Abstract
Background
At present, there are no available genetic data on the AGCU EX22 Kit from the Wuhu Han population.
Aim
This study investigates the applicability of the AGCU EX22 kit, designed for the Chinese population for forensic analysis and population genetics of the Wuhu Han population.
Subjects and methods
Bloodstains from 1565 unrelated healthy individuals in Wuhu city, Anhui Province, were collected for analysis. The AGCU EX22 kit was used for amplification, and capillary electrophoresis was used to separate the amplification products. Allele frequencies and forensic parameters were determined. The Wuhu Han population was compared to 10 reference populations through genetic distance, a phylogenetic neighbor-joining tree and principal component analysis.
Results
In total, 281 alleles and 1187 genotypes were observed. No significant deviations from Hardy-Weinberg equilibrium at any locus were found after Bonferroni’s correction. The 21 autosomal short tandem repeat (STR) genetic markers exhibited high informativeness and polymorphism. The cumulative power of discrimination and power of exclusion were 0.999999999999999999999999913380 and 0.999999996752339, respectively. Population comparisons revealed a genetic affinity between Wuhu Han and southern Han populations, except for the Guangdong Han population, which aligned with the traditional geographical division in China.
Conclusion
The AGCU EX22 Kit, containing 21 STR loci, is suitable for forensic application and population genetics studies in the Wuhu Han population.
Introduction
Short tandem repeats (STRs) or microsatellites, consisting of repeat motifs of 2 bps∼6 bps within small fragments of ≤1 kb, are highly abundant in the human genome. These STRs are well-suited for the polymerase chain reaction (PCR) methodology and have a high degree of polymorphism at a single locus. Consequently, STRs are widely employed in forensic individual identification (Nwawuba Stanley et al. Citation2020), paternity testing (Song W et al. Citation2022), population genetics (Mei et al. Citation2021; Zhang et al. Citation2023) and genomic analyses (Cong et al. Citation2022). The 5-dye AGCU EX22 Kit (AGCU ScienTech, Jiangsu, China), which was originally developed for paternity tests in a special campaign against human trafficking in China, includes amelogenin loci and 21 autosomal genetic markers, including D3S1358, D13S317, D7S820, D16S539, Penta E, D2S441, TPOX, TH01, D2S1338, CSF1PO, Penta D, D10S1248, D19S433, vWA, D21S11, D18S51, D6S1043, D8S1179, D5S818, D12S391 and FGA.
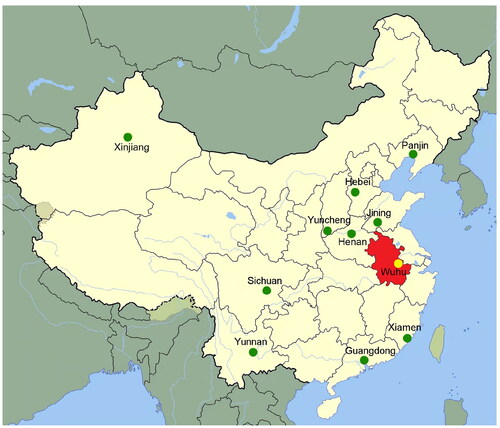
Wuhu, a prefecture-level city in Anhui Province and a central city in the Yangtze River Delta, covers 6009.02 km2 in the southeast of Anhui Province, near the lower reaches of the Yangtze River (). As of the 2020 census, the total population of Wuhu is 3,644,420, predominantly consisting of the Han population with a small number of ethnic minorities such as Hui, Miao, Zhuang, Yi, Tujia and Manchu (http://www.stats.gov.cn/sj/pcsj/rkpc/7rp/indexch.htm).
Figure 1. Geographic positions of the Wuhu Han population and other additional reference Chinese Han populations. The map highlights Anhui Province with a red background.
At present, there are no available genetic data on the AGCU EX22 Kit from the Wuhu Han population. In this study, we used the AGCU EX22 Kit to obtain 21 autosomal-STR genotypes from 1565 unrelated Han individuals in Wuhu city, East China, and calculated forensic genetic parameters of the data. Additionally, to better comprehend the genetic structural background, we compared the studied population with 10 other reference Han populations.
Materials and methods
Samples
Human blood samples were collected from 1565 unrelated healthy Han individuals in Wuhu city, Anhui Province, between July 2021 and December 2022. The sample comprised 850 males and 715 females, all from families residing in Wuhu city for at least three generations. Informed consent was obtained from all participants, and the study followed the Helsinki Declaration and its subsequent amendments and was approved by the Medical Ethics Committee of Wannan Medical College (No. 2021-2).
PCR amplification and capillary electrophoresis
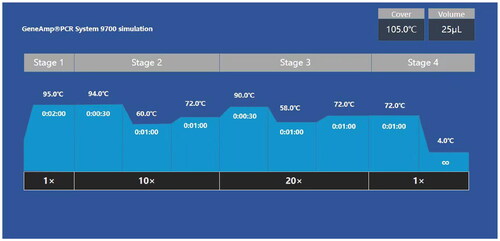
Genomic DNA was extracted using the Chelex-100 protocol (Walsh et al. Citation1991) and subsequently quantified by the Epoch 2 microplate spectrophotometer (BioTek, USA). PCR amplification was performed using GeneAmp® PCR 9700 (Thermo Fisher Scientific, MA, USA) with the AGCU EX22 Kit and a total reaction volume of 25 μL comprising 10 μL of reaction mix, 5 μL of EX22 primers, 1.0 μL of C-Taq polymerase and 9 μL of template with 0.5 ∼ 2 ng of DNA. The thermal cycling conditions were as follows: the initial incubation at 95 °C for 2 min; 10 cycles of 94 °C for 30 s, 60 °C for 1 min and 72 °C for 1 min; 20 cycles of 90 °C for 30 s, 58 °C for 1 min and 72 °C for 1 min; followed by a final extension step at 72 °C for 10 min (). 9947A and ddH2O were used as the positive and negative controls, respectively.
The amplification products were separated and detected on Applied Biosystems™ 3500 Series Genetic Analyser (Thermo Fisher Scientific, MA, USA). 1 μL of allelic ladder or PCR products was added to a 10 μL mixture of Hi-Di formamide SIZ500 (Hi-Di formamide: SIZ500 = 1000:30). Samples were injected at 3 kV for 6 s and electrophoresed at 3 kV for 1400s at 60 °C. Raw data analysis was performed using GeneMapper ID-X Software (Thermo Fisher Scientific, MA, USA). Allelic nomenclatures were assigned by comparing the fragment sizes of PCR products with allelic ladders provided in AGCU EX22 Kit.
Data statistics
Allele frequencies of the 1565 samples were calculated using the direct counting method. The chi-squared tests for Hardy-Weinberg equilibrium (HWE) and forensic parameters, including the observed heterozygosity (H), matching probability (MP), power of discrimination (PD), polymorphism information content (PIC), power of exclusion (PE) and typical paternity index (TPI), were evaluated using the Modified PowerStats software (Zhao et al. Citation2003).
Pairwise genetic distances (Fst) and corresponding p values were calculated using Arlequin v3.5.1.3 (Excoffier and Lischer Citation2010) based on the allele frequencies of 19 overlapped autosomal STRs (excluding D10S1248 and D2S441) between our data and 10 other published populations: Hebei Han (Sun et al. Citation2017), Yuncheng Han (Chen et al. Citation2019), Guangdong Han (Qu et al. Citation2019), Yunnan Han (Lu et al. Citation2021), Henan Han (Wu et al. Citation2022), Jining Han (Dang et al. Citation2020), Panjin Han (Wang et al. Citation2022), Xinjiang Han (Adnan et al. Citation2021), Xiamen Han (Lu et al. Citation2017) and Sichuan Han (He et al. Citation2017). Nei’s genetic distances (Saitou and Nei Citation1987) were obtained using the PHYLIP 3.69 package (Felsenstein Citation2009). Phylogenetic trees based on the Nei’s genetic distance matrix were constructed using the neighbor-joining method with MEGA 11 software (Tamura et al. Citation2021). A principal component analysis (PCA) scatter plot was generated using MVSP software (version 3.22).
Results and discussion
Allele frequencies and forensic parameters
The allele frequencies and corresponding forensic parameters of 21 autosomal-STR loci in the Wuhu Han population are presented in Supplementary Table S1. HWE p values for the 21 STR loci ranged from 0.0056 (Penta D) to 0.9466 (D19S433), with most loci (except for D3S1358 and Penta D) having p values greater than 0.05 indicating compliance with HWE in the Wuhu Han population. After Bonferroni’s correction (p = .05/21 = .0024), no significant deviations from HWE were observed at any locus. In total, 281 alleles and 1187 genotypes were identified in the 1565 Wuhu Han individuals from Anhui Province.
Among the 21 STR loci, TPOX exhibited the lowest allele number (7) and genotype number (20), whereas the Penta E locus exhibited the highest values (26 and 165, respectively). The allele frequencies were 0.0003–0.5188, with TPOX showing the highest value (allele 8). H values varied from 0.5962 (TPOX) to 0.9137 (Penta E). In a previous study, Dávila et al. (Citation2009) suggested that STR loci with H values exceeding 0.5 are appropriate for genetic diversity studies.
The panel’s MP ranged from 0.0136 (Penta E) to 0.2009 (TPOX), the TPI spanned 1.2381 (TPOX) to 5.7963 (Penta E), and the PIC values varied from 0.5564 (TPOX) to 0.9112 (Penta E). Loci with PIC >0.5 are considered highly informative and polymorphic (Marshall et al. Citation1998; Botstein et al. Citation1980), indicating that the 21 autosomal STR genetic markers included in the AGCU EX22 Kit are suitable for genetic characterisation and diversity studies.
Regarding forensic parameters, TPOX exhibited the least PD value (0.7991) and PE value (0.2863), whereas Penta E had the highest PD (0.9864) and PE (0.8263) values. The cumulative PD and PE values were calculated as 0.999999999999999999999999913380 and 0.999999996752339, respectively, demonstrating the suitability of 21 STR loci for individual identification and parentage testing in the Wuhu Han population.
Population comparisons among Chinese Han populations
To investigate the genetic structure of the Wuhu Han population and its genetic relationships with other Chinese Han populations, Fst and corresponding p values based on 19 overlapping STR loci were calculated between Wuhu Han and 10 other Chinese Han populations (). After Bonferroni’s correction (p = .05/19 = .0026), the results revealed statistically significant differences between Wuhu Han and Guangdong Han at 8 out of 19 STR loci, followed by Henan Han and Xinjiang Han at 1 STR locus. However, no significant differences were observed between Wuhu Han and Hebei Han, Yuncheng Han, Yunnan Han, Jining Han, Panjin Han, Xiamen Han and Sichuan Han in the shared 19 STR loci. These results suggest that the Wuhu Han population exhibits the most genetic differences from the Guangdong Han population, which can be attributed to their distant geographical location.
Table 1. Pairwise genetic distance (Fst) values and p values calculated for the Wuhu Han population and 10 other Han populations.
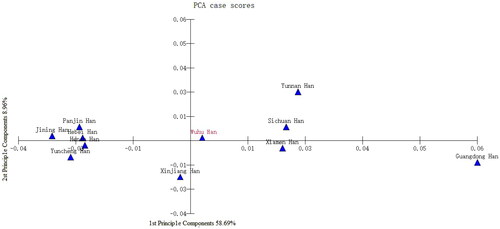
PCA results based on the allele frequencies of the 19 shared STR loci are illustrated in . PCA captured approximately 67.65% of the genetic variations among 11 Chinese Han populations, with the first and second component accounting for 58.69% and 8.96%, respectively. Based on the first principal component, the 11 Chinese Han populations were categorised into the northern Han group (Hebei Han, Yuncheng Han, Henan Han, Jining Han, Panjin Han and Xinjiang Han) and the southern Han group (including the studied Wuhu Han population, Guangdong Han, Yunnan Han, Xiamen Han and Sichuan Han). This division aligns with the traditional geographical boundary of the Qinling-Huaihe Line in China. Additionally, Wuhu Han was positioned between the northern Han group and the other four southern Han populations but was significantly distant from the Guangdong Han population. Previous research based on array-based genome-wide data, also demonstrated that Guangdong Han and Anhui Han (including Wuhu Han) belonged to two different clusters (Cao et al. Citation2020). Regarding the second principal component, the northern Han populations were closely grouped in the second and third quadrants except for Xinjiang Han, which exhibited scattered distribution, indicating a geographic distance association. However, the southern Han populations were relatively dispersed in the first and fourth quadrants, suggesting greater variation within the southern Han group. This phenomenon suggests the presence of small migration movements among the southern Han populations, likely influenced by geographical factors, which is consistent with findings from a previous study using Y-chromosome haplogroups (Lang et al. Citation2019).
Figure 3. Principal component analysis (PCA) plots of the Wuhu Han population and 10 other reference Han populations. The first and second principal components account for 58.69% and 8.96% of the variance, respectively.
Nei’s standard genetic distances were calculated to explore the genetic similarities and differences between the newly studied Wuhu Han population and the 10 other reference Han populations (). The results revealed that Wuhu Han exhibited the closest genetic distance to Sichuan Han (0.001544), followed by Xiamen Han (0.001596), whereas the largest genetic distance (0.005793) was observed between Wuhu Han and Guangdong Han. Nei’s genetic distance matrix was then used to construct a phylogenetic tree using the neighbor-joining method (). In the neighbor-joining (N-J) tree, Wuhu Han initially clustered with the six northern Han populations, and subsequently formed a cluster with the Sichuan Han population, followed by the Yunnan Han population. These populations constituted the first branch, whereas Guangdong Han and Xiamen Han constituted the second branch. Overall, the genetic differences between the Wuhu Han population and the 10 other Chinese Han populations mainly aligned with their geographic distributions and were basically in accordance with the findings from PCA.
Figure 4. Neighbor-joining phylogenetic tree based on the allele frequencies of 19 shared STRs between the Wuhu Han population and 10 other Han populations. The Wuhu Han population is represented in red font.
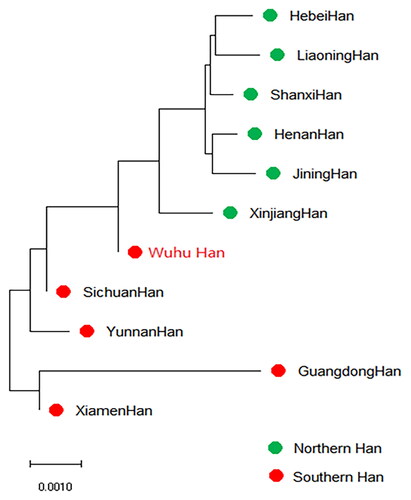
Table 2. Nei’s genetic distances between the Wuhu Han population and 10 other Han populations.
Conclusions
In this study, we presented a comprehensive dataset of 21 autosomal STR loci for the Wuhu Han population using the AGCU EX22 Kit, along with the calculation of relevant forensic parameters. The 21 STR panel was highly informative and polymorphic, making it suitable for forensic applications. Additionally, population comparisons revealed that the Wuhu Han population exhibited closer genetic distances with the southern Han populations (excluding Guangdong Han) compared with the northern Han populations, aligning roughly their geographical positions. The AGCU EX22 Kit also provides valuable insights for population genetics studies.
Supplemental Material
Download MS Excel (17.9 KB)Acknowledgements
We would like to thank all donors for this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are available from the corresponding author, [FH], upon reasonable request.
Additional information
Funding
References
- Adnan A, Anwar A, Simayijiang H, Farrukh N, Hadi S, Wang C-C, Xuan J-F. 2021. The heart of silk road “Xinjiang,” its genetic portray, and forensic parameters inferred from autosomal STRs. Front Genet. 12:1. doi: 10.3389/fgene.2021.760760.
- Botstein D, White RL, Skolnick M, Davis RW. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 32(3):314–6.
- Cao Y, Li L, Xu M, Feng Z, Sun X, Lu J, Xu Y, Du P, Wang T, Hu R, et al. 2020. The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals. Cell Res. 30(9):717–731. doi: 10.1038/s41422-020-0322-9.
- Chen P, Wu J, Luo L, Gao H, Wang M, Zou X, Li Y, Chen G, Luo H, Yu L, et al. 2019. Population genetic analysis of modern and ancient DNA variations yields new insights into the formation, genetic structure, and phylogenetic relationship of Northern Han Chinese. Front Genet. 10:1045. doi: 10.3389/fgene.2019.01045.
- Cong PK, Bai WY, Li JC, Yang MY, Khederzadeh S, Gai SR, Li N, Liu YH, Yu SH, Zhao WW, et al. 2022. Genomic analyses of 10,376 individuals in the Westlake BioBank for Chinese (WBBC) pilot project. Nat Commun. 13(1):2939. 26 doi: 10.1038/s41467-022-30526-x.
- Dang Z, Liu Q, Zhang G, Li S, Wang D, Pang Q, Yang D, Li C, Cui W, Wang Y. 2020. Population genetic data from 23 autosomal STR loci of Huaxia Platinum system in the Jining Han population. Mol Genet Genomic Med. 8(4):e1142.
- Dávila SG, Gil MG, Resino-Talaván P, Campo JL. 2009. Evaluation of diversity between different Spanish chicken breeds, a tester line, and a White Leghorn population based on microsatellite markers. Poult Sci. 88(12):2518–2525. doi: 10.3382/ps.2009-00347.
- Excoffier L, Lischer HE. 2010. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x.
- Felsenstein J. 2009. PHYLIP (phylogeny inference package) version 3.69. Seattle: University of Washington.
- He G, Li Y, Wang Z, Liang W, Luo H, Liao M, Zhang J, Yan J, Li Y, Hou Y, et al. 2017. Genetic diversity of 21 autosomal STR loci in the Han population from Sichuan province, Southwest China. Forensic Sci Int Genet. 31:e33–e35. doi: 10.1016/j.fsigen.2017.07.006.
- Lang M, Liu H, Song F, Qiao X, Ye Y, Ren H, Li J, Huang J, Xie M, Chen S, et al. 2019. Forensic characteristics and genetic analysis of both 27 Y-STRs and 143 Y-SNPs in Eastern Han Chinese population. Forensic Sci Int Genet. 42:e13–e20. doi: 10.1016/j.fsigen.2019.07.011.
- Lu X, Yang S, Jiang Y, Li K, Zhao F, Yang X, Zhang N, Zhang S, Xia S, Wu L, et al. 2021. Genetic variation of 20 autosomal STR loci in the Han nationality in Central Yunnan, Southwest China. Leg Med (Tokyo)). 48:101807. doi: 10.1016/j.legalmed.2020.101807.
- Lu Y, Song P, Huang JC, Wu X. 2017. Genetic polymorphisms of 20 autosomal STR loci in 5141 individuals from the Han population of Xiamen, Southeast China. Forensic Sci Int Genet. 29:e31–e32. doi: 10.1016/j.fsigen.2017.03.023.
- Marshall TC, Slate J, Kruuk LE, Pemberton JM. 1998. Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol. 7(5):639–655. doi: 10.1046/j.1365-294x.1998.00374.x.
- Mei S, Liu Y, Zhao C, Xu H, Li S, Zhu B. 2021. The polymorphism analyses of short tandem repeats as a basis for understanding the genetic characteristics of the Guanzhong Han Population. Biomed Res Int. 2021:8887244. doi: 10.1155/2021/8887244.
- Nwawuba Stanley U, Mohammed Khadija A, Adams Tajudeen B, Omusi Precious I, Ayevbuomwan Davidson E. 2020. Forensic DNA proffling: autosomal short tandem repeat as a prominent marker in crime investigation. Malays J Med Sci. 27(4):22–35.
- Qu N, Zhang X, Liang H, Ou X. 2019. Analysis of genetic polymorphisms and mutations at 23 autosomal STR loci in Guangdong Han population. Forensic Sci Int Genet. 38:e16–e17. doi: 10.1016/j.fsigen.2018.11.003.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4(4):406–425.
- Song W, Xiao N, Zhou S, Yu W, Wang N, Shao L, Duan Y, Chen M, Pan L, Xia Y, et al. 2022. Non-invasive prenatal paternity testing by analysis of Y-chromosome mini-STR haplotype using next-generation sequencing. PLoS One. 17(4):e0266332. doi: 10.1371/journal.pone.0266332.
- Sun L, Shi K, Tan L, Zhang Q, Fu L, Zhang X, Fu G, Li S, Cong B. 2017. Analysis of genetic polymorphisms and mutations at 19 STR loci in Hebei Han population. Forensic Sci Int Genet. 31:e50–e51. doi: 10.1016/j.fsigen.2017.07.016.
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Walsh PS, Metzger DA, Higuchi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 10(4):506–513.
- Wang H, Xin C, Meng X, Xing S, Guo B, Chen Y, Wang BJ, Yao J. 2022. Genetic polymorphism and forensic application of 23 autosomal STR loci in the Han population of Panjin City, Liaoning Province, Northeastern China. Ann Hum Biol. 49(5-6):254–259. doi: 10.1080/03014460.2022.2100479.
- Wu H, Wang K, Zhang L, Fan A. 2022. Genetic and structural characterisation of 20 autosomal STR loci from the Henan Han population of Central China. Ann Hum Biol. 49(1):80–86. doi: 10.1080/03014460.2022.2030406.
- Zhang X, Li K, Duan Y. 2023. Population data and phylogenetic analysis of 37 Y-STR loci in the Hui population from Yunnan Province, Southwest China. Ann Hum Biol. 50(1):196–199. doi: 10.1080/03014460.2023.2188258.
- Zhao F, Wu XY, Cai GQ, Xu CC. 2003. Application of modified powerstates software in forensic biostatistics. China J Forensic Med. 18(5):297–298.