Abstract
Over the past several years, strawberry production in Canada and the United States has been severely affected by strawberry acute decline symptoms. The decline disease is apparently caused by synergistic infections of several viruses, including Strawberry mild yellow edge virus (SMYEV) and Strawberry mottle virus (SMoV). To better understand the causal viral pathogens, we cloned and sequenced the entire genome of a SMYEV isolate, AB5-2, from a strawberry field in Prince Edward Island. The viral genome of AB5-2 consists of 5969 nucleotides excluding the 3ʹ terminal poly(A) tail. Searches of the NCBI database identified two SMYEV isolates (MY-18 from the USA and D74 from Germany) whose complete genome sequences were documented and 28 SMYEV isolates with determined coat protein (CP) sequences. At the genome level, the Canadian SMYEV isolate shares 86% and 90% nucleotide sequence identities to MY-18 and D74, respectively. Phylogenetic analysis of all CP sequences available clustered 28 SMYEV isolates into five phylogroups, including AB5-2 alone as a new group, suggesting this Canadian isolate is distinct from SMYEV isolates of other origins. This report represents the first molecular characterization of a SMYEV isolate from Canada.
Résumé
Au cours des dernières années, la production de fraises au Canada et aux États-Unis a été gravement touchée par des symptômes de dépérissement aigu. La maladie du dépérissement est apparemment causée par l’infection synergique de plusieurs virus, y compris du virus du jaunissement du bord des feuilles du fraisier (SMYEV) et du virus de la marbrure du fraisier (SMoV). Afin de mieux comprendre les virus pathogènes, nous avons cloné et séquencé tout le génome d’un isolat de SMYEV, AB5-2, provenant d’une fraisière de l’Île-du-Prince-Édouard. Le génome viral d’AB5-2 comporte 5969 nucléotides, à l’exception de la queue poly(A) en 3ʹ. Des recherches effectuées dans la base de données du National Center for Biotechnology Information (NCBI) ont permis de trouver 2 isolats de SMYEV (MY-18, des États-Unis, et D74, d’Allemagne) dont les génomes entiers étaient documentés, et 28 isolats qui possédaient des séquences déterminées de protéine capsidiaire (CP). À l’échelle du génome, l’isolat canadien de SMYEV partage 86% et 90% de l’identité des séquences des nucléotides avec MY-18 et D74, respectivement. L’analyse phylogénétique de toutes les séquences de CP disponibles a rassemblé les 28 isolats de SMYEV en 5 phylogroupes, y compris AB5-2 comme nouveau groupe unique, ce qui suggère que cet isolat canadien est différent des isolats de SMYEV provenant d’ailleurs. Ce rapport constitue la première caractérisation moléculaire d’un isolat canadien de SMYEV.
Introduction
Strawberry (Fragaria × ananassa, Duch.) is an economically important berry crop in Canada and the United States. However, over the past several years, strawberry production in both countries has been severely affected by acute decline symptoms. A recent study has indicated that the causal viral pathogens in strawberry samples from Nova Scotia, Canada included two major viruses – Strawberry mild yellow edge virus (SMYEV) and Strawberry mottle virus (SMoV) (Martin & Tzanetakis Citation2013). Currently, the distribution and epidemiology of these two viruses in Canada still remains poorly understood. SMYEV is a common and widespread virus infecting strawberry around the world (Hepp & Martin Citation1992; Thompson & Jelkmann Citation2004). The virus was first reported in California nearly a century ago (Horne Citation1922) and subsequently found in Europe in 1933 (Harris, Citation1933). Symptoms of SMYEV-infected strawberry include dwarfing, marginal chlorosis, leaf distortion and small fruit size (Martin & Tzanetakis Citation2006). SMYEV can cause up to 30% yield loss (Tzanetakis & Martin Citation2013). SMYEV is a member of the genus Potexvirus in the family Alphaflexiviridae. This is the only known vector-transmitted potexvirus which is transmitted by a strawberry aphid in the genus Chaetosiphon in a persistent but non-propagative manner (Craig & Stultz Citation1964; Tzanetakis & Martin Citation2013). The viral genome is a single-stranded positive-sense RNA of approximately 6.0 kb in length and has a 3ʹ polyA tail (Jelkmann et al. Citation1992). There are two complete genomic sequences available in the NCBI database with GenBank accession D12517 (for isolate MY18) and AJ577359 (for isolate D74). A full-length infectious cDNA derived from MY18 was produced (Lamprecht & Jelkmann Citation1997). On an indicator strawberry cultivar, this clone induced mild symptoms similar to those induced by the parental virus and the viral particles were filamentous (Lamprecht & Jelkmann Citation1997).
The objective of this study was to clone and sequence the complete genome of an SMYEV isolate from Prince Edward Island, Canada. We further analysed the genomic sequence of the Canadian isolate and found it to be distinct from all known isolates at both nucleotide (nt) and amino acid (aa) sequence levels.
Materials and methods
Plant materials and RNA purification
Five strawberry plants (cultivar ‘Jewell’ or ‘Mira’; first year harvest in 2013) positive for SoMV or SMYEV in an RT-PCR screening were collected from a field in Prince Edward Island, Canada. The plants were maintained in Pacific Agri-Food Research Centre in Summerland, British Columbia, Canada. Two plants, AB5-1 and AB5-2, were used for this study. Total RNA was extracted from leaf tissues using the CTAB method as described previously (Cui et al. Citation2012). Since our initial RT-PCR test only confirmed the presence of SMYEV in AB5-2, subsequent studies were carried out using the total RNA isolated from this plant.
cDNA synthesis, PCR and RACE
cDNA synthesis was performed using Superscript III reverse transcriptase and an oligo(dT)12–18 primer (Invitrogen, Burlington, ON) following the supplier’s instructions. Primers () used for PCR amplification were designed based on the two published SMYEV genome sequences (accession numbers AJ577359 and D12517). PCR reactions were performed using Phusion DNA polymerase (NEB, Pickering, ON). The PCR program consisted of an initial denaturation at 94°C for 3 min, followed by 35 cycles of 30 s at 94°C, 30 s at 56°C and 1–4 min (depending on the putative fragment length) at 72°C. A final extension step of 10 min at 72°C terminated the program. For amplification of the 3ʹand 5ʹ termini, the 5ʹ and 3ʹ RACE system for rapid amplification of cDNA ends (Invitrogen) was utilized.
Table 1. Primers used in this study.
Cloning, sequencing of PCR products and sequence comparison
The PCR products were cloned using the Zero Blunt TOPO PCR Cloning Kit (Invitrogen) and were introduced into Escherichia coli Top 10. Ten randomly selected colonies were subjected to plasmid purification, and positive clones containing correct inserts were confirmed by PCR. The sequence of the cloned viral genomic fragments was determined by Sanger sequencing (Eurofins, Louisville, KY, USA).
Sequencing results were assembled and analysed using the DNAStar suite (Lasergene version 7.0). Sequence was confirmed by similarity search using BLAST computer program in GenBank. Multiple nucleotide alignments were conducted using CLUSTALX 1.8. Phylogenetic trees were generated from nt or aa alignments of the complete CP sequences of SMYEV isolates by using the neighbour-joining method with the Molecular Evolutionary Genetics Analysis (MEGA) software program version 4.1. Robustness of inferred evolutionary relationships was assessed by 1000 bootstrap replicates. The names and accession numbers of the isolates included for this analysis are listed in .
Table 2. The names, accession numbers, countries of origin and hosts of SMYEV isolates with full-length coat protein sequences available in the NCBI database.
Results and discussion
The complete genome sequence of the Canadian SMYEV isolate AB5-2
To confirm the presence of SMYEV in AB5-2, a cDNA fragment, including the entire CP gene and flanking region, was amplified by RT-PCR with primer set SMYEV-F3/ SMYEV-R1 (). The resulting cDNA fragment (~812 bp) was purified and sequenced. Blast search against the NCBI database (http://www.ncbi.nlm.nih.gov/) showed that AB5-2 shares 81 ~ 92% nucleotide sequence identities with the other SMYEV isolates deposited in the NCBI database, including 314CP2cav (accession AJ 577351) as the most similar isolate, and sy03 (GenBank accession EU107085) as the most distant one.
A cDNA fragment containing the near-full-length genome sequence of AB5-2 was produced using a forward primer SMYEV-F1, homologous to the 5ʹ-UTR region of SMYEV, and reverse primer SMYEV-R4, complementary to the N-terminus of the CP region. The 5ʹ or 3ʹ terminal cDNAs were obtained using a 5ʹ or 3ʹ RACE kit, with primer sets SMYEV-5ʹrace-1/SMYEV-5ʹrace-2/SMYEV-5ʹrace-3 and SMYEV-3ʹ race/SMYEV-3ʹ race2 (), which were designed based on the previously documented genomic sequences of MY18 and D74. The resulting PCR products were cloned and sequenced, and the complete cDNA sequences of SMYEV isolate AB5-2 were assembled. The full-length genomic cDNA sequence of AB5-2 was deposited in GenBank database (GenBank accession KR707814).
The full-length genomic cDNA sequence of AB5-2 is 5969 nucleotides (nts) in length excluding the 3ʹ terminal poly(A). Blast searches against the NCBI database identified two complete genomic sequences of isolates D74 and MY18, with 90% and 86% nt sequence identity to AB5-2, respectively. Notably, the first six nts of the 5ʹ-terminal sequence of AB5-2 are ‘GAAACA’, which differ from ‘GAAATC’ of isolate MY18 and ‘GAAAAC’ of isolate D74. The nt sequence ‘GAAAAC’ at the 5ʹ terminus is conserved for the majority of potexviruses (Thompson & Jelkmann Citation2004). Whether the difference of the 5ʹ-terminal sequence has an impact on viral pathogenicity needs to be further investigated.
Analysis of the genomic sequence of isolate AB5-2
The 5ʹ-UTR and 3ʹ-UTR are 77 and 125 nts in length, respectively (). Similar with other potyviruses, the viral genome of AB5-2 contains five open reading frames (ORF), ORF1 through ORF5 (). ORF1, nt ORF1, nt 78 to 4049, codes for a putative viral replicase of 1323 aa with a predicted molecular mass of ~149.7 kDa. The AB5-2 ORF1 has an aa sequence identity of 97% with the corresponding part of D74, and 97% with that of MY-18. BLASTp searches showed that this protein contains a conserved methyltransferase domain (MET, aa 40–337), a viral RNA helicase domain (aa 601–834), and an RNA-dependent RNA Polymerase (RdRp, aa 986–1254) domain, which are characteristic of the replicase of many positive-strand RNA viruses of the Alphavirus-like supergroup (). Three conserved motifs, motif I (H67, H69, K73, E76), motif II (D123, R126), and motif IV (Y214), were identified in the MET domain (Rozanov et al. Citation1992). For RdRp, eight conserved motifs, including motif I (K1031), motif IV (D1110, T1112, D1115), motif V (G1164, T1168, N1172, T1173) and motif VI (GDD1196-1198), were identified (Koonin Citation1991). Conserved motifs present in putative helicase domain include motif I (G608, GKS610-612), motif II (D672), motif III (G699, D700, Q703) and motif V (Q796, G797) (Koonin et al. Citation1993).
Fig. 1 Schematic representation of the genomic organization of Strawberry mild yellow edge virus (SMYEV) isolate AB5-2. ORF1 through ORF5 are defined by the vertical number. ORF1 codes for a putative viral replicase, which contains the conserved domains for methyltransferase (MET), viral RNA helicase, and RNA-dependent RNA polymerase (RdRp). The AB5-2 TGBp1 has a conserved helicase domain (nt 4171–4761).

ORF2, ORF3 and ORF4 encode triple gene block proteins, TGBp1, TGBp2 and TGBp3, respectively, which are involved in cell-to-cell movement of viral genomes of potexviruses (Morozov & Solovyev Citation2003; Zamyatnin et al. Citation2004). Phylogenetically, TGBp1 is a diverged duplicate of the viral replication-related NTPase/helicase (Morozov et al. Citation1989). Indeed, the TGBp1 of AB5-2 is 229 aa in length with a predicted molecular mass of ~25.5 kDa, and contains a conserved helicase domain (aa 25–221). The conserved helicase motifs identified in this domain include: motif I (G32, GKS34-36), motif II (D80), motif III (G97/D98) and motif VI (R212) (Koonin et al. Citation1993). The putative non-AUG start codon, AUU, of this ORF is the same as MY18 but different from that of D74 (AUC) (Thompson & Jelkmann Citation2004); however, both codons code for the same aa, isoleucine. The TGBp1 of AB5-2 has an aa sequence identity of 97% with the counterpart of D74 and MY-18. The TGBp2 of AB5-2 consists of 108 aa with a predicted molecular mass of ~11.6 kDa and shares 98% and 97% aa sequence identities with the counterpart of D74 and MY-18, respectively. Like D74, a conserved motif Px30-31 GDxxHx*PxGGxYxDGxKx*xY, which contains the conserved motif identified in potex-, benyi- and hordeiviruses, was identified in the AB5-2 TGBp2 (Thompson & Jelkmann Citation2004). The AB5-2 TGBp3 has 75 aa with a predicted molecular mass of ~8.1 kDa, sharing aa sequence identities of 96% and 96% aa with the TGBp3 of D74 and MY-18, respectively.
The coat protein (CP) gene (ORF5) encodes a protein of 242 amino acids with a predicted molecular mass of 25.7 kDa. It has an aa sequence identity of 97% and 94% with the corresponding CPs of D74 and MY-18, respectively.
Phylogenetic relationship of the Canadian isolate with isolates from different origins
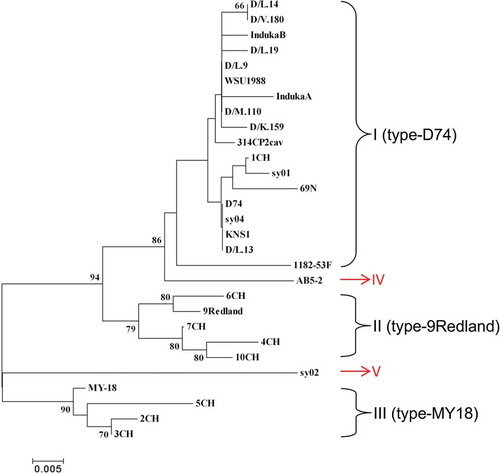
Previous phylogenetic analysis suggested all SMYEV isolates from different origins were clustered into three groups, i.e. I (type-D74), II (type-9Redland) and III (type-MY18) (Thompson & Jelkmann Citation2004). Here, the complete CP sequences of AB5-2 (from this study) and 28 SMYEV isolates were retrieved from the NCBI database and used for the phylogenetic analysis (). A majority (~62%) of SMYEV isolates (18 out of 29) such as D74, 1CH and 69 N, sharing >96% up to 100% sequence identities at both nt and aa sequence levels, were placed into group I (type-D74) (, ). Five isolates, i.e. 4CH, 6CH, 7CH, 10CH and 9Redland, sharing 97 ~ 99% nt aa sequence identities, were classified into group II (type-9Redland). Four isolates, i.e. MY-18, 2CH, 3CH and 5CH, sharing 97.9 ~ 98.2% nt and 97.5 ~ 99.6% aa identity, were clustered into group III (type-MY18). Interestingly, AB5-2 characterized in this study was clustered alone as a new group, designated as IV (type AB5-2). It shares 79 ~ 91% nt and 91 ~ 97% aa sequence identity with the other 28 isolates. It is worth noting that the SMYEV isolate sy02 originating from China, which is 77 ~ 84% and 90 ~ 94% identical to the other 28 isolates at the nt and aa sequence level, respectively, was also grouped into a new cluster. These data suggest that both AB5-2 and sy02, either at nt or aa levels, are quite distinct from each other and from the other 27 isolates as well. These data may help to understand SMYEV evolution and diversity. It would be interesting to determine if the isolates in these five phylogroups differ in their pathogenicity to strawberry.
Fig. 2 (Colour online) A phylogenetic tree based on the coat protein (CP) nucleotide sequences of 28 Strawberry mild yellow edge virus (SMYEV) isolates retrieved from the NCBI database and AB5-2 determined in this study. The tree was reconstructed with MEGA 4.1 using the neighbour-joining method. The numbers at the nodes indicate the percentage of 1000 bootstraps occurring in this group. The scale bar represents the number of substitutions per nucleotide.
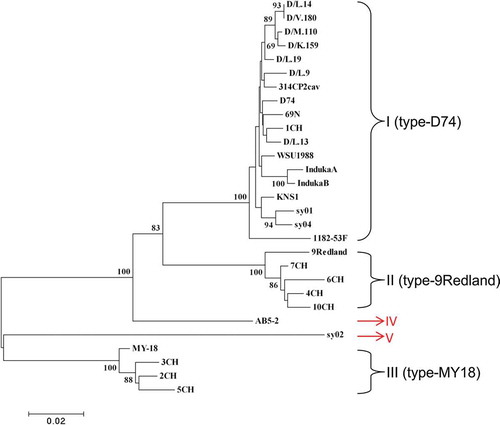
Fig. 3 (Colour online) Phylogenetic tree based on the coat protein (CP) amino acid sequences of 28 Strawberry mild yellow edge virus (SMYEV) isolates retrieved from the NCBI database and the Canadian isolate AB5-2 determined in this study. The tree was reconstructed with MEGA 4.1 using the neighbour-joining method. The numbers at the nodes indicate the percentage of 1000 bootstraps occurring in this group. The scale bar represents the number of substitutions per amino acid.
Acknowledgements
The authors are indebted to Chris Jordan (Department of Agriculture and Forestry, Prince Edward Island) for sample collection, and to Jamie McNeil and Xiaofei Cheng (Agriculture and Agri-Food Canada) for assistance.
Additional information
Funding
References
- Craig DL, Stultz HT. 1964. Aphid dissemination of strawberry viruses in Nova Scotia. Can J Plant Sci. 44:235–239.
- Cui H, Hong N, Wang G, Wang A. 2012. Detection and genetic diversity of Prunus necrotic ringspot virus in the Niagara Fruit Belt, Canada. Can J Plant Pathol. 34:104–113.
- Harris RV. 1933. The strawberry “yellow edge” disease. J Pomol Hortic Sci. 11:56–76.
- Hepp RF, Martin RR. 1992. Occurrence of strawberry mild yellow-edge associated virus in wild Fragaria chiloensis in South America. Acta Hort. 308:57–59.
- Horne WT. 1922. Strawberry troubles. Calif Agric Exp Stn Rep. 1921–22:122–123.
- Jelkmann W, Maiss E, Martin RR. 1992. The nucleotide sequence and genome organization of strawberry mild yellow edge-associated potexvirus. J Gen Virol. 73:475–479.
- Koonin EV. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 72:2197–2206.
- Koonin EV, Dolja VV, Morris TJ. 1993. Evolution and taxonomy of positive-strand RNA viruses: Implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 28:375–430.
- Lamprecht S, Jelkmann W. 1997. Infectious cDNA clone used to identify strawberry mild yellow edge associated potexvirus as causal agent of the disease. J Gen Virol. 78:2347–2353.
- Martin RR, Tzanetakis IE. 2006. Characterization, detection and management of strawberry viruses. Plant Dis. 90:384–396.
- Martin RR, Tzanetakis IE. 2013. High risk strawberry viruses by region in the United States and Canada: Implications for certification, nurseries and fruit production. Plant Dis. 97:1358–1362.
- Morozov SY, Dolja VV, Atabekov JG. 1989. Probable reassortment of genomic elements among elongated RNA-containing plant viruses. J Mol Evol. 29:52–62.
- Morozov SY, Solovyev AG. 2003. Triple gene block: Modular design of a multifunctional machine for plant virus movement. J Gen Virol. 84:1351–1366.
- Rozanov MN, Koonin EV, Gorbalenya AE. 1992. Conservation of the putative methyltransferase domain: A hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol. 73:2129–2134.
- Thompson JR, Jelkmann W. 2004. Strain diversity and conserved genome elements in Strawberry mild yellow edge virus. Arch Virol. 149:1897–1909.
- Tzanetakis IE, Martin RR. 2013. Expanding field of strawberry viruses which are important in North America. Inter J Fruit Sci. 13:184–195.
- Zamyatnin Jr AA, Solovyev AG, Savenkov EI, Germundsson A, Sandgren M, Valkonen JP, Morozov SY. 2004. Transient coexpression of individual genes encoded by the triple gene block of Potato mop-top virus reveals requirements for TGBp1 trafficking. Mol Plant-Microbe Interac. 17:921–930.

