Abstract
Aim
The effect of liposomes bi-functionalized with phosphatidic acid and with a synthetic peptide derived from human apolipoprotein E has been evaluated on the aggregation features of different amyloidogenic proteins: human Amyloid β1–40 (Aβ1–40), transthyretin (TTR) variant S52P, human β2microglobulin (β2m) variants ΔN6 and D76N, Serum Amyloid A (SAA).
Methods
The formation of fibrillar aggregates of the proteins was investigated by ThioflavinT fluorescence assay and validated by Atomic Force Microscopy.
Results
The results show that liposomes are preventing the transition of non-aggregated forms to the fibrillar state, with stronger effects on Aβ1–40, β2m ΔN6 and SAA. Liposomes also induce disaggregation of the amyloid aggregates of all the proteins investigated, with stronger effects on Aβ1–40, β2 D76N and TTR.
SPR assays show that liposomes bind Aβ1–40 and SAA aggregates with high affinity (KD in the nanomolar range) whereas binding to TTR aggregates showed a lower affinity (KD in the micromolar range). Aggregates of β2m variants showed both high and low affinity binding sites. Computed Structural analysis of protein fibrillar aggregates and considerations on the multidentate features of liposomes allow to speculate a common mechanism of action, based on binding the β-stranded peptide regions responsible for the amyloid formation.
Conclusion
Thus, multifunctional liposomes perform as pharmacological chaperones with anti-amyloidogenic activity, with a promising potential for the treatment of a number of protein-misfolding diseases.
Amyloidosis is a group of diseases, each due to a specific protein misfolding.
Anti-amyloidogenic nanoparticles have been gaining the utmost importance as a potential treatment for protein misfolding disorders.
Liposomes bi-functionalized with phosphatidic acid and with a synthetic peptide derived from human apolipoprotein E showed anti-amyloidogenic activity.
Key message
Introduction
There are several different types of Amyloidosis, either genetic or acquired, each due to a specific protein misfolding [Citation1]. These include both localised amyloidosis, which affect only one body organ or tissue type, e.g. Alzheimer’s disease (AD) or Cerebral Amyloid Angiopathy (CAA), and systemic amyloidosis, which affect more than one body organ or system, e.g. amyloidosis involving the misfolding of serum Amyloid A, β2-Microglobulin or Transthyretin [Citation2–4]. Although amyloidogenic proteins have heterogeneous structures and functions and the causes of amyloid-associated diseases vary, all these proteins can generate amyloid deposits in various organs. Some amyloidoses are life-threatening diseases, due to the cellular toxicity of the aggregates, as well as the fact that accumulated deposits are a burden that obstructs the proper functioning of the affected tissues. Without treatment, average life expectancy varies based on the type of amyloid, how much the organs are involved and the stage at diagnosis [Citation2,Citation5].
Some therapies are available for the treatment of amyloidosis (e.g. chemotherapy for light chain amyloidosis, anti-inflammatory therapy in serum A amyloidosis, tetramer stabilising drugs, such as tafamidis, and antisense RNA therapy in transthyretin amyloidosis) and others are emerging from research. Unfortunately, in a significant proportion of patients, organ damage is irreversible at the time of diagnosis and solid organ transplantation is often the last option [Citation6,Citation7].
One of the routes to develop effective therapeutic strategies for the treatment/prevention of Amyloidosis [Citation8] is relying on affecting the process of pathological amyloid formation. In the last years, disaggregation or inhibition of amyloid aggregates formation using small molecules, peptidomimetics and antibodies have been investigated; however, the results, even if promising, allowed only scarce translation to the clinics and, even less to the commercialization [Citation9]. Within this frame, nanomedicines are investigated as a possible innovative approach, due to their peculiar physicochemical features and mainly to the possibility of their multi-functionalization, which gives them the capability of multiple interactions [Citation10].
The present investigation stems from our previous research on AD that led us to design and synthesise liposomes composed of sphingomyelin/cholesterol and functionalized with phosphatidic acid and mApoE (a synthetic peptide derived from human apolipoprotein E). The double functionalization conferred to liposomes a higher affinity for Aβ1–42 fibrils and a higher ability to oppose fibrils formation and to induce their disaggregation in vitro and in vivo [Citation11–14]. In the present investigation, we tested the ability of Amyposomes to interact in vitro with amyloidogenic proteins other than Aβ1–42, specifically serum amyloid A (SAA), Aβ1–40, β2-microglobulin (β2m) and transthyretin (TTR). We show that Amyposomes strongly interact with these proteins, which are unrelated in reference to their biological function or to their amino acid sequences, slowing down or preventing their aggregation into fibrils or inducing the disassembly of their fibrillary aggregates.
Materials and methods
Reagents
The following reagents were purchased from Sigma-Aldrich: Thioflavin T (ThT, code T3516-25G), cholesterol (Chol, code C8667-5G), brain sphingomyelin (SM, code 860062 P), dimyristoylphosphatidic acid (PA, code 830845 P), distearoyl-phospatidylethanolamine-Polyetyleneglycl-maleimide (DSPE-PEG-MAL, code 880126 P), 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP, code 105228), phenylmethylsulfonyl fluoride (PMSF, code 10837091001). All other common reagents, resins for columns, solvent, reagents for electrophoresis and syntheses were also purchased from Sigma-Aldrich. The peptide CWGLRKLRKRLLR-NH2 (mApoE, Code 822594, SEQ ID No. 1) was purchased by KareBay Biochem – NJ USA. Trypsin Gold-Mass Spec Grade was purchased by Promega (code V5280).
Amyloid proteins
Human β − amyloid (1–40) was purchased from AnaSpect Inc. Recombinant human TTR variant TTR S52P, recombinant human β2m variants ΔN6β2-microglobulin (ΔN6β2m) and D76Nβ2-microglobulin (D76Nβ2m) were expressed, purified and characterized as described previously [Citation15,Citation16]. Details about the synthesis, purification and characterization are in Supplementary material (Figures S1–S7); Serum amyloid A, fragments 1–76, 1–13 and 2–13 were synthesized, purified and characterized as described in Supplemental material (Figures S8–S11, Table S1).
Preparation and characterization of Amyposomes
Amyposomes were synthesized as described [Citation15]. Briefly, SM and Chol (1:1 molar ratio) were mixed with 2.5 molar % of DSPE-PEG-MAL and with 5 molar % of PA in chloroform/methanol (2:1, v/v) and dried under a gentle stream of nitrogen followed by a vacuum pump for 3 h to remove traces of organic solvent. The resulting lipid film was rehydrated in phosphate-buffered saline containing 150 mM NaCl, pH 7.40 (PBS), vortexed and then extruded 10 times at 65 °C through a stack of two polycarbonate filters (100-nm pore size diameter). mApoE peptide was added to liposomes in PBS to give a final peptide: DSPE-PEG-MAL molar ratio of 1.2:1, and incubated overnight at room temperature to form a thioether bond with maleimide of lipid and cysteine of the peptide. The unbound peptide was removed by the PD-10 column and the yield of coupling of the peptide to liposomes was measured as described [Citation17]. Amyposomes size and ζ-potential were characterised as described previously [Citation17]. Liposomes composed of SM/Chol (1:1, molar ratio) were used as controls and named as PLAIN liposomes.
Amyposomes effect on formation of amyloid aggregates by ThT fluorescence intensity
The effect of Amyposomes on the formation of amyloid aggregates and on their disaggregation was studied at different protein:total lipids molar ratio ranging from 1:2 to 1:50 by ThT fluorescence assay, which identifies amyloid-containing β-sheet structures, as described in [Citation13,Citation18] with small modifications. ThT is a weakly fluorescent probe in water, but its fluorescence increases when it intercalates among the stacked β-sheets of aggregated amyloid protein molecules. The increase of ThT fluorescence during time in the presence of an amyloid protein can be taken as a parameter related to the extent of aggregation, while the decrease of fluorescence can be taken as a parameter related to the extent of disaggregation [Citation19]. To monitor the possible inhibitory effect of Amyposomes on protein aggregation, the non-aggregated form of proteins was added with 10 μM ThT and different amounts of Amyposomes (protein:total lipids molar ratio ranging from 1:2 to 1:50) directly in Costar 96-well black plates. The change in ThT fluorescence was monitored continuously during time by a spectrofluorometer (Victor 2, Perkin Elmer) excited at 445 nm, at 37 °C under agitation. As a control, instead of continuously following the fluorescence, the non-aggregated form of proteins was added with different amounts of Amyposomes and, at different times of incubation at 37 °C under agitation, an aliquot of the samples was withdrawn, added with 10 μM ThT, and the ThT fluorescence measured. The results were almost comparable to the continuous monitoring approach, that was therefore routinely utilised.
To monitor the disaggregating effect of Amyposomes, the pre-aggregated form of proteins was added with 10 μM ThT and with different amounts of Amyposomes (protein:total lipids molar ratio ranging from 1:2 to 1:50), then the time course of fluorescence was followed as above described. The conditions to obtain aggregated forms of the proteins are described below.
The fluorescence intensities of the samples were corrected against fluorescence signals of the buffer containing the same amount of Amyposomes. All the experiments were carried out under stirring conditions in order to avoid sedimentation of proteins.
Experimental protocol for Aβ1–40 peptide
The non-aggregated form of Aβ1–40 was obtained as previously described [Citation20]. The lyophilized peptide was solubilized in HFIP at a concentration of 1 mM. After evaporation of HFIP, the peptide was solubilized in DMSO at 5 mM protein concentration, then diluted in PBS to obtain a 25 μM peptide concentration and used immediately to evaluate the effect of Amyposomes on aggregation.
Aβ1–40 aggregates were prepared by incubating 25 μM Aβ1–40 in PBS at 37 °C and monitoring amyloid formation by ThT assay over time as described above. At the time point of maximum fluorescence, Amyposomes were added to evaluate their disaggregating effect.
Experimental protocol for TTR S52P protein
It has been previously shown that, under physiological conditions, trypsin cleaves human TTR in a mechano-enzymatic mechanism that generates abundant amyloid fibrils in vitro, which are morphologically similar to natural fibrils extracted from human amyloidotic tissue [Citation21,Citation22]. TTR S52P aggregates were prepared by dissolving 35 µM of recombinant protein in PBS and adding trypsin, in order to release the amyloidogenic fragment [Citation23]. Then, the mixture was incubated at 37 °C under stirring in order to allow the amyloid formation that started after 5h, as monitored by ThT fluorescence intensity. At this time, 1.5 mM of trypsin inhibitor PMSF was added to prevent the possible hydrolysis of mApoE peptide, followed by the addition of Amyposome to monitor their effect on the aggregation. To evaluate the disaggregating effect of Amyposomes, TTR S52P amyloid aggregates were prepared as above described, at the time point of maximum ThT fluorescence, PMSF was added to the incubation mixture followed by the addition of Amyposomes.
Experimental protocol for ΔN6β2m and D76N β2m proteins
In order to obtain a preparation of non-aggregated ΔN6β2m or D76N β2m, the lyophilized peptides were dissolved in PBS to obtain a 40 μM peptide concentration and used immediately to evaluate the effect of Amyposomes on amyloid formation, monitored by ThT fluorescence intensity.
Amyloid aggregates were prepared by incubating 40 μM of ΔN6β2m or D76N β2m protein in PBS at 37 °C under constant stirring [Citation24]. At the time point of maximum ThT fluorescence, Amyposomes were added to evaluate their disaggregating effect.
Experimental protocol for SAA peptides
In order to obtain a preparation of non-aggregated SAA1-76, the lyophilized peptide was dissolved in PBS to obtain 8 μM peptide concentration and used immediately to evaluate the effect of Amyposomes on amyloid formation, by ThT assay.
Amyloid aggregates were prepared by incubating 8 μM of SAA1-76 in PBS at 37 °C under constant stirring. At the time point of maximum ThT fluorescence, Amyposomes were added to evaluate their disaggregating effect. Concerning the SAA fragments 1–13 and 2–13, they immediately aggregated upon suspension in PBS. Thus, only disaggregation was investigated using the same conditions.
Atomic force microscopy
AFM technique is able to detect also other types of aggregates, besides amyloids, thus it has been used as a complementary technique in the present investigation. Amyloid proteins were obtained following the protocols described above. After aggregation, samples were centrifuged at 16.000 g to pellet insoluble aggregates [Citation25]. Aliquots of pellets were allowed to adhere onto freshly cleaved mica for 10 min. The samples were washed 3 times with 200 μl Milli-Q water and air-dried overnight.
After the deposition on the mica, the fibrillary sample were investigated immediately (see ) and after the addition of Amyposomes to see the effect of their interaction with the aggregates after 3 d (see ).
Figure 1. Characterization of amyloid aggregates by AFM. The aggregates of different amyloid proteins deposited on mica surfaces were characterised by AFM. Representative images in false colours of (A,B) Aβ1-40, (C,D) TTR S52P, (E,F), D76N B2M and (G,H) ΔN6 B2M are shown (see the lateral vertical scale of the different heights measured with respect to the mica surface). Images were obtained before (A,C,E,G) or after adding (B,D,F,H) Amyposomes. All the images are obtained according to the procedure described in Material and Methods.
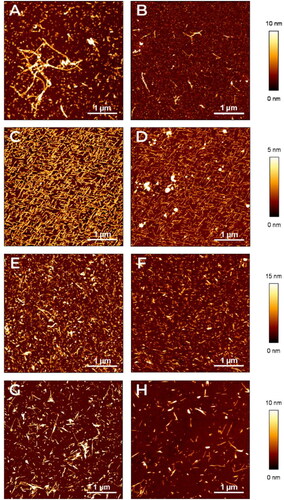
All the Atomic Force Microscopy (AFM) measurements were performed in Tapping Mode in air using stiff silicon cantilevers (RTESP-Veeco, resonant frequencies ∼300 kHz, spring constant ∼40 N/m). AFM images were acquired with a Nanowizard II (JPK Instruments, Berlin, Germany) at a scan rate of 1 Hz with 512 × 512 pixels resolution. Each image was analyzed using JPKs software. Samples were exhaustively examined to confirm their homogeneity.
Binding of amyloid aggregates with Amyposomes studied by surface Plasmon Resonance
These studies were carried out using the ProteOn XPR36 SPR apparatus (Bio-Rad Laboratories). This system is characterized by the presence of six flow channels which can uniformly immobilize up to six ligands on parallel strips of the same sensor surface. For the present study we prepared channels coated with the different protein aggregates and one channel used as a reference. The flow channels can be rotated 90° so that up to six concentrations of Amyposomes can be flowed in parallel, creating a 36-spot interaction array. The different protein aggregates were immobilized in parallel-flow channels of a GLC sensor chip (Biorad) using amine coupling chemistry, as in previous studies [Citation11].
Preliminary ‘preconcentration’ scouting assays, without covalent immobilization, were carried out in order to define the optimal conditions for the immobilization of the aggregates, in terms of buffer, pH and protein concentration. The following conditions were selected: Aβ1–40: 30 µg/mL, in acetate buffer, pH 4.0; ΔN6β2m: 30 µg/mL, in acetate buffer, pH 4.5; D76Nβ2m: 30 µg/mL, in acetate buffer, pH 4.0; TTR: 30 µg/mL, in acetate buffer, pH 4.5; SAA: 30 µg/mL, in acetate buffer, pH 4.5; BSA, used as internal reference: 30 µg/mL, in acetate buffer, pH 4.5.
The aggregates were immobilized using the classical protocols for amine coupling. Briefly, sensor surfaces were activated with sulfo-NHS/EDC according to the manufacturer’s recommendation, proteins were diluted in the immobilization buffer as indicated above and these solutions were flowed for 5 min at a rate of 30 µL/min over the activated chip surface. The remaining activated groups were blocked with ethanolamine, pH 8.0. After rotation of the fluidic system, different concentrations of Amyposomes were flowed over the immobilized protein aggregates and the reference surface. The running buffer of the SPR instrument was 10 mM phosphate buffer containing 150 mM NaCl and 0.005% Tween 20 (PBST pH 7.4). Analytes flowed over immobilized ligands for three min at a rate of 30 µL/min. Dissociation was measured in the following 11 min. All of these assays were carried out at 25 °C. The sensorgrams (time course of the SPR signal in Resonance Units, RU) were normalized to a baseline value of 0. The signals observed in the surfaces immobilizing protein aggregates were corrected by subtracting the nonspecific response observed in the reference surfaces, as indicated. When appropriate, the sensorgrams were fitted using the ProteOn analysis software to obtain the association and dissociation rate constants (ka and kd) and the equilibrium dissociation constant (KD).
Structural analyses
The experimentally-solved structure of Aβ1–40 (PDB ID: 2M4J; ssNMR) [Citation26], Aβ1–42 (PDB ID: 2NAO; sNMR) [Citation27], SAA (PDB ID: 6MST; EM) [Citation28] and TTR (PDB ID: 6SDZ; EM) [Citation29] amyloid fibrils were retrieved from the RCSB Protein Data Bank. The β2m amyloid fibril structure has not yet been solved. 3D structures were then verified and prepared using the MOE Structure Preparation Module (Molecular Operating Environment 2020.09, Chemical Computing Group, Quebec, Canada), to fix experimental related issues, adding hydrogen atoms and/ or filling up any unresolved residues/atoms. The 3D structures were then submitted to energy minimization, down to a Root Mean Square (RMS) gradient of 0.05 kcal/mol/Å2, with the Amber10:EHT force field and the reaction field (R-field) for electrostatics treatment.
Since the average identity of primary structures of selected proteins is approx. 5% (excluding the obvious identity level between Aβ1-40 and Aβ1–42) a structural alignment was carried out based on the 3D structures of selected amyloid fibrils, using the MOE Sequence Editor Module.
In order to investigate in depth the Aβ1–40 and Aβ1–42 structural similarities, we also analyzed two recent experimentally-solved structures of amyloid fibrils containing Aβ1–40 and Aβ1–42 in a paired (PDB ID: 6TI6; ssNMR) [Citation30] or staggered (PDB ID: 6TI7; ssNMR) [Citation30] cross-beta structure. Both structures were submitted to the same refinement, minimization and superposition procedures, as described above.
Statistical analysis
Data of ThT assays were normalized to the ThT signal of reference samples (PBS alone or Amyposomes alone). At least three independent experiments were conducted, each of them in triplicates. Statistical analysis was carried out by Student t-test or two-way ANOVA.
Results
Characterization of Amyposomes
The average diameter of Amyposomes used for these studies was 160 ± 20 nm with a PDI of 0.088 ± 0.04 and a ζ-potential value of −18 ± 1 mV. The content of PA was 4.2 ± 0.5 mol % and the content of mApoE was 1.7 ± 0.1 mol %.
Characterization of amyloid aggregates by AFM
AFM was employed to validate the protocol of aggregation used for the proteins under study and, subsequently, to confirm the results obtained by ThT fluorescence intensity measurements experiments.
In panels A, C, E, G we report some representative AFM images of aggregates of the different proteins under investigations. All the images are obtained following the protocols described in the Materials and Methods section.
shows the presence of unbranched, slightly curved, and elongated fibril chains in the aggregated form of the Aβ1–40 peptide. Such elongated fibrils exhibit an apparent height of 6 nm. Imaged by AFM, TTR S52P produces morphologically typical mature amyloid fibrils, 4 nm in height, geometrically ordered probably as a consequence of the interaction of the fibrils with the ordered crystalline substrate, where the fibrils were deposited ().
Analogously, show AFM images of D76N β2m and ΔN6 β2m fibrils, which have similar characteristics; even if in these cases, a slightly lateral aggregation seems present and the fibrils look thicker. In all the investigated samples reported in , the images clearly show the formation of fibrillary or elongated aggregates and demonstrate the effectiveness of the fibrils preparation procedures.
Furthermore AFM images reveal the effectiveness of Amyposomes in disaggregating preformed amyloid fibrils. Indeed show images, acquired after the effects of the presence of Amyposomes, where the concentration, length and thickness of fibrils is clearly reduced in all the situations. No effect was detected when fibrils were incubated with PBS (Figure S17).
In the Supplementary Material we added an AFM image (Figure S18) of the liposomes which gives an idea about their size, despite the inevitable alteration of liposomes due to their deposition on a flat surface.
In order to use another approach to test the Amyposomes and Aβ interactions, we performed DLS measurements (see Figure S19 and S20) which showed the increment of Amyposome radius, as a consequence of Aβ fibrils binding on nanoparticle surface.
Effect of Amyposomes on formation and disaggregation of Aβ1-40 peptide amyloid aggregates monitored by ThT fluorescence assay
As first, the behaviour of Aβ1–40 alone suspended in PBS in non-fibrillar form, was monitored. ThT fluorescence intensity displays a rise during the first 24 h of incubation, reaching a steady-state after 48 h, then remaining stable for 7 d.
In the presence of Amyposomes at different peptide:lipids ratios, a biphasic behaviour was observed. After an initial phase (24 h) during which an increase of ThT fluorescence was observed, the intensity decreased constantly during the time at all ratios tested. The lowest fluorescence intensity of ThT was detected starting from the 1:30 ratio. No changes in ThT fluorescence intensity were detected in presence of control SM/Chol liposomes ().
Figure 2. Amyposomes effect on Aβ1-40 aggregation and disaggregation. The effect of Amyposomes on Aβ1-40 aggregation and disaggregation at a different peptide:lipids (M:M) ratios was investigated by measuring the ThT fluorescence intensity. (A) Time course of ThT fluorescence starting from the non aggregated form of Aβ1-40. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 81.72 p < .0001). (B) Time course of ThT fluorescence starting from Aβ1-40 aggregates. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 39.71 p <.0001). AMYPO: amyposomes; PLAIN: liposomes composed of SM/Chol (1:1, molar ratio) used as a control.
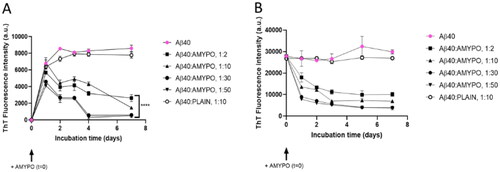
The influence of Amyposomes on the disaggregation of fibrillar Aβ1–40 was evaluated. The fibrillar form of the peptide alone in PBS, evaluated by ThT fluorescence, was stable for at least 7 d in the absence of Amyposomes. In the presence of Amyposomes at different peptide:lipids ratios, the ThT fluorescence intensity decreased at all ratios tested, following a seemingly exponential decreasing course. At 1:30 ratio and 1:50, 40% residual of aggregates was present after 24 h and only 10% after 5 d ().
Effect of Amyposomes on formation and disaggregation of TTR S52P amyloid aggregates monitored by ThT fluorescence assay
To study the effect on aggregation, as first the behaviour of non-aggregated TTR S52P, alone in PBS was studied. ThT fluorescence intensity started to increase 5 h after the addition of trypsin and reached a maximum after 12 h, then remaining constant. As a control, the trypsin inhibitor PMSF, added after 5 h of incubation did not influence the aggregation. The presence of Amyposomes strongly hindered the increase of ThT fluorescence at all ratios, except 1:2. At protein:lipids ratio of 1:50 a 40% lower fluorescence with respect to the protein alone was measured. No changes in ThT fluorescence intensity, which remained comparable to that of protein alone, was detected in presence of non-functionalized SM/Chol liposomes ().
Figure 3. Amyposomes effect on TTR S52P aggregation and disaggregation. The effect of Amyposomes on TTR S52P aggregation and disaggregation at a different peptide:lipids (M:M) ratios was investigated by measuring the ThT fluorescence intensity. (A) Time course of ThT fluorescence starting from the non-aggregated form of TTR S52P. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 4.47 p < .021). (B) Time course of ThT fluorescence starting from TTR S52P aggregates. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 7.94 p < .0001). AMYPO: amyposomes; PLAIN: liposomes composed of SM/Chol (1:1, molar ratio) used as a control.
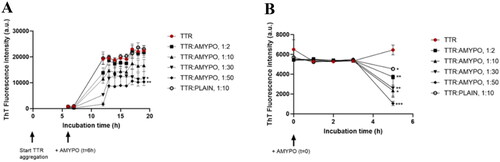
Considering that the control liposomes used were tested at 1:10 molar excess at which even Amyposomes were not effective, the effect of non-functionalized SM/Chol liposomes at a 1:50 ratio was also measured. The results (Figure S12A) showed that control liposomes did not affect the aggregation of TTR S52P even at a higher dose.
The influence of Amyposomes on the disaggregation of TTR S52P was evaluated. The protein in the fibrillar form was stable at least for 5 h. In the presence of Amyposomes, ThT fluorescence intensity decreases on increasing the amount of Amyposomes. In the case of the 1:50 protein:lipids ratio, only 5% of initial fluorescence was measured after 5 h of incubation ().
Effect of Amyposomes on formation and disaggregation of β2m D76N and ΔN6 amyloid aggregates monitored by ThT fluorescence assay
Two variants of the protein (D76N and ΔN6) were investigated. To study the effects on fibrillar aggregation, as first D76N and ΔN6 alone in solution were monitored. D76N followed a multi-step behaviour. The ThT fluorescence started to increase after 10 h of incubation, increasing slowly up to 19 h, later on it abruptly increased reaching a maximum at 21 h of incubation, and then remained constant (). On the other side, kinetics for the ΔN6 variant were slower, the aggregation reaching a maximum after 48 h, then remaining constant (). Concerning the influence on fibrillar aggregation, when Amyposomes were incubated with the protein, they hindered the increase of ThT fluorescence intensity at all protein:lipid ratios tested. At D76N:lipids ratio of 1:50, the final ThT fluorescence intensity was 36% with respect to that of the protein alone (). In the case of ΔN6, at a protein:Amyposomes ratio of 1:50, the aggregation extent was almost undetectable upon incubation ().
Figure 4. Amyposomes effect on β2microglobulin aggregation and disaggregation. The effect of Amyposomes on D76N and ΔN6 proteins aggregation and disaggregation at a different peptide:lipids (M:M) ratios was investigated by ThT fluorescence intensity. (A) Time course of ThT fluorescence starting from the non-aggregated form of D76N. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 207.6 p < .0001). (B) Time course of ThT fluorescence starting from D76N aggregates. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 50.19 p < .0001). (C) Time course of ThT fluorescence starting from the non-aggregated form of ΔN6. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 59.49 p < .0001). (D) Time course of ThT fluorescence starting from ΔN6 aggregates. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 9.10 p < .0001). AMYPO: amyposomes; PLAIN: liposomes composed of SM/Chol (1:1, molar ratio) used as a control.
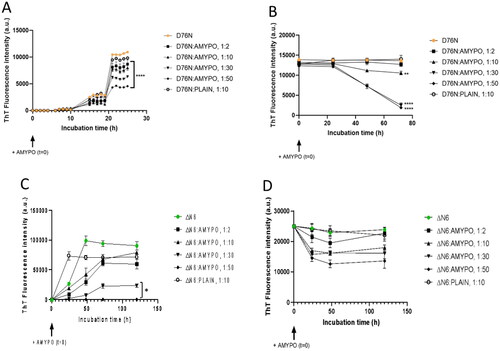
Considering that the control liposomes used were tested at 1:10 molar excess at which even Amyposomes were not effective on ΔN6, the effect of non-functionalized SM/Chol liposomes at 1:50 ratio was also measured. The results (Figure S12B) showed that control liposomes did not affect the aggregation of β2m ΔN6 even at a higher dose.
The influence of Amyposomes on the disaggregation of D76N and ΔN6 proteins was evaluated on preparations of fibrillar proteins. In the case of D76N, the protein in the fibrillar form was stable for at least 3 d. In the presence of Amyposomes, a decrease of fluorescence was observed starting after 24 h of incubation and decreased constantly up to 72 h. This effect was not evident at the 1:2 protein:lipids ratio, was minimal at the 1:10 ratio, and was stronger and comparable at 1:30 and 1:50 ratios, leading to only a 15% residual ThT fluorescence after 3 d of incubation ().
In the case of ΔN6, a decrease of fluorescence was observed starting immediately after incubation in the presence of Amyposomes reaching a minimum after 48 h. The effect increased with the amount of Amyposomes and at 1:50 ratios, 50% residual fluorescence intensity was present ().
Effect of Amyposomes on formation and disaggregation of SAA amyloid aggregates monitored by ThT fluorescence assay
To study the effect of Amyposomes on aggregation, as first SAA1-76 alone was monitored. ThT fluorescence intensity of SAA1-76 started to increase after 48 h of incubation. Contrarily, in the presence of Amyposomes at all protein:lipid ratios the increase of ThT fluorescence intensity was prevented, and at a protein:lipids ratio of 1:50, almost no fluorescence signal was detectable ().
Figure 5. Amyposomes effect on SAA fragments aggregation and disaggregation. The effect of Amyposomes on SAA fragments aggregation and disaggregation at a different peptide:lipids (M:M) ratios was investigated by measuring the ThT fluorescence intensity. (A) Time course of ThT fluorescence starting from non-aggregated form of SAA1-76. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 705.3 p < .0001). (B) Time course of ThT fluorescence starting from SAA1-76 aggregates. Data are expressed as the mean ± SD of triplicates and analyzed by two-ANOVA (F = 34.88 p < .0001). AMYPO: amyposomes; PLAIN: liposomes composed of SM/Chol (1:1, molar ratio) used as a control.
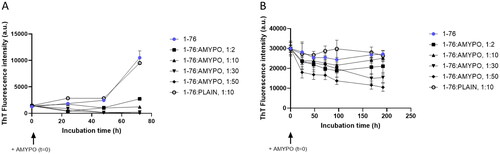
The effect of Amyposomes on disaggregation of SAA1-76 was evaluated on preparations of previously aggregated protein. A decrease of ThT fluorescence was observed with Amyposomes at all ratios tested. At the protein:lipids ratio of 1:50 a 35% residual fluorescence was measured after 8 d of incubation ().
In the case of SAA fragments 1–13 and 2–13, the effects on aggregation were not studied since these peptides were already aggregated immediately after their suspension in the buffer. The ThT disaggregation assay was performed on the aggregated forms of the peptides in the presence of different Amyposomes concentrations. The fluorescence intensity decreased over time and after 8 days, 25% residual fluorescence was present in the case of 1–13 fragment starting from a 1:10 ratio and 50% for 2–13 fragments, at all ratios tested (data not shown).
Validation of ThT fluorescence intensity data by AFM technique
AFM experiments were performed to confirm the fact that Amyposomes are actually disaggregating amyloid fibrils and not merely hindering thioflavin T fluorescence. AFM technique allows a qualitative validation of the fluorescence intensity results since proteins are immobilized on the mica surface in the case of AFM, while are suspended in a buffer in the case of the ThT technique.
Therefore, samples of aggregated proteins after incubation with Amyposomes were analyzed by AFM (at 1:30 ratio) since at this value all the proteins under study undergo disaggregation. The representative AFM images, reported in , Panel B, D, F, H show that the length and concentration of fibrils on the mica surface is reduced after the incubation with Amyposomes. Overall, the imaging of the incubation mixture is mirroring the results obtained by ThT, confirming the validity of the ThT technique in monitoring the aggregation state of fibrils. In order to further confirm the disaggregating influence of Amyposomes on mature fibrils, we have also compared the effect of Amyposomes (Figure S17, Panel B, D, F, H which are identical to ) with the effect of the incubation buffer (PBS) used for dispersing the nanoparticles (Figure S17 Panel A, C, E, G). As apparent from Figure S17, without the presence of Amyposomes the structure and density of fibrils appear stable on the mica surface, or at least increasing in the case of ΔN6.
It is worth to note that the agreement between the results obtained by using AFM and ThT fluorescence technique is just qualitative, given the different experimental conditions of the two methods. Namely, for the AFM, the proteins are immobilised on the mica surface, while for the ThT fluorescence technique they are suspended in a buffer.
Binding of amyloid aggregates with Amyposomes by surface Plasmon Resonance
A first experimental session was carried out by immobilizing Aβ1–40, β2m ΔN6, D76N, TTR S52P and BSA in five parallel surfaces of the same chip, while the sixth surface was left empty as reference. The conditions identified for the immobilization procedure allowed the immobilization of a suitable amount of proteins on the chip surfaces, with the following immobilization levels (in RU): Aβ1–40: 4619; β2m ΔN6: 3632; β2mD76N: 2860; TTR: 2736; BSA: 4850 RU.
The injection of different concentrations of Amyposomes resulted in some non-specific SPR signal with the empty surface, very similar to the SPR signal observed on BSA, used here as an unrelevant protein. The raw sensorgrams are shown in Supplementary, Figure S13. After subtraction of the signal measured on the empty surface, used as a reference, it appears that Amyposomes have no ‘specific’ binding signal on BSA, but a concentration-dependent specific binding to all the protein aggregates, although with different features ().
Figure 6. SPR sensorgrams showing the specific binding of different concentrations of Amyposomes (amyp) to the indicated immobilized protein aggregates. The Amyposomes concentrations, expressed as µM of the exposed PA, were 1.56, 3.125, 6.25 and 12.5 µM, and are indicated with increased thickness of the sensorgram. Amyposomes (amyp) flowed for three min as indicated. Black dashed lines shows the results of the global fitting of these sensorgrams (i.e. the four concentrations fitted together) according to the (A) Langmuir equation (1:1 interaction model) for Aβ1–40 and the heterogeneous ligand model for (B) TTR S52P, (C) β2m D76 and (D) β2m ΔN6. The parameters of the fittings are shown in the table below (ka1 and ka2: association rate constants; kd1 and kd2: dissociation rate constants; KD1 and KD2: equilibrium dissociation constants).
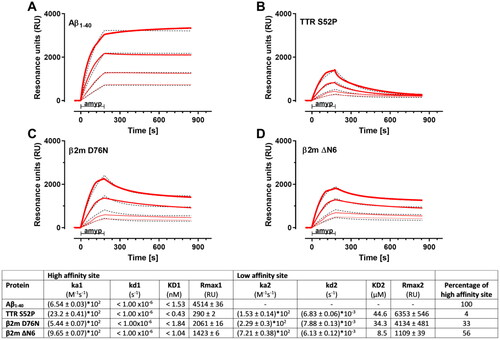
The binding of Amyposomes to immobilized Aβ1–40 was very stable, with no apparent dissociation during the 11-min dissociation phase, indicating a very high affinity. All the sensorgrams obtained with the four tested concentrations () could be globally fitted using the simplest Langmuir model (1:1 interaction) with a KD value of 1.5 nM or even lower (the uncertainty is due to the instruments specification, which only allows a dissociation rate constant (kd) higher than 1 × 10−6 s−1 to be reliably determined)
The binding of Amyposomes to TTR, β2m ΔN6 and β2mD76N () could not be well described by the Langmuir 1:1 equation. In fact, the dissociation phase indicated the presence of at least two components, one with a pseudo-irreversible binding (dissociation rate constant kd1 fixed to 1 × 10−6 s−1) as for Aβ1-40 , and an additional one with a faster dissociation (kd2 = 6 – 8 × 10−3 s−1). Thus, a ‘two-sites’ ligand model was used to fit these sensorgrams ().
The parameters, summarized in the table below , show similar KD values (nM range) of the high binding affinity site for all different protein aggregates, whereas the low binding affinity site is in the µM range. The main difference is the amount of high affinity binding sites compared to the low affinity binding sites, indicated by Rmax1 and Rmax2 values (Rmax being the maximal feasible SPR signals estimated by the fitting). Aβ1–40 aggregates contain only high-affinity binding sites, whereas TTR aggregates mostly showed low affinity binding sites. The percentage of high affinity binding sites for β2m ΔN6 and β2mD76N are 56% and 33% respectively.
The binding behaviour of Amyposomes to protein aggregates was confirmed in a second experiment, where a shorter contact time was used. The same models and binding parameters previously determined () described well the sensorgrams (Supplementary Figure S14).
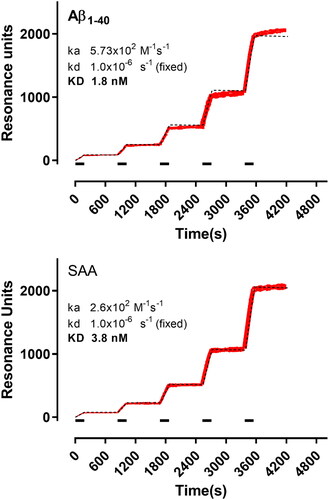
A second experimental session was carried out by immobilizing Aβ1–40, SAA and BSA in three parallel surfaces of the same chip. In this case, we used the so-called ‘kinetic titration’ approach with serial injections of increasing concentrations of the Amyposomes, without intermediate regeneration. shows the corresponding sensorgrams, already subtracted for the signal obtained in parallel on the reference surface (BSA). Global fitting (i.e. all the concentrations together) with the 1:1 interaction model taking into account the cumulative binding, well described the sensorgrams (dashed lines in ). The estimated KD values for the binding of Amyposomes to Aβ1–40 and SAA aggregates were in the low nM range (1.8 nM for Aβ1–40 and 3.8 nM for SAA), due to the very low dissociation rate constants.
Figure 7. SPR sensorgrams showing the specific binding of Amyposomes to the immobilised protein aggregates (as indicated). Increasing concentrations of Amyposomes (1.56, 3.125, 6.25 and 12.5 µM of the exposed PA) were injected for three min (bars), in sequence, and are indicated in red with increased thickness of the sensorgram. Black dashed lines show the results of the global fitting of these sensorgrams (i.e. the five concentrations fitted together) according to the Langmuir equation (1:1 interaction model). For these fittings, Rmax (i.e. the maximum binding) was fixed for each protein to the corresponding immobilisation level, in order to take into account the differences on this parameter. The results of the fittings are shown. ka: association rate constant; kd: dissociation rate constant; KD: equilibrium dissociation constant.
Discussion
Amyposomes were originally designed and developed as a therapeutic approach for Alzheimer’s disease by targeting Aβ1–42 [Citation12,Citation14]. Amyposomes were shown in vitro and in vivo to oppose the formation of Aβ1–42 fibrils and to induce their disaggregation [Citation11–14].
Starting from these observations, this study was carried out to test in vitro the efficacy of this nanomedicine on the aggregation paradigm of other amyloidogenic/amyloid proteins, i.e. to evaluate their capability to prevent protein aggregation or to induce the disaggregation of amyloid aggregates. The study has been carried out utilising a spectrofluorometric technique, based on the Thioflavin T (ThT) fluorescence assay, which identifies amyloid-containing β-sheet structures [Citation19] increasing its fluorescence when it intercalates among the stacked β-sheets of aggregated proteins. The minimal binding site on the fibril surface has been suggested to span four consecutive β-strands [Citation18]. Protocols for the preparation of aggregated forms of the proteins were validated by AFM. We note that the opposite experimental approach, namely to prepare planar lipid membranes and to study by AFM the interaction of such membranes with fibrils, has been reported (see for instance [Citation31]). However, considering a possible translational medical application, the presented approach seems to better simulate the interaction between nanostructured particles with amyloid fibrils.
SPR technique was utilized to assess the affinity of binding of Amyposomes with the proteins studied, and in silico molecular simulation studies of amyloid structures were carried out for supporting the mechanism of action.
We focused our attention on some proteins responsible of major Amyloidoses in humans:
β-amyloid 1–40 (Aβ1–40) a proteolytic product of the amyloid precursor protein (APP). Its deposition in arterioles and/or capillaries in the cerebral cortex and leptomeninges is the main molecular feature of cerebral amyloid angiopathy (CAA). CAA is the second disease (after hypertension) causing cerebral haemorrhage in the elderly, with a mortality of 30–50%. In addition, CAA is commonly found in Alzheimer’s disease (AD) and nearly 80% of AD cases is associated with CAA.
Transthyretin (TTR) an homo-tetrameric protein produced in the liver or in the cerebral spinal fluid (CSF), associated with two Amyloidoses: senile systemic amyloidosis (SSA), characterized by massive deposition of TTR mainly in the heart, and familial amyloid polyneuropathy (FAP), characterized by deposition at the peripheral nerves and tissues. In the case of TTR we specifically focused on the S52P variant, causing the most aggressive phenotype of hereditary systemic TTR amyloidosis.
β2 microglobulin (β2m) the light chain of class I major histocompatibility complex; β2m in vivo aggregation is responsible for dialysis-related amyloidosis. In the case of β2m we focused on the variants D76N associated with a familial form of the disease and on the variant ΔN6, a ubiquitous constituent of β2m amyloid deposits in patients affected by dialysis-related amyloidosis.
Protein serum amyloid A (SAA), forming amyloid deposits in organs like the spleen, liver, and kidneys, originating from a condition called reactive amyloidosis or Amyloid A (AA) amyloidosis. Reactive amyloidosis generally accompanies other conditions that induce chronic inflammation such as rheumatoid arthritis and atherosclerosis.
The results show that Amyposomes are able either to prevent or to slow down the transition of the above said proteins from non-aggregated soluble forms to the aggregated state. Amyposomes are also able to induce disaggregation of pre-existing aggregates of the proteins investigated. Amyposomes affect all the proteins investigated, however with differences in the extent of the aggregation/disaggregation or in their kinetics. The strongest effects of Amyposomes on disaggregation are recorded with Aβ1–40, β2m D76N and TTR with almost total disruption of aggregates; concerning the effects on the aggregation, almost complete inhibition is recorded with Aβ1–40, β2m ΔN6 and SAA. In addition, the time of incubation necessary to exert the effect showed relevant differences, with a minimum of only a few hours required for TTR, while the time required in other cases is in the order of 1–3 d.
SPR data showed that Amyposomes bind to a similar extent both the empty sensor surface and BSA and that a much higher, specific binding occurred to the aggregates of all the proteins tested. The binding of Amyposomes to Aβ1–40 and SAA amyloid aggregates is very stable with estimated KD in the low nanomolar range, similar to the high-affinity binding of Amyposomes to Aβ1–42 fibrils [Citation11]. This high-affinity binding site is also present at a lower percentage on β2m D76N and β2m ΔN6 (occurrence = 33–56% of total binding sites) and is almost absent on TTR aggregates. β2m D76N, β2m ΔN6 and TTR aggregates additionally contain a second binding site with a KD in the micromolar range. Altogether, SPR results show that Amyposomes interact with all the amyloids tested, although with different binding properties.
Since the protein tested are unrelated in reference to their amino acid sequences or to their biological function, it is likely that the ability of Amyposomes to affect the aggregation process of proteins resides in the ability to interact with their amyloidogenic regions, as it was previously suggested in the case of Aβ1–42 [Citation12,Citation13,Citation30].
This hypothesis is based on the observation that the proteins under examination display common aggregation features due to the presence of β-stranded regions. Such regions stick together forming fibrils, stabilised by hydrogen bonds between β-sheet structures. Non-polar amino acid residues are favoured to be found in β-strands within β-sheets.
To further support this hypothesis, we selected from the Protein Data Bank the structures of Aβ1–40, Aβ1–42, SAA and TTR with an amyloid fibril conformation. Supplementary Figure S15 reports the selected proteins in both their native and amyloid fibril conformations. Since these three different proteins share a very low sequence identity (Supplementary Figure S15, bottom), as pointed out by their multiple alignment, but they share quite common structural features after fibril formation, a 3D superposition was produced and used to generate a structural alignment. In fact, a β-strand/turn/β-strand (β/t/β) conserved secondary structure features were identified in all the Aβ1–40, Aβ1–42, SAA and TTR structures. Moreover, the quaternary structure of all the studied proteins shows a cross-β structure characterised by a network of intermolecular hydrogen bonds (Supplementary Figure S15).
The multiple structural alignment between Aβ1–40 and Aβ1–42 (PDB ID: 6TI6, 6TI7) pointed out their low overall root mean square deviation (RMSD) (0.83 Å), confirming that Aβ1–40 and Aβ1–42, in these experimental conditions, have the same folding pattern. Differently, the RMSD of Aβ1–40 and Aβ1–42 from 2M4J and 2NAO shows a reduced value in the turn (0.15 Å) and a significantly higher value (5.9 Å) considering the β/t/β pattern (from Leu17 to Gly37), suggesting that the β/t/β pattern is more conserved folding than the N- and C-terminal remaining residues (26 Å). Since the turn organisation of 6TI6, 6TI7 Aβ1–4x forms is significantly different from the other commonly investigated turns (SAA, TTR), 2M4J will be used as a representative of Aβ1–4x for further structural analysis.
The multiple structure alignment of Aβ1–4x, SAA and TTR points out that these three proteins share the same β/t/β pattern (RMSD of 5.9 Å) in which the most structural conserved 3D conformation is localised on the turn, with an RMSD of 0.16 Å (). The β/t/β pattern is characterised by an average distance of approx. 9 Å between the two opposite β-strands and by a length of 30 Å () and it is also shared by several amyloid and prionic proteins [Citation32].
Figure 8. Structural superpositions of selected amyloid fibrils. (A) Structural superposition of TTR, SAA and Aβ1–4x, coloured according to their RMSD: in green the loop with associated the lowest RMSD value, in red the regions with associated the highest RMSD. 3D structures are represented as liquorice. (B) Structural superposition of TTR (light blue), SAA (yellow) and Aβ1–4x (orange). 3D structures are represented as ribbon.
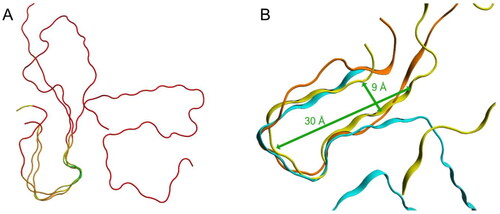
These data strengthen the hypothesis that the mechanism of action of Amyposomes is based on the interaction of their components with the cross-β structure of the β/t/β patterns that are forming a network of intermolecular hydrogen bonds. The synergistic interaction of Amyposomes active components (phosphatidic acid and mApoE peptide) with the proteins is likely weakening the inter-molecular interactions maintaining the fibrillar structures, either preventing their formation or inducing their destabilisation. It is possible to speculate that the high-affinity sites present on protein aggregates coincide with β/t/β structures and low-affinity sites with the cross-β structure. The evaluation of this intriguing possibility will deserve further experimental studies.
Moreover, it can be observed that, for a given protein, aggregation and disaggregation are occurring on a comparable time scale. Thus, it is also possible to speculate that the efficacy of Amyposomes may be dependent on at least two factors: on the one hand, a specific binding toward the aggregates of all the proteins tested, although with different binding properties. On the other hand, their ability to affect the amount of aggregates but not the kinetics of aggregation. It should be noted that, in some instances, destabilisation could not be the ideal option; for example, in the case of β-amyloid peptides, it has been hypothesised that the destabilisation of aggregates may release toxic oligomeric species [Citation33]. Other anti-amyloidosis nanomedicines have been employed to mitigate the aggregation and toxicity of amyloid peptides and proteins such as tau, alpha-synuclein, human islet amyloid polypeptide, associated with AD, PD and T2D, with effects of various extents in vitro and in vivo [Citation34–36]. Contrary to these investigations, our study has not been focused on a single protein but on a broad range of amyloidogenic proteins (Aβ1–42 included [Citation11–14]), unrelated in reference to their biological function and to their amino acid sequences. This approach allows us to infer that Amyposomes are an all-in-one nanotechnology with broad anti-amyloidogenic activity. In addition, since Amyposomes display interesting features, such as biocompatibility and the capacity of systemic circulation [Citation12–14], makes them a promising tool for the treatment of different forms of amyloidosis.
Author contributions
M.M. and M.S. designed research experiments, supervised the study, and wrote the manuscript. F.R., S.S. and L.R. performed ThT experiments and analyzed the data. V.C. and F.M. performed AFM experiments and analyzed the data. I.E. and L.P. performed a structural analysis of amyloids and analyzed the data. M.B. and M.G. performed SPR experiments and analyzed the data. S.G., B.B. and S.S. performed the synthesis and purification of amyloid proteins. M.M., M.S., F.R. and F.M. contributed intellectual input and data analysis. All authors discussed the data and contributed to the manuscript revision. M.M., F.R. and M.S. conceived the study and provided the funding. All authors agree to be accountable for all aspects of the work meet the criteria for authorship.
Supplemental Material
Download MS Word (4.6 MB)Disclosure statement
The authors declare that they have no conflicts of interest with the contents of this article.
Data availability statement
Data available within the article or its supplementary materials.
Additional information
Funding
References
- Benson MD, Buxbaum JN, Eisenberg DS, et al. Amyloid nomenclature 2020: update and recommendations by the international society of amyloidosis (ISA) nomenclature committee. Amyloid. 2020;27(4):1–15.
- Hazenberg BP. Amyloidosis: a clinical overview. Rheum Dis Clin North Am. 2013;39(2):323–345.
- Sipe JD, Benson MD, Buxbaum JN, et al. Nomenclature 2014: amyloid fibril proteins and clinical classification of the amyloidosis. Amyloid. 2014;21(4):221–224.
- Pepys MB. Amyloidosis. Annu Rev Med. 2006;57:223–241.
- Wechalekar AD, Gillmore JD, Hawkins PN. Systemic amyloidosis. Lancet. 2016;387(10038):2641–2654.
- Nevone A, Merlini G, Nuvolone M. Treating protein misfolding diseases: therapeutic biologicalSuccesses against systemic amyloidoses front. Pharmacol. 2020;11:1024.
- Theodorakakou F, Fotiou D, Dimopoulos MA, et al. Solid organ transplantation in amyloidosis. Acta Haematol. 2020;143(4):352–364.
- Mitra A, Sarkar N. Sequence and structure-based peptides as potent amyloid inhibitors: a review. Arch Biochem Biophys. 2020;695:108614–108619.
- Rajan R, Ahmed S, Sharma N, et al. Review of the current state of protein aggregation inhibition from a materials chemistry perspective: special focus on polymeric materials. Mater Adv. 2021;2(4):1139–1176.
- De Crozals G, Bonnet R, Farre C, et al. Nanoparticles with multiple properties for biomedical applications: a strategic guide. Nano Today. 2016;11(4):435–463.
- Gobbi M, Re F, Canovi M, et al. Lipid-based nanoparticles with high binding affinity for amyloid-beta1-42 peptide. Biomaterials. 2010;31(25):6519–6529.
- Balducci C, Mancini S, Minniti S, et al. Multifunctional liposomes reduce brain β-amyloid burden and ameliorate memory impairment in Alzheimer’s disease mouse models. J Neurosci. 2014;34(42):14022–14031.
- Bana L, Minniti S, Salvati E, et al. Liposomes bi-functionalized with phosphatidic acid and an ApoE-derived peptide affect Aβ aggregation features and cross the blood-brain-barrier: implications for therapy of Alzheimer disease. Nanomedicine. 2014;10(7):1583–1590.
- Mancini S, Balducci C, Micotti E, et al. Multifunctional liposomes delay phenotype progression and prevent memory impairment in a presymptomatic stage mouse model of Alzheimer disease. J Control Release. 2017;258:121–129.
- Valleix S, Gillmore J, Bridoux F, et al. Hereditary systemic amyloidosis due to Asp76Asn variant β2-microglobulin. N Engl J Med. 2012;366(24):2276–2283.
- Esposito G, Michelutti R, Verdone G, et al. Removal of the N-terminal hexapeptide from human β2-microglobulin facilitates protein aggregation and fibril formation. Protein Sci. 2000;9(5):831–845.
- Re F, Cambianica I, Sesana S, et al. Functionalization with ApoE-derived peptides enhances the interaction with brain capillary endothelial cells of nanoliposomes binding amyloid-beta peptide. J Biotechnol. 2011;156(4):341–346.
- Xue C, Lin TY, Chang D, et al. Thioflavin T as an amyloid dye: fibril quantification, optimal concentration and effect on aggregation. R Soc Open Sci. 2017;4(1):160696.
- Nardo L, Re F, Brioschi S, et al. Fluorimetric detection of the earliest events in amyloid β oligomerization and its inhibition by pharmacologically active liposomes. Biochim Biophys Acta. 2016;1860(4):746–756.
- Dahlgren KN, Manelli AM, Stine WBJr, et al. Oligomeric and fibrillar species of amyloid-beta peptides differentially affect neuronal viability. J Biol Chem. 2002;277(35):32046–32053.
- Mangione PP, Verona G, Corazza A, et al. Plasminogen activation triggers transthyretin amyloidogenesis in vitro. J Biol Chem. 2018;293(37):14192–14199.
- Raimondi S, Mangione PP, Verona G, et al. Comparative study of the stabilities of synthetic in vitro and natural ex vivo transthyretin amyloid fibrils. J Biol Chem. 2020;295(33):11379–11387.
- Cantarutti C, Raimondi S, Brancolini G, et al. Citrate-stabilized gold nanoparticles hinder fibrillogenesis of a pathological variant of β2-microglobulin. Nanoscale. 2017;9(11):3941–3951.
- Mok Y-F, Howlett GJ. Sedimentation velocity analysis of amyloid oligomers and fibrils. Methods Enzymol. 2006;413:199–217.
- Lu JX, Qiang W, Yau WM, et al. Molecular structure of β-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154(6):1257–1268.
- Wälti MA, Ravotti F, Arai H, et al. Atomic-resolution structure of a disease-relevant Aβ(1-42) amyloid fibril. Proc Natl Acad Sci U S A. 2016;113(34): e4976–e4984.
- Liberta F, Loerch S, Rennegarbe M, et al. Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids. Nat Commun. 2019;10(1):1104.
- Schmidt M, Wiese S, Adak V, et al. Cryo-EM structure of a transthyretin-derived amyloid fibril from a patient with hereditary ATTR amyloidosis. Nat Commun. 2019;10(1):5008.
- Cerofolini L, Ravera E, Bologna S, et al. Mixing Aβ(1-40) and Aβ(1-42) peptides generates unique amyloid fibrils. Chem Commun. 2020;56(62):8830–8833.
- Ahyayauch H, Raab M, Busto JV, et al. Binding of β-amyloid (1-42) peptide to negatively charged phospholipid membranes in the liquid-ordered state: modeling and experimental studies. Biophys J. 2012;103(3):453–463.
- Lin H, Bhatia R, Lal R. Amyloid beta protein forms ion channels: implications for Alzheimer’s disease pathophysiology. Faseb J. 2001;15(13):2433–2444.
- Galkin AP, Velizhanina ME, Sopova Y, et al. Prions and non-infectious amyloids of mammals – similarities and differences. Biochemistry. 2018;83:1184–1195.
- Sakono M, Zako T. Amyloid oligomers: formation and toxicity of Aβ oligomers. FEBS J. 2010;277(6):1348–1358.
- Sonawane SK, Ahmad A, Chinnathambi S. Protein-capped metal nanoparticles. Inhibit tau aggregation in Alzheimer’s disease. ACS Omega. 2019;4(7):12833–12840.
- D’Onofrio M, Munari F, Assfalg M. Alpha-synuclein-nanoparticle interactions: understanding, controlling and exploiting conformational plasticity. Molecules. 2020;25(23):5625.
- Cabaleiro-Lago C, Lynch I, Dawson KA, et al. Inhibition of IAPP and IAPP(20-29) fibrillation by polymeric nanoparticles. Langmuir. 2010;26(5):3453–3461.

