc.es/snipper/index.php) was used to calculate the cross-validation estimation success ratios and informativeness for assignment (In) values. Heatmap was generated using “pheatmap” in R. Principal component analysis (PCA) was performed using the “stats” package in R. Multidimensional scaling (MDS) analysis was performed with the R package “cmdscale.”
3. Results and discussion
3.1. The HWE tests and LD analysis of 57 A-InDels in Yunnan Yi group
Population genetics is an important theoretical basis for individual identification and paternity testing in forensic genetics. Hardy-Weinberg equilibrium and the independence of each InDel locus was first assessed in the Yunnan Yi group. The results showed that the distribution of all InDel loci genotype fit Hardy-Weinberg equilibrium (p > .05) (Supplementary Table 1). After Bonferroni correction, no linkage disequilibrium was observed among the 57 InDels (p > .000 031 33) (Supplementary Table 2). Therefore, these loci were in accordance with the law of genetic balance and could be used as independent genetic markers in the Yunnan Yi group, indicating the samples in this study could represent the general population.
3.2. Analysis of 57 A-InDels genetic polymorphisms and forensic parameters in Yunnan Yi group
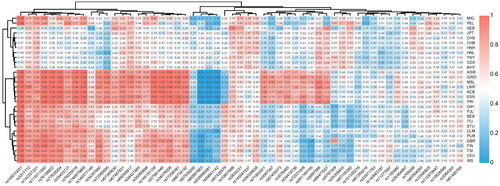
Clearly understanding the genetic polymorphism and forensic characteristics of Yunnan Yi group is indispensable in its forensic applications, especially in paternity testing and individual identification. In the present study, 57 InDels allele frequencies were counted in 122 samples from the Yunnan Yi group (Supplementary Table 3). The frequencies of these alleles ranged from 0.279 (rs67264216) to 0.742 (rs5897566), with a mean value of 0.485. The allele frequencies were then compared and visualised. The heat map showed that African and European populations clustered together, respectively, and East Asian populations also had their own distinct cluster. American populations were involved in the clusters of European and East Asian populations. The Yunnan Yi group clustered together with the populations from East Asia (). These results demonstrated that the allele frequency distribution of populations from the same continent was essentially consistent, whereas there were obvious differences among populations from different continents. The 57 InDels could be applied to distinguish samples from different continental populations.
Figure 1. Heat map of insertion allele frequencies at 57 A-InDels of the Yunnan Yi group and 29 reference populations conducted by R.
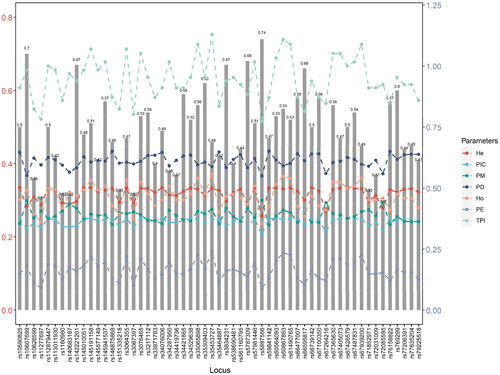
To assess the application value of these genetic markers for individual identification and paternity testing in forensic genetics, the corresponding forensic parameters were calculated based on the genotyping data (Supplementary Table 4, ). Ho and He ranged from 0.352 (rs5897566) to 0.557 (rs35453727) and 0.385 (rs5897566) to 0.502 (rs10590825, rs112879447, rs66739142, rs67426579) with mean values of 0.466 and 0.477, respectively, indicating a relatively high heterozygosity of the 57 InDels. The PIC was between 0.310 (rs5897566) − 0.3750 (rs67426579) with a mean value of 0.362, implying each locus was informative. The PD ranged from 0.5492 (rs5897566) to 0.6516 (rs59841142), with an average value of 0.612, indicating the high efficacy of 57 InDels for forensic personal identification. PE was between 0.0875 (rs5897566) and 0.2428 (rs35453727) with a mean value of 0.163, PM was between 0.3484 (rs59841142) and 0.4508 (rs5897566) with a mean value of 0.388, and TPI ranged from 0.7722 (rs5897566) to 1.1296 (rs35453727) with a mean value of 0.945. The combined power of discrimination (CPD) and combined power of exclusion (CPE) were 0.999 999 999 999 999 996 69 and 0.999 962 965, respectively. These results demonstrated that the 57 InDels system not only had strong individual identification ability, but also had greater practical value for forensic paternity identification. The AGCU InDel 60 kit was improved and refined on the basis of the AGCU InDel 50 kit, adding 18 newly polymorphic A-InDels and removing 8 poorly polymorphic A-InDels (Chen X et al. Citation2022), and the efficacy of individual identification and paternity identification was further improved. For samples from the Yunnan Yi group, the CPD and CPE of the Investigator® DIPplex kit (Zhang YD et al. Citation2015) were found to be 0.999 999 999 957 13 and 0.977 46, respectively, which was lower than those of the AGCU InDel 60 kit.
3.3. Principal component analysis and multidimensional scaling analysis based on samples from Yunnan Yi and 29 reference populations
The results of the PCA based on allele frequencies showed that 70.28% of the variance among the 30 populations was explained by principal component 1 (PC1, 46.7%) and PC2 (23.58%), indicating the two principal components could fully reflect the overall information. According to the scatter plot, the first two PCs could distinguish the populations in different continents (except the American continent). Populations from Africa and Europe clustered separately, and were located in the lower right and upper top, respectively. Additionally, nine populations from East Asia clustered together in the lower left (). Yunnan Yi group showed the closest genetic relationship with the populations from East Asia. These results demonstrated that the genetic differences of these loci among different intercontinental populations were greater than those within populations from the same continent, and could be used to distinguish different intercontinental populations.
Figure 3. Principal component analysis (A) and multidimensional scaling analysis (B) among 30 populations conducted by R.
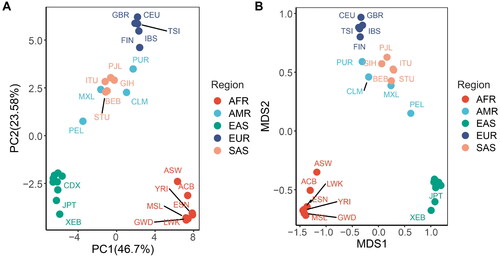
MDS was further performed. The results were similar to that of PCA and demonstrated the close genetic relationship between Yunnan Yi group and other populations from East Asia ().
3.4. Phylogenetic analysis of Yunnan Yi and 29 reference populations
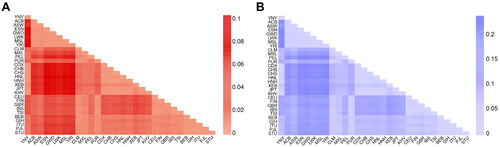
Several studies have documented the forensic and phylogenetic analyses of A-STR (Zhang X et al. Citation2017; Cheng et al. Citation2021), X-STR (He et al. Citation2017) and Y-STR (Fan et al. Citation2019) in Yi populations, but InDel markers have been rarely reported. To explore the genetic differences and the degree of genetic differentiation among populations, and further assess the genetic relationships between the Yunnan Yi group and other reference populations, we measured the FST and DA values among paired populations, which were visualised by heat maps. The results showed that the change trend of FST was consistent with that of DA (Supplementary Table 5, ).
Figure 4. Heat map of pairwise DA and FST of Yunnan Yi group and 29 reference populations on the basis of allele frequency by R.
The results of the N-J phylogenetic trees based on genetic distance DA divided into three major branches, which revealed that populations from Africa combined into one branch, European populations clustered into one branch, while American, South Asian, and East Asian populations clustered into one branch (), with each of the South Asian populations forming a subbranch. The Yunnan Yi were located in the East Asian subbranch. Dai in Xishuangbanna were first clustered with Hainan Li, then gathered with Yunnan Yi, Hunan Han and other East Asian populations, which indicated that Yunnan Yi exhibited close genetic relationships with East Asian populations. Yunnan Yi were similar to Dai in Xishuangbanna and Hainan Li not only because of the geographical proximity, but also because they belong to the Sino-Tibetan Zhuang-Dong language group. Although Dingjie Sherpas and Yunnan Yi belong to the same Sino-Tibetan language family, expected genetic homogeneity was not observed. This might be due to the fact that Dingjie Sherpas live on the Qinghai-Tibetan plateau, with unique topography, geographic isolation, and less genetic exchange. Compared with Dingjie Sherpas, there were frequent gene exchanges between the Han and Yi during their history, which influenced the genetic differentiation of Yunnan Yi group (Yao et al. Citation2017). Thus, more in-depth archaeological study and the populations or ethnic groups for genetic relationship analysis are required to obtain more accurate results of the complex genetic structure of Yunnan Yi group.
Figure 5. Phylogenetic trees showing phylogenetic relationships among the Yunnan Yi group and 29 reference populations on the basis of DA using MEGA Software (v11.0).
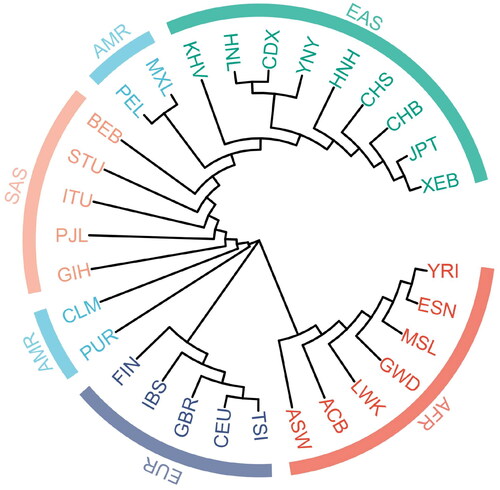
FST is widely used as a measure of genetic differentiation among and within populations. The larger the FST value is, the higher the genetic differentiation between pairwise populations is, and vice versa. The FST values between Yunnan Yi group and other East Asian populations ranged from 0.0025 to 0.0392, indicating a small degree of genetic differentiation. However, the FST values between Yunnan Yi group and African populations were larger than those between Yunnan Yi group and the East Asian populations (0.1835 − 0.2391), indicating a larger genetic differentiation. Additionally, moderate genetic differentiations were observed between Yunnan Yi group and South Asian (0.0679 − 0.0874), European (0.1431 − 0.1495) and American (0.0751 − 0.1169) populations. These results were consistent with the results of PCA, MDS and phylogenetic trees described above, implying that the level of genetic differentiation was correlated with geographical location.
3.5. Genetic structure analysis among Yunnan Yi and 29 reference populations
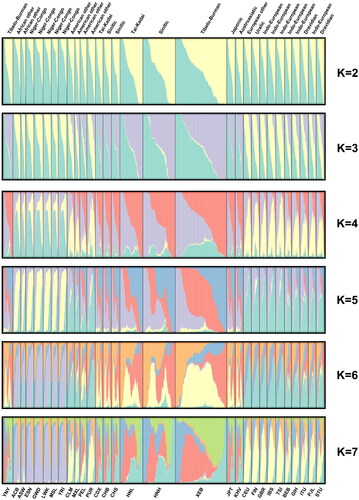
Structural clustering analyses were performed based on the 57 InDels genotyping data to assess the ancestral composition in Yunnan Yi group and another 29 populations. Different colour compositions reflected different ancestral components (). The results of STRUCTURE HARVESTER analysis reveal that the optimal K value was 3. When K = 2, the ancestral component of all African populations was predominantly green, while the other intercontinental populations were dominated by yellow. When K = 3, the ancestral components of the African, East Asian, and European populations showed a predominance of green, yellow, and purple, respectively. The distribution of ancestral components in Yunnan Yi group was most similar to that of the East Asian population (a mixture of green, yellow, and purple, but with a predominance of green-yellow). In conclusion, different intercontinental populations have different ancestral components. In contrast, the composition colours of the South Asian and the American populations were relatively balanced, suggesting a mixed origin with more genetic exchanges between their ancestors.
Figure 6. STRUCTURE analysis on the individual levels of 30 populations when assuming K = 2–7 with STRUCTURE program v2.3.4.
Next, allele frequency differences (δ) between different intercontinental populations and informativeness for assignment (In) values were calculated to evaluate genetic differentiation of 57 InDels. The details of these results are described in Supplementary Tables 7 and 8. A panel of 30 InDels was screened out by δ ≥ 0.3 and In ≥ 0.1. Finally, the success rates of the continental population classifications were estimated using the method of cross-validation. Based on the 57 InDels, when only three continental populations were included, the estimation of success rates for the East Asians, Europeans, and Africans were 99.95%, 99.60%, and 98.79%, respectively. When it comes to five continental populations, the estimation of success rates of the East Asians, Europeans, Africans, South Asians, and Americans were 98.34%, 90.06%, 98.64%, 83.23% and 66.28%, respectively. On the other hand, success rates of the selected 30 InDels for the East Asians (99.90%), Europeans (99.40%), and Africans (98.49%) were not much different to those of the 57 InDels. Similar results could also be observed when five continental populations were included (the East Asians [97.62%], Europeans [83.50%], Africans [98.49%], South Asians [74.64%], and Americans [62.54%]). The above results revealed that the efficiency for continental population classifications of the selected 30 InDels was basically consistent with that of 57 InDels, suggesting that the 30 InDels were more suitable for forensic ancestry analysis as candidates of ancestry informative markers (AIMs). Although these InDels could be used to differentiate the ancestors of admixed populations to some extent, it is necessary to explore more effective markers for identification of ancestry contributions in future studies.
There have been several studies investigating the genetic background of the Yi group. Genetic heterogeneity and homogeneity between the Yi group and populations from different continents has been revealed by multiple genetic markers including STRs and InDels (Zhang YD et al. Citation2015; Liu et al. Citation2020; Zou et al. Citation2020; Song et al. Citation2021). In brief, the results demonstrated a strong association between genetic clustering of the Yi group and its phylogeographic division and language. There were noticeable variations between the Yi group and populations from different continents, which were consistent with our research results. The Yi people have resided in the Tibetan-Yi corridor for a long time. The Tibetan-Yi corridor is not only an important channel of prehistoric ethnic migration but also a significant channel of cultural intercommunion and goods trade, profoundly influencing the gene flow of the Yi group (Zhang Z et al. Citation2022). Recently, there have been several studies focusing on the genetic associations and differences between the Yi and other ethnic groups in China (Fan et al. Citation2019; Cheng et al. Citation2021; Fan GY et al. Citation2021; Song et al. Citation2021). However, the ethnic origin of the Yi group could hardly be revealed by a small number of markers. Whole genome sequencing of large numbers of individuals from the Yi group could provide a wealth of information about genetic structure and population histories and help to identify specific AIMs for the Yi, which might be an effective tool.
4. Conclusion
In this study, we assessed the efficacy of the 57 InDels system for forensic applications in the Yi ethnic group in Yunnan, China, and analysed the genetic relationships between Yi and other reference populations. The results revealed that these loci were highly polymorphic in the Yunnan Yi group, and 30 InDels were found to be candidate AIMs for ancestry. In conclusion, the 57 InDels could be used for individual identification and paternity testing, and distinguishing intercontinental populations, with the potential of biogeographic ancestry inference. This study provides good genetic background data for the establishment of a database of InDels in Chinese populations and the analysis of genetic relationships, which will be helpful for future population genetic diversity studies.
Supplemental Material
Download MS Excel (87.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Budowle B, Moretti TR, Baumstark AL, Defenbaugh DA, Keys KM. 1999. Population data on the thirteen CODIS core short tandem repeat loci in African Americans, U.S. Caucasians, Hispanics, Bahamians, Jamaicans, and Trinidadians. J Forensic Sci. 44(6):1. doi: 10.1520/JFS14601J.
- Chen X, Nie S, Hu L, Fang Y, Cui W, Xu H, Zhao C, Zhu BF. 2022. Forensic efficacy evaluation and genetic structure exploration of the Yunnan Miao group by a multiplex InDel panel. Electrophoresis. 43(16–17):1765–8. doi: 10.1002/elps.202100387.
- Chen L, Pan X, Wang Y, Du W, Wu W, Tang Z, Xiao C, Han X, Liu C, Liu C. 2021. Development and validation of a forensic multiplex system with 38 X-InDel loci. Front Genet. 12:670482. doi: 10.3389/fgene.2021.670482.
- Cheng J, Song B, Fu J, Zheng X, He T, Fu J. 2021. Genetic polymorphism of 19 autosomal STR loci in the Yi ethnic minority of Liangshan Yi autonomous prefecture from Sichuan province in China. Sci Rep. 11(1):16327. doi: 10.1038/s41598-021-95883-x.
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x.
- Fan GY, An YR, Peng CX, Deng JL, Pan LP, Ye Y. 2019. Forensic and phylogenetic analyses among three Yi populations in Southwest China with 27 Y chromosomal STR loci. Int J Legal Med. 133(3):795–797. doi: 10.1007/s00414-018-1984-4.
- Fan H, He Y, Li S, Xie Q, Wang F, Du Z, Fang Y, Qiu P, Zhu B. 2021. Systematic evaluation of a novel 6-dye direct and multiplex PCR-CE-based InDel typing system for forensic purposes. Front Genet. 12:744645. doi: 10.3389/fgene.2021.744645.
- Fan GY, Zhang ZQ, Tang PZ, Song DL, Zheng XK, Zhou YJ, Liu MN. 2021. Forensic and phylogenetic analyses of populations in the Tibetan-Yi corridor using 41 Y-STRs. Int J Legal Med. 135(3):783–785. doi: 10.1007/s00414-020-02453-3.
- Fang Y, Zhao C, Jin X, Lan Q, Lan J, Xie T, Zhu B. 2022. Genetic characterization evaluation of a novel multiple system containing 57 deletion/insertion polymorphic loci with short amplicons in Hunan Han population and its intercontinental populations analyses. Gene. 809:146006. doi: 10.1016/j.gene.2021.146006.
- He G, Li Y, Zou X, Li P, Chen P, Song F, Gao T, Liao M, Yan J, Wu J. 2017. Forensic characteristics and phylogenetic analyses of the Chinese Yi population via 19 X-chromosomal STR loci. Int J Legal Med. 131(5):1243–1246. doi: 10.1007/s00414-017-1563-0.
- Jakobsson M, Rosenberg NA. 2007. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 23(14):1801–1806. doi: 10.1093/bioinformatics/btm233.
- LaRue BL, Ge J, King JL, Budowle B. 2012. A validation study of the Qiagen Investigator DIPplex(R) kit; an INDEL-based assay for human identification. Int J Legal Med. 126(4):533–540. doi: 10.1007/s00414-012-0667-9.
- Li CT, Zhang SH, Zhao SM. 2011. Genetic analysis of 30 InDel markers for forensic use in five different Chinese populations. Genet Mol Res. 10(2):964–979. doi: 10.4238/vol10-2gmr1082.
- Li C, Zhao S, Zhang S, Li L, Liu Y, Chen J, Xue J. 2011. Genetic polymorphism of 29 highly informative InDel markers for forensic use in the Chinese Han population. Forensic Sci Int Genet. 5(1):e27-30–e30. doi: 10.1016/j.fsigen.2010.03.004.
- Liu Y, Zhang H, He G, Ren Z, Zhang H, Wang Q, Ji J, Yang M, Guo J, Yang X, et al. 2020. Forensic features and population genetic structure of Dong, Yi, Han, and Chuanqing human populations in Southwest China inferred from insertion/deletion markers. Front Genet. 11:360. doi: 10.3389/fgene.2020.00360.
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 70(12):3321–3323. doi: 10.1073/pnas.70.12.3321.
- Nei M, Tajima F, Tateno Y. 1983. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol. 19(2):153–170. doi: 10.1007/BF02300753.
- Pereira R, Phillips C, Alves C, Amorim A, Carracedo A, Gusmão L. 2009. A new multiplex for human identification using insertion/deletion polymorphisms. Electrophoresis. 30(21):3682–3690. doi: 10.1002/elps.200900274.
- Pimenta JR, Pena SD. 2010. Efficient human paternity testing with a panel of 40 short insertion-deletion polymorphisms. Genet Mol Res. 9(1):601–607. doi: 10.4238/vol9-1gmr838.
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155(2):945–959. doi: 10.1093/genetics/155.2.945.
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. 2002. Genetic structure of human populations. Science. 298(5602):2381–2385. doi: 10.1126/science.1078311.
- Santos C, Phillips C, Oldoni F, Amigo J, Fondevila M, Pereira R, Carracedo A, Lareu MV. 2015. Completion of a worldwide reference panel of samples for an ancestry informative Indel assay. Forensic Sci Int Genet. 17:75–80. doi: 10.1016/j.fsigen.2015.03.011.
- Santos C, Fondevila M, Ballard D, Banemann R, Bento AM, Børsting C, Branicki W, Brisighelli F, Burrington M, Capal T, EUROFORGEN-NoE Consortium, et al. 2015. Forensic ancestry analysis with two capillary electrophoresis ancestry informative marker (AIM) panels: results of a collaborative EDNAP exercise. Forensic Sci Int Genet. 19:56–67. doi: 10.1016/j.fsigen.2015.06.004.
- Song M, Wang Z, Lyu Q, Ying J, Wu Q, Jiang L, Wang F, Zhou Y, Song F, Luo H, et al. 2022. Paternal genetic structure of the Qiang ethnic group in China revealed by high-resolution Y-chromosome STRs and SNPs. Forensic Sci Int Genet. 61:102774.doi: 10.1016/j.fsigen.2022.102774.
- Song Z, Wang Q, Zhang H, Tang J, Wang Q, Zhang H, Yang M, Ji J, Ren Z, Wu Y, et al. 2021. Genetic structure and forensic characterization of 36 Y-chromosomal STR loci in Tibeto-Burman-speaking Yi population. Mol Genet Genomic Med. 9(2):e1572.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 28(10):2731–2739. doi: 10.1093/molbev/msr121.
- Tillmar A, Sturk-Andreaggi K, Daniels-Higginbotham J, Thomas JT, Marshall C. 2021. The FORCE panel: an all-in-one SNP marker set for confirming investigative genetic genealogy leads and for general forensic applications. Genes (Basel). 12(12):1968. doi: 10.3390/genes12121968.
- Walsh PS, Metzger DA, Higuchi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 10(4):506–513.
- Wang M, Du W, Tang R, Liu Y, Zou X, Yuan D, Wang Z, Liu J, Guo J, Yang X, et al. 2022. Genomic history and forensic characteristics of Sherpa highlanders on the Tibetan Plateau inferred from high-resolution InDel panel and genome-wide SNPs. Forensic Sci Int Genet. 56:102633. doi: 10.1016/j.fsigen.2021.102633.
- Yao HB, Tang S, Yao X, Yeh HY, Zhang W, Xie Z, Du Q, Ma L, Wei S, Gong X, et al. 2017. The genetic admixture in Tibetan-Yi Corridor. Am J Phys Anthropol. 164(3):522–532. doi: 10.1002/ajpa.23291.
- Zhang X, Hu L, Du L, Zheng H, Nie A, Rao M, Bo Pang J, Nie S. 2017. Population data for 20 autosomal STR loci in the Yi ethnic minority from Yunnan Province, Southwest China. Forensic Sci Int Genet. 28:e43–e44. doi: 10.1016/j.fsigen.2017.02.017.
- Zhang X, Shen C, Jin X, Guo Y, Xie T, Zhu B. 2021. Developmental validations of a self-developed 39 AIM-InDel panel and its forensic efficiency evaluations in the Shaanxi Han population. Int J Legal Med. 135(4):1359–1367. doi: 10.1007/s00414-021-02600-4.
- Zhang YD, Shen CM, Jin R, Li YN, Wang B, Ma LX, Meng HT, Yan JW, Dan Wang H, Yang ZL, et al. 2015. Forensic evaluation and population genetic study of 30 insertion/deletion polymorphisms in a Chinese Yi group. Electrophoresis. 36(9–10):1196–1201. doi: 10.1002/elps.201500003.
- Zhang Y, Yu Z, Mo X, Zhao X, Li W, Liu H, Liu C, Wu R, Sun H. 2021. Development and validation of a new 18 X-STR typing assay for forensic applications. Electrophoresis. 42(6):766–773. doi: 10.1002/elps.202000168.
- Zhang Z, Zhang Y, Wang Y, Zhao Z, Yang M, Zhang L, Zhou B, Xu B, Zhang H, Chen T, et al. 2022. The Tibetan-Yi region is both a corridor and a barrier for human gene flow. Cell Rep. 39(4):110720. doi: 10.1016/j.celrep.2022.110720.
- Zhao GB, Ma GJ, Zhang C, Kang KL, Li SJ, Wang L. 2021. BGISEQ-500RS sequencing of a 448-plex SNP panel for forensic individual identification and kinship analysis. Forensic Sci Int Genet. 55:102580. doi: 10.1016/j.fsigen.2021.102580.
- Zhou Y, Cui W, Wu B, Zhu B. 2022. Development and validation of a new multiplex Y-STR panel designed to increase the power of discrimination. Electrophoresis. 43(18–19):1899–1910. doi: 10.1002/elps.202100313.
- Zou X, He G, Wang M, Huo L, Chen X, Liu J, Wang S, Ye Z, Wang F, Wang Z, et al. 2020. Genetic diversity and phylogenetic structure of four Tibeto-Burman-speaking populations in Tibetan-Yi corridor revealed by insertion/deletion polymorphisms. Mol Genet Genomic Med. 8(4):e1140.