ABSTRACT
Soil extracellular enzymes play an important role in regulating nitrogen (N) mineralization processes in paddy fields. Therefore, in this work, we determine the influence of a long-term fertilizer regime on rhizosphere soil N fertilization rates, soil enzymes, soil microbial community (bacterial ureolytic community (ureC), bacterial chitinolytic community (chiA)) under the double-cropping rice field in southern of China. The field experiment included the following fertilizer regimes: inorganic fertilizer alone (IF), rice straw and inorganic fertilizer (RF), 30% organic manure and 70% inorganic fertilizer (OM), without any fertilizer input as a control (CK). The results showed that rhizosphere soil N transformation rates in the paddy fields with OM and RF treatments were increased, compared with IF and CK treatments. Soil urease, β-glucosaminidase and arginase activities were significantly increased under OM and RF treatments. This result indicated that functional gene abundances of sub, npr and chiA were significantly greater under RF and OM treatments compared to IF and CK treatments. Soil bacterial ureolytic and chitinolytic communities were significantly changed by the different fertilization strategies. The practice of applying rice straw and organic manure seems beneficial for increasing rhizosphere soil N mineralization rates and microbial community diversity in the double-cropping rice fields.
Introduction
It is generally accepted that soil nitrogen (N) is the main component in agricultural soil and provides the main nutrient for crop growth. In recent years, excessive use of inorganic N fertilizers has led to problems in agricultural production, such as soil degradation, irrigation water pollution, increase in nitrate leaching and greenhouse gas emission (Spiertz Citation2010). Soil health is defined as the continued capacity of soil to function as a vital living ecosystem that sustains plants, animals and humans, which also significantly affected by the different fertilizer managements. Therefore, the practice of improving N fertilizer use efficiency and controlling amount of N fertilizer seems beneficial for increasing ecological environment and soil health in the paddy fields (Zhang et al. Citation2023; Zhu et al. Citation2023). There is a close relationship of soil N mineralization and microbial community composition with fertilizer managements. Some results showed that the practice of applying organic manure and crop residue beneficial for increasing soil quality and fertility in the paddy fields (Huang et al. Citation2009).
Soil N mineralization plays an important role in regulating N turnover processes in paddy fields, which in turn is influenced by different soil management techniques, including tillage, cropping system and fertilizer regime. Previous results showed that soil anaerobic and aerobic N mineralization rates in paddy fields were increased following organic matter additions (Carpenter-Boggs et al. Citation2000; Cheng et al. Citation2016; Kader et al. Citation2017). Findings by Li et al. (Citation2016) confirmed that the rate of soil N mineralization was closely related to enzymes activities in estuarine and tidal wetlands. These results proved that soil N mineralization were influenced by soil physical properties, soil pH, moisture and soil microbial food web (Whalen et al. Citation2013; Jalali et al. Citation2014).
Soil enzyme activities play an important role in improving of soil quality and soil fertility, and are generally considered an early evaluation indicator in varied soil environment, soil fertility and soil health, which was sensitive to different fertilizer managements (Gou et al. Citation2023; Yang et al. Citation2023). Meanwhile, soil enzyme activities are significantly influenced by soil physical properties, and soil N mineralization is mainly regulated by soil extracellular enzymes activities (Tabatabai et al. Citation2010). It is generally accepted that the breakdown of complex organic N compounds into easily hydrolysable N fractions is the first step of soil N mineralization, and soil arylamidase and β-glucosaminidase activities play a vital regulating function in this process (Acosta-Martínez and Tabatabai Citation2000). Soil β-glucosidase activity is usually regarded as an indicator for nutrient or energy flow, which is closely linked with carbon (C) turnover processes, such as cellobiose hydrolase. Soil L-glutaminase activity is closely linked with ammonification (Muruganandam et al. Citation2009), and soil urease plays an important role in catalyzing hydroxyurea, urea, dihydroxyurea and semicarbazide into hydrogen nitride (NH3) and carbon dioxide (CO2) (Tabatabai and Bremner Citation1972). It is generally accepted that there is a close link between soil urease, L-asparaginase, amidohydrolase activities and soil mineralizable N (Khorsandi and Nourbakhsh Citation2008). However, there is still a lack of knowledge regarding the specific enzymes directly involved in soil N mineralization processes, the link between the composition of the soil microbial community and the production of key enzymes.
Rice is the main grain crop in Asia, with early rice and late rice (double-cropping rice system), the main cropping system in South China (Zhang et al. Citation2023). The practice of applying organic manure is beneficial for maintaining or improving soil quality, soil fertility and soil health in paddy fields. Soil chemical and physical properties, including soil bulk density, soil pH, soil organic carbon (SOC) and N content, in the 0–20 cm layer in paddy fields are known to vary with different fertilization treatments (Tang et al. Citation2020), thus the absorption of soil N and soil fertility is also significantly affected. However, there is still a lack of knowledge around the effects of long-term fertilizer treatments on rhizosphere soil microbial community composition (bacterial ureolytic (ureC) and chitinolytic (chiA)), the abundance of functional genes involved in N mineralization, N transformation rate and soil enzyme activity in the double-cropping rice fields. A long-term (37-years) field fertilization experiment was set up in a paddy field under double-cropping rice system. Therefore, the purpose of this study was (1) to explore the characteristics of rhizosphere soil N transformation processes and enzyme activities in paddy fields under different fertilizer conditions; (2) to investigate the response of rhizosphere soil microbial community structure and function to different fertilizer managements in a double-cropping paddy field.
Materials and methods
Site and cropping system
This fertilizer field experiment was located in Ningxiang City (28°07′ N, 112°18′ E) of Hunan Province, China. The annual mean precipitation, evapotranspiration and monthly mean temperature of field experiment region are 1553 mm, 1354 mm and 17.2°C, respectively. At beginning of this experiment, soil characteristics at plough layer (0–20 cm) were shown as follows: soil organic carbon (SOC) 29.4 g kg−1, total nitrogen (N) 2.0 g kg−1, available N 144.1 mg kg−1, total phosphorous (P) 0.59 g kg−1, available P 12.87 mg kg−1, total potassium (K) 20.6 g kg−1 and available K 33.0 mg kg−1. The cropping system of this experiment was barley (Hordeum vulgare L.), early rice and late rice (Oryza sativa L.) in a year.
Experimental design
This field trial, started in 1986, included four fertilizer treatments: inorganic fertilizer alone (IF), rice straw and inorganic fertilizer (RF), 30% organic manure and 70% inorganic fertilizer (OM), without any fertilizer input as a control (CK). A randomized block design was applied with each plot, with three replications of each fertilizer treatment. The area of each plot was 66.7 m2 (10 m × 6.67 m). The field experiment ensured that the total amount of N, phosphorus pentoxide (P2O5), potassium oxide (K2O) in the IF, RF and OM treatments remained the same throughout the growth period of early rice and late rice, respectively. The chemical fertilizers included urea, ordinary superphosphate and potassium chloride. The kind of organic manure was decomposed chicken manure. The N, P and K contents of early rice and late rice straw were 6.5 g kg−1, 1.3 g kg−1, 8.9 g kg−1 and 6.8 g kg−1, 1.5 g kg−1, 9.1 g kg−1, respectively. And N, P and K contents of decomposed chicken manure were 17.7 g kg−1, 8.0 g kg−1 and 11.2 g kg−1, respectively. For early and late rice, 70% and 60% of mineral N fertilizer were applied at seedling, and the remaining N fertilizer was applied at 7–10 days after transplanting during crop growth, respectively. All the P2O5 and K2O fertilizers were applied at soil tillage before rice transplanting. More detail information about fertilizer regimes and other field managements were described by Tang et al. (Citation2018).
Soil sampling
Soil samples were collected at maturity stage of late rice in October 2022. Rhizosphere soil was carefully collected adhering to the total root of rice after gentle shaking. In order to obtain enough rhizosphere soil for multiplicating, 20 plants were randomly selected from each plot and then pooled to form one composite sample. Thus, three composite soil samples of each fertilizer treatment were collected at sampling time, and the total number of 12 composite soil samples were collected at maturity stage of late rice. After removing the visible organic material, small stones and rice roots by hand, then the soil samples were divided into two parts. One part of the fresh soil samples was passed through a 2-mm mesh sieve and was kept at 4°C for measurement of soil N transformation rate, and the other part soil sample was stored at − 20°C for deoxyribonucleic acid (DNA) extraction and soil enzyme activity analysis.
Soil laboratory analysis
Soil N transformation rate
Gross N transformation rates of soil samples were investigated using a15N pool dilution. Three well mixed 40 g dry weight equivalent subsamples were weighed into plastic specimen cups. Then, 1.6 ml of 15NH4+ solution were added to the soils and carefully mixed, creating final soil water content of 0.18 kg kg−1. The quantity of15N added approximately double the soil ammonium nitrogen (NH4+) or nitrate nitrogen (NO3−) pool. Immediately following soil mixing, one subsample was harvested and extracted with 2 M KCl to determine NH4+ or NO3− content and15N enrichment at time 0. The other subsamples were placed in 1-l Mason jars with lids containing butyl rubber septa and with 1 ml of water at the bottom of the jar to minimize loss of moisture from the soil. Jars were incubated for 48 h at 25°C before the soil was extracted with 2 M KCl. Soil NH4+ or nitrite (NO2−) plus NO3− contents were measured with a flow injection analyzer. The extracts were prepared for15N analyses using a diffusion procedure described by Stark and Hart (Citation1996), and the15N enrichment was determined by continuous-flow direct dry combustion and mass spectrometry with an ANCA 2020 system (Europa Scientific, Cincinnati, OH). Net mineralization and nitrification of soil samples were measured by 21 d incubation. Headspace carbon dioxide (CO2) was measured at 3 d, 7 d, 14 d and 21 d by a gas chromatograph with a thermal conductivity detector to determine soil respiration rate.
Soil enzyme activity
Soil extracellular enzyme activities were quantified after a one week pre-incubation period following the method described by Zhang et al. (Citation2016), including soil protease (EC 3.4.21), arginase (EC 3.5.3.1), urease (EC 3.5.1.5) and β-glucosaminidase (EC 3.21.30). Briefly, soil samples were incubated at 37°C with 0.6% casein for protease assay. Soil protease activity were calculated from the difference between amino acid concentrations over 2 h. Soil slurries were incubated with final concentration of 1.0 mM L-arginine at 37°C for 1 h to investigate soil arginase activity. Fresh soils were incubated at 37°C with 0.2 M urea solution for 2 h for determined soil urease activity. Fresh soils were mixed with sodium acetate buffer (pH 5.5) and p-nitrophenyl-N-acetyl-β-D-glucosaminide solution in 50ml centrifuge tubes and kept at 37°C for 1 h for investigate soil β-glucosaminidase activity. 3,5-dinitro salicylic acid as substrate was added and then soil enzyme reaction was stopped as the control after culture experiment.
Soil DNA extract and real-time quantitative polymerase chain reaction (PCR)
Soil DNA was extracted using a MoBio PowerSoil DNA isolation kit (MoBio Laboratories Inc, Carlsbad, CA). DNA extracts were quantified using a Quant-iT PicoGreen dsDNA BR assay kit (Molecular Probes, Inc., Eugene, OR) according to the manufacturer’s protocol. PCR of gene encoding enzyme involved in soil N mineralization were conducted using SsoAdvanced SYBR green Supermix and CFX Connect real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA). Meanwhile, the abundances of gene encoding subtilisin (sub), neutral metalloprotease (npr), chitinase (chiA) and urease (ureC) of soil samples were measured. Briefly, primer pair of npr was amplified using Fp nprl and Rp nprll, respectively. Primer pair of sub was amplified using Fp subla and Rp subll, primer pair of chiA was amplified using GA1F and GA1R, primer pair of ureC was amplified using ureC1F and ureC2R, respectively. Real-time amplification of npr and sub PCR conditions was conducted with an initial denaturation at 95°C for 20 s, annealing at 30°C, elongation at 72°C for 30 s. Real-time amplification of chiA PCR conditions were conducted with an initial denaturation at 95°C for 60 s, annealing at 60°C, elongation at 72°C for 60 s. Real-time amplification of ureC PCR conditions were conducted with an initial denaturation at 95°C for 60 s, annealing at 60°C and elongation at 72°C for 120 s. More detail information about efficiency and calibration standard of bacterial isolate (ureC, npr, sub and chiA) were described by Ouyang et al. (Citation2020).
Soil metagenome processing and gene-targeted assembly
Soil DNA was sequenced on an Illumina HiSeq 2500 platform with a 2- by 150-bp paired-end format. Quality-filtered metagenomes were downloaded and used for gene-targeted assembly. Four genes involved in N mineralization (ureC, npr, sub and chiA) were included for the assembly. For each gene of interest, seed sequences, hidden Markov models (HMMs), and nucleotide and protein reference sequences were downloaded from FunGene (Fish et al. Citation2013). Default assembly parameters were used, and sequences were clustered at 95% amino acid similarity. A representative sequence from each cluster was searched against the reference gene database and the nonredundant database (nr) from National Center for Biotechnology Information (NCBI) using BLAST. Overall, the top hit of these representative sequences to the reference gene database had a similarity higher than 49% and an E value higher than 1.5 E_46.
Illumina sequencing and data analysis for ureC and chiA
The same ureC and chiA primers described above were used for high-throughput sequencing. Linkers were added to the primers for the ureC and chiA genes, while tags were added to separate different soil samples (Herbold et al. Citation2015). The same amount of soil DNA were used for ureC and chiA amplifications, and then the PCR products were further purified using size selection (Agencourt Ampure XP PCR purification). Pooled purified products were sequenced on an Illumina MiSeq platform (Illumina, Inc., San Diego, CA) using V3 chemistry (2- by 300-bp paired-end reads).
High-quality ureC and chiA sequences were extracted from merged reads in each soil sample using the Ribosomal Database Project (RDP) SeqFilters with a read Q score cutoff of 25. Chimera sequences were removed using UCHIME with the ureC and chiA nucleotide reference databases downloaded from the FunGene (Fish et al. Citation2013). The remaining quality-screened protein sequences in each sample were aligned based on ureC and chiA hidden Markov models using HMMER3. Operational taxonomic units (OTUs) were clustered at 95% amino acid similarity using the RDP clustering tool. A maximum-likelihood phylogenetic tree was constructed from representative sequences using FastTree with default parameters (Price et al. Citation2010). OTUs table and taxonomy files were further organized for diversity analysis using the R package phyloseq (McMurdie et al. Citation2013).
Illumina sequencing of 16S rRNA
The variable V3-V4 region of the 16S gene was amplified with 515F and 816 R universal primers for soil bacterial community. The 16S amplicon sequencing was conduct on an Illumina MiSeq instrument (Illumina Inc., San Diego, CA). The Illumina raw reads were processed using a custom pipeline. Briefly, raw reads were first quality filtered and then the high-quality sequences were clustered into OTUs based on 97% identity for a prokaryotic data set using the USEARCH pipeline (Edgar Citation2013). Taxonomies were assigned to each OTU using the RDP Classifier with a confidence threshold of 0.60. All data files were then organized using the R package phyloseq (McMurdie et al. Citation2013). Illumina sequence data for ureC and chiA were deposited in NCBI under BioProject accession number PRJNA597781. Illumina sequence data for 16S rRNA were available under NCBI project ID 1,061,777.
Statistical analysis
The survey indexes of all fertilizer treatments were expressed by mean and standard deviation. These data of each measured items with different fertilizer treatments means were compared using one-way analysis of variance (Anova) following standard procedures at the 5% probability level. Alpha diversity and beta diversity were calculated. Nonmetric multidimensional scaling (NMDS) and PERMANOVA were conducted to visualize and assess the distance matrices using the R package vegan. Two-way Anova were used to analyze the effects of fertilizer treatment on functional gene abundance and alpha diversity of prokaryotic community. All the data of each measured items in this present paper were analyzed with SAS 9.3 software package (SAS Citation2008).
Results
Soil N transformation rate and enzyme activity
The effects of different long-term fertilizer managements on rhizosphere soil nitrogen (N) transformation rate in the double-cropping rice field are shown in . Our results indicated that gross N mineralization rate with 30% organic manure and 70% inorganic fertilizer (OM) treatment were significantly higher (p < 0.05) than that of inorganic fertilizer alone (IF), rice straw and inorganic fertilizer (RF), and without any fertilizer input (CK) treatments. Compared with CK treatment, soil gross N mineralization rate with OM treatment increased by 37.78%. Meanwhile, soil gross ammonium consumption, gross nitrification, gross nitrate consumption, net mineralization and net nitrification rates with OM and RF treatments were significantly higher (p < 0.05) than that of IF and CK treatments, and these rates were significantly different (p < 0.05) between OM and RF treatments. Soil respiration rates with OM and RF treatments were significantly higher (p < 0.05) than that of IF and CK treatments, but did not have any difference (p < 0.05) between OM and RF.
Table 1. Effects of different long-term fertilizer treatments on rhizosphere soil N transformation rates in the double-cropping rice fields.
The effects of different long-term fertilizer managements on rhizosphere soil enzymes activities (soil urease, protease, β-glucosaminidase and arginase) in the double-cropping rice field are shown in . Our results indicated that soil urease activity with RF and OM treatments were significantly higher (p < 0.05) than that of IF and CK treatments, but there was no difference (p < 0.05) between RF and OM treatments in soil urease activity. Compared with IF treatment, soil urease activity with RF and OM treatments increased by 48.40% and 37.78% (). Soil protease, β-glucosaminidase and arginase activities with OM treatment were significantly higher (p < 0.05) than that of RF, IF and CK treatments ().
Figure 1. Effects of different long-term fertilizer treatments on rhizosphere soil enzyme activities in a double-cropping rice field (a) was soil urease; (b) was soil protease; (c) was soil β-glucosaminidase; (d) was soil arginase.
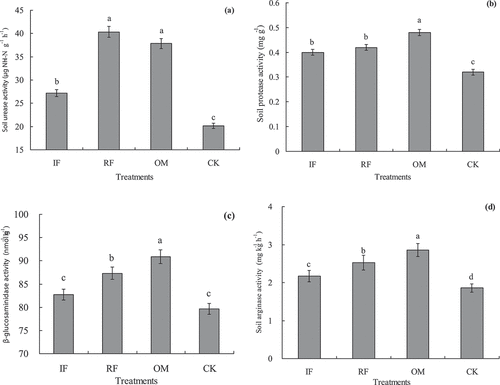
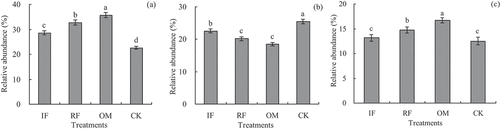
Figure 2. Effects of different long-term fertilizer treatments on rhizosphere soil relative abundances of Proteobacteria, Acidobacteria and Actinobacteria in the double-cropping rice fields (a) was Proteobacteria; (b) was Acidobacteria; (c) was Actinobacteria.
Abundance of gene in soil N mineralization
Our results indicate that across all treatments the abundance of the subtilisin (sub) gene ranged from 2.35 to 3.96 × 107 copies g−1 soil with abundances for RF and OM treatments significantly greater (p < 0.05) than those for IF and CK treatments. Abundances of neutral metalloprotease (npr) and chitinase (chiA) with all fertilizer treatments ranged from 2.24 to 2.95 × 105, and 2.21 to 3.01 × 108 copies g−1 soil, respectively. Meanwhile, abundances of npr and chiA with RF and OM treatments were significantly higher (p < 0.05) than that of IF and CK treatments. Our results showed that thebrange of abundances of urease (ureC) with all fertilizer treatments was from 7.69 to 10.74 × 107 copies g−1 soil, abundances of ureC with OM treatment were significantly higher (p < 0.05) than that of RF, IF and CK treatments. The mean values of the abundances of sub, npr, chiA and ureC with all fertilizer treatments were 3.23 × 107, 2.61 × 105, 2.66 × 108 and 9.22 × 107 copies g−1 soil, respectively ().
Table 2. Effects of different long-term fertilizer treatments on abundance of sub, npr, chiA and ureC in rhizosphere soil under the double-cropping rice system.
Bacterial community composition
Our results showed that Actinobacteria, Acidobacteria, Bacteroidetes, Gemmatimonadetes and Proteobacteria were the five most abundant phyla, which comprised more than 80% of the relative abundance of soil bacterial community. Meanwhile, our results indicated that relative abundances of Proteobacteria, Actinobacteria and Acidobacteria were significantly changed by different fertilizer treatments (). Actinobacteria and Proteobacteria abundances with OM and RF treatments were significantly higher (p < 0.05) than that of IF and CK treatments, but Acidobacteria abundance with RF and OM treatments were significantly lower (p < 0.05) than that of CK and IF treatments. However, Acidobacteria abundance with CK and IF treatments were significantly higher (p < 0.05) than that of RF and OM treatments.
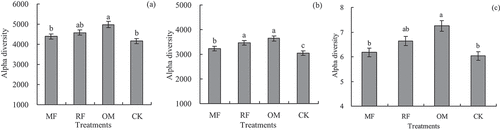
Our results indicated that alpha diversity of rhizosphere soil bacterial community was influenced with different long-term fertilizer treatments (). Compared with CK treatment, Chao 1 and Shannon diversity with OM treatment were significantly increased. But there were no differences (p < 0.05) in Chao 1 and Shannon diversity between IF and CK treatments. Our results showed that observed operational taxonomic units (OTUs) diversity with RF and OM treatment were significantly increased, compared with IF and CK treatments.
Figure 3. Effects of different long-term fertilizer treatments on alpha diversity of rhizosphere soil bacterial community in the double-cropping rice fields (a) was Chao 1; (b) was observed OTUs; (c) was Shannon.
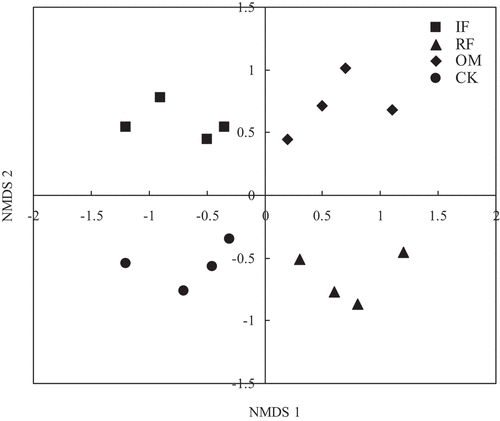
Soil bacterial community structure with all fertilizer treatments through weighted UniFrac distance was revealed. Our results indicated that bacterial community structure was distinct from RF, IF, OM and CK treatments (). Two-way permutational multivariate analysis of variance (PERMANOVA) further confirmed that soil bacterial community structure was significantly affected by fertilizer treatments (p = 0.005).
Figure 4. Nonmetric multidimensional scaling (NMDS) ordination (stress = 0.1) of the weighted UniFrac distance for rhizosphere soil bacterial community with different long-term fertilizer treatments.
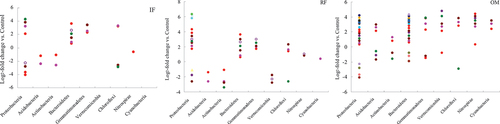
Based on log2-fold change of the relative abundance of OTUs, many OTUs responded significantly to different fertilizer practices (). Most of these responsive OTUs, mainly from Proteobacteria and Bacteroidetes, were increased following the application of rice straw and organic manure. Our results showed that there were 30, 52 and 75 responsive OTUs in rhizosphere soil treated with IF, RF and OM treatments, respectively. Soil treated with RF and OM treatments shared half of their responsive OTUs, while less than 7% responsive OTUs were shared with CK treatment.
Figure 5. Log2-fold change in relative abundance of OTUs compared with those of the control treatment in rhizosphere soil.
Ureolytic community composition
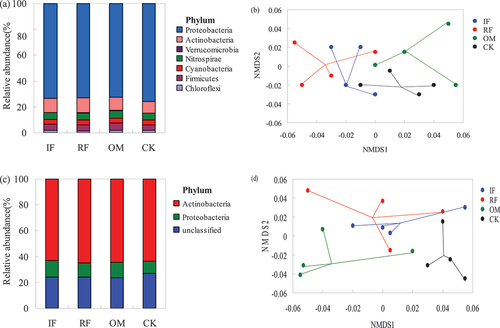
Based on the nearest match to reference taxonomy, we detected 11 distinct prokaryotic phyla although only seven bacterial phyla were among the most prevalent ( < 1%) (). The majority of sequences were assigned to Proteobacteria (75%), Actinobacteria (10%) and Nitrospirae (6%). Thaumarchaeota was also detected, but their relative abundance was very low (0.04%). There were significant difference (p < 0.05) in the relative abundance of phyla among different fertilizer-treated rhizosphere soil. The ureolytic community composition as revealed by weighted UniFrac distance matrices were significantly changed by fertilizer treatments (p = 0.005) (). Pairwise comparison demonstrated that result from CK treatment were significantly different from those with RF (p = 0.025) and OM (p = 0.030) treatments.
Figure 6. Effects of different long-term fertilizer treatments on relative abundance of rhizosphere soil bacterial community in the double-cropping rice fields (a) relative abundances of the dominant phyla (>1%) for bacterial ureC. (b) Nonmetric multidimensional scaling (NMDS) ordination (stress = 0.09) of the weighted UniFrac distance for bacterial ureC under four fertilizer treatments. (c) Relative abundances of the dominant phyla (>1%) for bacterial chiA. (d) Nonmetric multidimensional scaling (NMDS) ordination (stress = 0.05) of the weighted UniFrac distance for bacterial chiA under four fertilizer treatments.
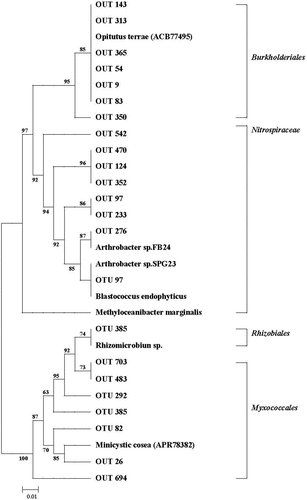
Most of these top ureC OTUs were assigned to Proteobacteria with family from the order Burkholderiales, Rhizobiales and Myxococcales. OTU 385 was closely affiliated with Rhizobiales and was not grouped together with other Proteobacteria taxa. OTU 124 and OTU 413 were affiliated with the family Nitrospiraceae and Myxococcales, respectively. Among the top ureC 50 OTUs, abundances of 15 OTUs were significantly changed by fertilizer treatments ().
Chitinolytic community composition
Based on the nearest match to the reference taxonomy, most of the OTUs were assigned to Actinobacteria and Proteobacteria (). The relative abundance of Actinobacteria was lowest, but abundance of Proteobacteria was highest in IF-treated soil. The chitinolytic community composition as revealed by weighted UniFrac distance matrices was significantly changed by fertilizer treatments (p = 0.01) (). Pairwise comparison demonstrated that result from CK treatment was significantly different from those with RF (p = 0.027) and OM (p = 0.031) treatments.
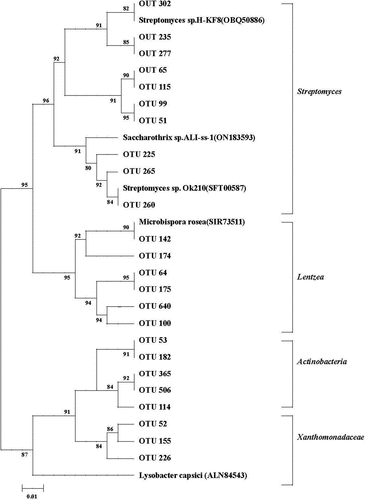
Most of these top 50 chiA OTUs were assigned to Streptomyces (e.g. OTUs 115, 225), Lentzea (e.g. OTUs 64, 175) and Actinoplanes (e.g. OTU 365) in the phylum Actinobacteria. The most abundant OTUs was 134 and 385, which were closest to Xanthomonadaceae. Among the top 50 chiA OTUs, abundances of 15 OTUs were significantly changed by fertilizer treatments ().
Discussions
Effects of fertilizer managements on soil N transformation rate and enzyme activity
There is a close link between soil nitrogen (N) component and organic matter chemical composition, soil microorganism characteristics (e.g. chemical, physical and biological characteristics), which provides main soil nutrient for rice growth (Mohanty et al. Citation2011). We demonstrate that rhizosphere soil N transformation rate (gross N mineralization rate (GMR), gross ammonium consumption rate (GACR), gross nitrification rate (GNR), gross nitrate consumption rate (GNCR), net mineralization rate (NMR)) in the double-cropping rice field with inorganic fertilizer alone (IF), rice straw and inorganic fertilizer (RF), 30% organic manure and 70% inorganic fertilizer (OM) treatments were higher than that without any fertilizer input (CK) treatment, because of Tang et al. (Citation2020) founding that soil N, soil organic matter (SOM) quality and soil chemical characteristics were increased with fertilizer input practices, which suggested that organic matter provides higher level of soil nutrient for paddy field than that of the other fertilizer treatments. On the other hand, suggesting that lack of N in paddy soil were due to poor quality of SOM and soil quality under long term without any fertilizer input condition. Meanwhile, our results showed that application of rice straw practice is beneficial increasing soil N mineralization rate compared to application of inorganic fertilizers, because of Tang et al. (Citation2018) founding that long-term application of inorganic fertilizers was also led to soil N deficiency or lower level of SOM quality and soil chemical properties. Therefore, if rice paddy farming incorporates fertilization regime organic manure, an increase in soil N mineralization rate can be expected, the reason may be attributed to that organic fertilizer contains a higher level of soluble N, then soil N pool in paddy fields were larger under long-term organic fertilizer input condition. On the other hand, SOM quality and soil chemical characteristics with long-term combined application of inorganic fertilizers were also increased (Tang et al. Citation2020). These results were similar with the previous studies (Hartz et al. Citation2000), who found that the degree of soil N mineralization with organic fertilizer treatment was higher than that with compost treatment, for that there had similar C:N ratio and N content among these fertilizer treatments. In this study, our results proved that there had significantly positive correlation between soil N mineralization rate and soil enzyme activity (p < 0.05), which were similar with previous results (Kader et al. Citation2013), who found that soil N mineralization rate was closely related with soil enzyme activity (e.g. soil urease, protease, β-glucosaminidase and arginase). Therefore, this suggests that soil enzyme activity differences between different fertilizer treatments in our study might be the main factor regulating soil N mineralization in paddy fields. Meanwhile, productivity of soil system and soil ecosystem in the paddy fields was increased for that there is higher soil bacterial community diversity and soil enzymes activity under long-term organic fertilizer input condition.
In the present study, our results indicated that application of fertilizer practice is beneficial increasing soil enzyme activity in the double-cropping rice field, because of Tang et al. (Citation2020) founding that SOM quality and soil chemical characteristics (e.g. soil N) with fertilizer input practices were increased, which suggested that organic matter provide higher level of soil nutrient for paddy field. Therefore, if rice paddy farming incorporates fertilization regime rice straw and organic manure, an increase in soil enzyme activity can be expected, the reason may be attributed to that stimulation of related activity of the extracellular enzyme-organo complexes (Nannipieri et al. Citation2012). Meanwhile, our results found that the abundance of functional genes involved in N mineralization were affected by fertilizer treatments and that abundance of functional genes involved in N mineralization correlate with their corresponding soil enzyme activity. The reason may be attributed to that functional genes (sub and npr) cover all members of the microbial community responsible for the specific enzyme functions, and then protein hydrolysis gene primers for soil microbial with long-term application of fertilizers were increased (Bach et al. Citation2001). On the other hand, soil extracellular enzymes were regulated by genes encoding the corresponding enzyme; their activities were promoted under good soil physical, chemical and soil health conditions (Nannipieri et al. Citation2012).
Effects of fertilizer managements on soil bacterial community
In this study, our results indicated that the application of organic matter (rice straw and organic manure) significantly changed the structure of rhizosphere soil bacterial community in the double-cropping rice field. Rice straw and organic manure managements strongly increase the richness and diversity of soil bacterial community; these results were consistent with previous studies (Sun et al. Citation2015). The increased diversity of soil bacterial community in paddy fields with RF and OM treatments was likely due to stimulation in the growth of native soil bacteria by high level of soil available nutrient (e.g. available N, available phosphorus (P) and available potassium (K)) and diverse organic carbon fraction (e.g. mineralizable carbon, microbial biomass carbon, dissolved organic carbon, particulate organic carbon, light fraction organic carbon and permanganate oxidizable carbon) (Tang et al. Citation2020). In addition, our results found that more than half of operational taxonomic units (OTUs) from the inorganic fertilizer were recovered in rice straw and organic manure-treated soils. Furthermore, Sun et al. (Citation2015) reported that direct introduction of exogenous species to paddy soil may have contributed to the increase of soil microbial diversity although the microbe originating from rice straw and organic manure might be less competitive with the native soil microbial community under long-term experiment conditions. Meanwhile, our results showed that diversity of soil bacterial community in paddy fields with IF treatments was decreased, compared with RF and OM treatments, which were consistent with Chaudhry et al. (2015) and Ding et al. (Citation2016) results. The reason may be attributed to long term applied with crop residue and organic manure fertilizer may homogenize soil microbial community in paddy fields, soil microorganisms that were beneficial for resisting change in mineral N fertilization (Chaudhry et al. 2015).
In the present study, our results showed that soil ureolytic community composition in paddy fields with IF treatment was significantly promoted. Meanwhile, soil ureolytic microbial community in paddy fields with RF and OM treatments was also increased. In our previous study, the result indicated that there had difference in the levels of soil organic C (SOC) between IF and RF, OM treatments (Tang et al. Citation2018). In this study, our results suggested that microorganisms inhabiting organic manure play a vital role in shaping soil bacterial ureolytic community composition under applied with rice straw and organic manure conditions. Meanwhile, our results showed that abundances of 5 of the affected 50 OTUs with RF and OM treatments were increased, but abundances of most of the affected OTUs with IF treatment were decreased. The reason may be attributed to that some ureolytic microorganisms were repressed under long-term application of chemical fertilizer conditions.
In this study, our results indicated that ureolytic communities were mainly affiliated with Proteobacteria, which were consistent with the previous result (Collier et al. Citation2009), who found that soil bacterial urease were most commonly found in Proteobacteria. Meanwhile, our results showed that many ureC OTUs had no identified matches to current references, which were suggested that primer-based amplicon sequencing provide some information on previously uncultured ureolytic organism. But soil ureC sequences were closely related to Thaumarchaeota based on amplicon sequencing, although their relative abundances were very low. Our previous result showed that soil ammonia-oxidizing archaea (AOA) were abundant in paddy fields (Tang et al. Citation2019), which often contain ureC. Meanwhile, Nitrospira were important potential urease producers for that several top ureC OTUs were from Nitrospira (Daims et al. Citation2015).
In the present study, our results indicated that the application of fertilizer practice is beneficial increasing soil bacterial chitinolytic community in the double-cropping rice field, because of Kielak et al. (Citation2013) founding that soil bacterial chitinolytic community were significantly changed by fertilizer managements. Therefore, if rice paddy farming incorporates fertilization regime rice straw and organic manure, an increase in soil chitinolytic communities can be expected, the reason may be attributed to that organic matter contain multiple organic N polymers. However, we demonstrate that abundance of several top chiA OTUs were significantly increased under applied with chemical fertilizer, rice straw and organic manure conditions (). The reason was mainly attributed to that high variability of chitinolytic community composition in RF and OM treatments. Meanwhile, our results showed that the percentage of chiA OTUs to total OTUs was 2.7% in IF treatment soil, indicating that chitinolytic microorganism in inorganic fertilizer was less competitive than the indigenous chitinolytic community and weakly survive in soil. Several top chiA OTUs were enriched by organic matter-based fertilizers (OTU 134 and OTU 385), which indicated that soil chitinolytic community in paddy fields may be organic matter stimulated. Therefore, it was beneficial for increasing soil microbial diversity and maintaining soil health in paddy fields with the application of rice straw and organic manure input practices.
Conclusions
In the present study, our results indicated that rhizosphere soil N transformation rates, abundances of rhizosphere soil sub, npr, chiA and ureC in the double-cropping rice field with rice straw and organic manure practices were increased, compared with inorganic fertilizer practice. Application of rice straw and organic manure to a double-cropping rice field leads to an increase in rhizosphere soil bacterial community diversity and soil enzymes activity. There is a close link between soil N mineralization and soil quality in a double-cropping rice field. Our results proved that application of rice straw and organic manure input practices beneficial for increasing rhizosphere soil ureolytic and chitinolytic bacterial community in a double-cropping rice field. The abundances of selected functional genes involved in N mineralization were increased under long-term fertilization conditions. There was close relationship between abundances of targeted N functional genes and corresponding soil enzyme activity. It was beneficial for maintaining and improving soil ecosystem and soil fertility in paddy fields with rice straw and organic manure input practices. However, there is still a lack of investigation relationship between soil microbial functional genes and their associated enzyme activity in a double-cropping rice field.
Acknowledgments
This study was supported by the Hunan Provincial Natural Science Foundation of China (2022JJ30352), the National Natural Science Foundation of China (U21A20187), the National Key Research and Development Project of China (2023YFD2301403), and the Hunan science and technology talent lift project (2022TJ-N07).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Acosta-Martínez V, Tabatabai MA. 2000. Arylamidase activity of soils. Soil Sci Soc Am J. 64(1):215–221. doi: 10.2136/sssaj2000.641215x.
- Bach HJ, Hartmann A, Schloter M, Munch JC. 2001. PCR primers and functional probes for amplification and detection of bacterial genes for extracellular peptidases in single strains and in soil. J Microbiol Methods. 44(2):173–182. doi: 10.1016/S0167-7012(00)00239-6.
- Carpenter-Boggs L, Pikul JL, Vigil MF, Riedell WE. 2000. Soil nitrogen mineralization influenced by crop rotation and nitrogen fertilization. Soil Sci Soc Am J. 64(6):2038–2045. doi: 10.2136/sssaj2000.6462038x.
- Cheng WG, Padre AT, Sato C, Shiono H, Hattori S, Kajihara A, Aoyama M, Tawaraya K, Kumagai K. 2016. Changes in the soil C and N contents, C decomposition and N mineralization potentials in a rice paddy after long-term application of inorganic fertilizers and organic matter. Soil Sci Plant Nutr. 62(2):212–219. doi: 10.1080/00380768.2016.1155169.
- Collier JL, Baker KM, Bell SL. 2009. Diversity of urea-degrading microorganisms in open-ocean and estuarine planktonic communities. Environ Microbiol. 11(12):3118–3131. doi: 10.1111/j.1462-2920.2009.02016.x.
- Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, et al. 2015. Complete nitrification by Nitrospira bacteria. Nature. 528(7583):504–509. doi:10.1038/nature16461.
- Ding J, Jiang X, Ma M, Zhou B, Guan D, Zhao B, Zhou J, Cao F, Li L, Li J. 2016. Effect of 35 years inorganic fertilizer and manure amendment on structure of bacterial and archaeal communities in black soil of northeast China. Appl Soil Ecol. 105:187–195. doi: 10.1016/j.apsoil.2016.04.010.
- Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 10(10):996–998. doi: 10.1038/nmeth.2604.
- Fish JA, Chai B, Wang Q, Sun Y, Brown CT, Tiedje JM, Cole JR. 2013. FunGene: the functional gene pipeline and repository. Front Microbiol. 4:291. doi: 10.3389/fmicb.2013.00291.
- Gou Z, Zheng H, He Z, Su Y, Chen S, Chen H, Chen G, Sun Y, Ma NL. 2023. The combined action of biochar and nitrogen-fixing bacteria on microbial and enzymatic activities of soil N cycling. Environ Pollut. 317:120790. doi: 10.1016/j.envpol.2022.120790.
- Hartz TK, Mitchell JP, Giannini C. 2000. Nitrogen and carbon mineralization dynamics of manures and composts. Hortic Sci. 35(2):209–212. doi: 10.21273/HORTSCI.35.2.209.
- Herbold CW, Pelikan C, Kuzyk O, Hausmann B, Angel R, Berry D, Loy A. 2015. A flexible and economical barcoding approach for highly multiplexed amplicon sequencing of diverse target genes. Front Microbiol. 6:731. doi: 10.3389/fmicb.2015.00731.
- Huang QR, Hu F, Huang S, Li HX, Yuan YH, Pan GX, Zhang WJ. 2009. Effect of long-term fertilization on organic carbon and nitrogen in a subtropical paddy soil. Pedosphere. 19(6):727–734. doi: 10.1016/S1002-0160(09)60168-5.
- Jalali M, Mahdavi S, Ranjbar F. 2014. Nitrogen, phosphorus and sulfur mineralization as affected by soil depth in rangeland ecosystems. Environ Earth Sci. 72(6):1775–1788. doi: 10.1007/s12665-014-3082-3.
- Kader MA, Sleutel S, Begum SA, Moslehuddin AZM, De Neve S. 2013. Nitrogen mineralization in sub-tropical paddy soils in relation to soil mineralogy, management, pH, carbon, nitrogen and iron contents. Eur J Soil Sci. 64(1):47–57. doi: 10.1111/ejss.12005.
- Kader MA, Yeasmin S, Solaiman ZM, De NS, Sleutel S. 2017. Response of hydrolytic enzyme activities and nitrogen mineralization to fertilizer and organic matter application in subtropical paddy soils. Eur J Soil Biol. 80:27–34. doi: 10.1016/j.ejsobi.2017.03.004.
- Khorsandi N, Nourbakhsh F. 2008. Prediction of potentially mineralizable N from amidohydrolase activities in a manure-applied, corn residue-amended soil. Eur J Soil Biol. 44(3):341–346. doi: 10.1016/j.ejsobi.2008.03.001.
- Kielak AM, Cretoiu MS, Semenov AV, Sørensen S, van Elsas JD. 2013. Bacterial chitinolytic communities respond to chitin and pH alteration in soil. Appl Environ Microbiol. 79(1):263–272. doi: 10.1128/AEM.02546-12.
- Li XF, Hou LJ, Liu M, Lin XB, Li Y, Li SW. 2016. Primary effects of extracellular enzyme activity and microbial community on carbon and nitrogen mineralization in estuarine and tidal wetlands. Appl Microbiol Biotechnol. 99(6):2895–2909. doi: 10.1007/s00253-014-6187-4.
- McMurdie PJ, Holmes S, Watson M. 2013. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS One. 8(4):e61217. doi: 10.1371/journal.pone.0061217.
- Mohanty M, Reddy SK, Probert ME, Dalal RC, Rao SA, Menzies NW. 2011. Modelling N mineralization from green manure and farmyard manure from a laboratory incubation study. Ecol Model. 222(3):719–726. doi: 10.1016/j.ecolmodel.2010.10.027.
- Muruganandam S, Israel DW, Robarge WP. 2009. Activities of nitrogen mineralization enzymes associated with soil aggregate size fractions of three tillage systems. Soil Sci Soc Am J. 73(3):751–759. doi: 10.2136/sssaj2008.0231.
- Nannipieri P, Giagnoni L, Renella G, Puglisi E, Ceccanti B, Masciandaro G, Fornasier F, Moscatelli MC, Marinari S. 2012. Soil enzymology: classical and molecular approaches. Biol Fertil Soils. 48(7):743–762. doi: 10.1007/s00374-012-0723-0.
- Ouyang Y, Norton JM, Parales RE. 2020. Short-term nitrogen fertilization affects microbial community composition and nitrogen mineralization functions in an agricultural soil. Appl Environ Microbiol. 86(5):e02278–19. doi: 10.1128/AEM.02278-19.
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2 – approximately maximum-likelihood trees for large alignments. PloS One. 5(3):e9490. doi: 10.1371/journal.pone.0009490.
- SAS. 2008. SAS software of the SAS system for windows. Cary, NC, USA: SAS Institute Inc.
- Spiertz JHJ. 2010. Nitrogen, sustainable agriculture and food security. A review. Agron Sustain Dev. 30(1):43–55. doi: 10.1051/agro:2008064.
- Stark JM, Hart SC. 1996. Diffusion technique for preparing salt solutions, Kjeldahl digests, and persulfate digests for nitrogen-15 analysis. Soil Sci Soc Am J. 60(6):1846–1855. doi: 10.2136/sssaj1996.03615995006000060033x.
- Sun R, Zhang XX, Guo X, Wang D, Chu H. 2015. Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol Biochem. 88:9–18. doi: 10.1016/j.soilbio.2015.05.007.
- Tabatabai MA, Bremner JM. 1972. Assay of urease activity in soils. Soil Biol Biochem. 4(4):479–487. doi: 10.1016/0038-0717(72)90064-8.
- Tabatabai MA, Ekenler M, Senwo ZN. 2010. Significance of enzyme activities in soil nitrogen mineralization, Commun. Soil Sci Plant Anal. 41(5):595–605. doi: 10.1080/00103620903531177.
- Tang HM, Li C, Xiao XP, Pan XC, Cheng KK, Shi LH, Li WY, Wen L, Wang K. 2020. Effects of long-term fertiliser regime on soil organic carbon and its labile fractions under double cropping rice system of southern China. Acta Agric Scand Sect B—Soil Plant Sci. 70(5):409–418. doi: 10.1080/09064710.2020.1758763.
- Tang HM, Xiao XP, Li C, Cheng KK, Pan XC, Li WY. 2019. Effects of rhizosphere and long-term fertilization practices on the activity and community structure of ammonia oxidisers under double-cropping rice field. Acta Agric Scand Sect B—Soil Plant Sci. 69(4):356–368. doi: 10.1080/09064710.2019.1576763.
- Tang HM, Xiao XP, Tang WG, Li C, Wang K, Li WY, Cheng KK, Pan XC. 2018. Long-term effects of NPK fertilizers and organic manures on soil organic carbon and carbon management index under a double-cropping rice system in Southern China. Commun Soil Sci Plan. 49(16):1976–1989. doi: 10.1080/00103624.2018.1492600.
- Whalen JK, Kernecker ML, Thomas BW, Ngosong C, Sachdeva V. 2013. Soil food web controls on nitrogen mineralization are influenced by agricultural practices in humid temperate climates. CAB Rev. 8:1–18. doi: 10.1079/PAVSNNR20138023.
- Yang S, Xiao J, Liang T, Tan H. 2023. Response of bacterial compositions to the use of slow-release fertilizers with long-acting agents and synergists. Appl Soil Ecol. 182:104699. doi: 10.1016/j.apsoil.2022.104699.
- Zhang Q, Liang G, Zhou W, Sun J, Wang X, He P. 2016. Fatty-acid profiles and enzyme activities in soil particle-size fractions under long-term fertilization. Soil Sci Soc Am J. 80(1):97–111. doi: 10.2136/sssaj2015.07.0255.
- Zhang J, Nie J, Cao W, Gao Y, Lu Y, Liao Y. 2023. Long-term green manuring to substitute partial chemical fertilizer simultaneously improving crop productivity and soil quality in a double-rice cropping system. Eur J Agron. 142:126641. doi: 10.1016/j.eja.2022.126641.
- Zhu X, Ros GH, Xu M, Cai Z, Sun N, Duan Y, De VW. 2023. Long-term impacts of mineral and organic fertilizer inputs on nitrogen use efficiency for different cropping systems and site conditions in Southern China. Eur J Agron. 146:126797. doi: 10.1016/j.eja.2023.126797.