Abstract
Background
NOP2/Sun RNA methyltransferase 5 (NSUN5) is an RNA methyltransferase that has a broad distribution and plays critical roles in various biological processes. However, our knowledge of the biological functions of NSUN5 in mammals is very limited. Therefore, in this study, we investigate the role of NSUN5 in mice.
Methods
In the present research, we built a mouse model (Nsun5-/-) using the CRISPR/Cas9 system to investigated the specific role of NSUN5.
Results
We observed that Nsun5-/- mice had a reduced body weight compared to wild-type mice. Additionally, their survival rate gradually decreased to 20% after postnatal day (PD) 21. Further examination revealed the Nsun5-/- mice had multiple organ damage, with the most severe damage occurring in the kidneys. Moreover, we observed glycogen deposition and fibrosis, along with a notable shorting of the primary foot processes of glomeruli in Nsun5-/- kidneys. Furthermore, we found that the kidneys of Nsun5-/- mice showed increased expression of the apoptotic signal Caspase-3 and accumulated stronger DNA damage at PD 21.
Conclusions
In our study, we found that mice lacking NSUN5 died before puberty due to kidney fatal damage caused by DNA damage and cell apoptosis. These results suggest that NSUN5 plays a vital role in preventing the accumulation of DNA damage and cell apoptosis in the kidney.
Introduction
RNA modifications play a crucial role in regulating gene expression and are implicated in the pathogenesis of various diseases [Citation1]. There are over 100 chemical modifications of RNA, RNA methylation is an important epigenetic modification. Recent studies on RNA methylation have mainly focused on the N6-methyladenosine (m6A) modification, but very little is known about the 5-methylcytosine (m5C) [Citation2]. m5C is a prevalent and conserved modification found in different types of RNA, including transfer RNA, mRNA, ribosomal (r) RNA and other types of noncoding RNA [Citation1,Citation3]. The modification process of m5C is mainly mediated by three types of proteins: methyltransferases (writers), demethylases (erasers), and binding proteins (readers). The methyltransferases of m5C use S-adenosylmethionine as the methyl donor and transfer the methyl group to cytosine, forming m5C [Citation4]. There are over 10 known RNA m5C methyltransferases, including members of the NOP2/Sun domain (NSUN) family [Citation5,Citation6]. These RNA m5C methyltransferases, which are well-known, have been found to regulate various conditions such as glial scaffolds, Williams-Beuren syndrome, and lifespan [Citation6–8].
Recently, there has been extensive and in-depth research on the NSUN protein family. This family consists of a total of 9 proteins in human [Citation9]. Studies have shown that seven members of this family (NSUN1 to NSUN7) have potential functional domains for m5C methyltransferases [Citation10]. In particular, the nucleolar protein NSUN1 is capable of binding directly to 60-80S preribosomal particles and catalyzing the methylation of C4447 to m5C in human 28S rRNA and C2870 in yeast 25S rRNA [Citation11]. Another member, NSUN2, is an RNA m5C methyltransferase for mRNA, tRNA and some noncoding RNA in eukaryotes [Citation12]. NSUN3 is responsible for catalyzing m5C at C34 of mt-tRNAMet, which is essential for f5C34 formation by the dioxygenase ABH1 in human and mouse mitochondria [Citation13,Citation14]. Subsequent studies have shown that NSUN4 can independently methylate C911 in 12S rRNA (m5C911) of human mitochondria [Citation15]. NSUN6 has also been identified as a potential protein methyltransferase, with reports of NSUN6-dependent and LncRNA-mediated methylation at the Lys59 position of MST1 (serine/threonine kinase) [Citation16]. Finally, NSUN7, an m5C RNA methyltransferases, is found to undergo epigenetic inactivation in liver cancer, leading to the incorrect methylation of mRNA [Citation17].
The m5C RNA methyltransferase NSUN5 is a conservative RNA methyltransferase belonging to the NSUN domain family [Citation18,Citation19]. Recent research findings that NSUN5 plays the role of the ‘writer’ of m5C methyltransferase, playing an important role in the regulation of mRNA splicing, translation, and stability [Citation20]. However, knowledge regarding the function and transcriptome-wide distribution of NSUN5 remains extremely limited. Zhang et al. recently reported that single-gene Nsun5 knockout homozygous mice exhibit spatial cognitive defects [Citation21]. A new study demonstrates that NSUN5 suppresses ferroptosis in bone marrow-derived mesenchymal stem cells through interaction with TRAP1 and m5C modification of FTH1 and FTL mRNA [Citation22]. However, the role of NSUN5 in the kidney remains unclear. Therefore, studies in different cell types, tissues, and organs are necessary to gain insight into possible functions of this modification and its implications for other regulatory processes.
In this study, in order to investigate the role of NSUN5 in mice, we utilized the CRISPR/Cas9 system to create a mouse model (Nsun5-/-). We studied the phenotype of the Nsun5-/- mice, as well as the mechanisms though which NSUN5 plays a role. Our findings highlight the crucial role of NSUN5 in preventing the accumulation of DNA damage and cell apoptosis in the kidney.
Materials and methods
Generation of Nsun5 null mice
The mice were maintained at a temperature range of 23 ± 2 °C, with a humidity range of 55 ± 5%, with a 12 h light/dark cycle in the Animal Center of Shandong Normal University. They were provided with unrestricted access to food and water. All animal experiments were carried out in compliance with the Guidelines for the Care and Use of Laboratory Animals of Shandong Normal University and were authorized by the Animal Ethics Committee of Shandong Normal University (No: AEECSDNU2021036).
The CRISPR/Cas9 genome editing system was used to generate the Nsun5 knockout mouse model (Nsun5-/-). We employed CRISPR technology to induce a targeted disruption of the protein coding region of the Nsun5 gene. In mouse zygotes, the DNA repair mechanism known as non-homologous end joining (NHEJ) was utilized, resulting in a deletion within the protein coding region. This deletion effectively inactivated the Nsun5 protein, thereby achieving the objective of knocking out the Nsun5 gene. The most recent version of the spCas9.1 enzyme was employed to cleave the genomic DNA. Specifically, exon3-4 (73–167 amino acids) spans a length of 284 base pairs, which is not divisible by three. This exon is situated within the conserved functional domain known as the Methyltr_RsmF_N superfamily (95–424 amino acids). Consequently, deleting the exon3-4 segment would result in the complete loss of the gene’s function. Therefore, Cas9 target sites were strategically designed flanking exon3-4 with the intention of removing approximately one kilobase of the genome, thereby facilitating the knockout of the Nsun5 gene. Subsequently, the guide RNA transcribed in vitro was injected into the cytoplasm of the zygote with a C57BL/6J genetic background to generate founder mice. The specific sequences of the guide RNAs used in this study are as follows:
Nsun5-L3: CCAGCATTACAAAAAGACGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTT; Nsun5-R2: GTGCAGTCTGTAATCTTAGTTTTAGAGCTAGAAATAGCAAGTTAAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTT
F: CAGGAAGGGCTAAGTAGGACGA;
Wild Type (WT)-R: CTTGTTGGAGACTCTGGGTTC;
R: TCCGTAACAAGATCTGACGC
The mice obtained after genotype identification were divided into an experimental group and a control group, each containing five cages, with a male to female ratio of 1:2. Subsequently, the birth and survival of mice were observed and recorded daily for a total of eight months. The number of surviving mice were counted at birth and then again at postnatal day (PD) 21. The ratio of the two was considered the survival rate. During this period, each mouse was individually weighed to obtain its body weight using an electronic scale. At PD 21, mice observed to be in extremely poor condition were euthanized by neck dislocation and then dissected. The kidneys, liver, lungs, and heart were weighed using an electronic scale, snap-frozen in liquid nitrogen, and stored in a −80 °C freezer for future tissue sectioning and protein extraction.
Hematoxylin-Eosin (H&E) staining analysis
Mouse kidneys were dissected and fixed overnight in 4% paraformaldehyde at 4 °C. The fixed samples were processed using ascending grades of alcohol for dehydration (55%, 65%, 75%, 85%, 95%, 100%) and xylol for clearing, followed by embedding in paraffin wax to obtain blocks. The fixed tissues were then embedded in paraffin and cut into slices. The sections were deparaffinized and rehydrated using a xylene-alcohol gradient before being equilibrated to room temperature. After dewaxing under the aforementioned conditions, tissue sections were then stained with hematoxylin stain solution (Servicebio, Wuhan, China) for 5s. Rinse in running tap water. Re-dyeing with eosin stain solution (Servicebio) for 10s. Subsequently, the samples were dehydrated using an alcohol gradient and transparentized twice using xylene prior to being mounted with neutral gum (Biosharp). The morphology of the kidney was observed using an automatic digital slice scanner (Panoramic MIDI II, 3Dhitech, Hungary).
Periodic acid-Schiff (PAS) staining analysis
After dewaxing under the aforementioned conditions, then 100 μl of Periodic Acid solution was added to each sample. After incubating for 10 min in a dark, humid chamber, the Periodic Acid solution was removed, and the sections were washed for 5 min on a shaker in distilled water. Next, 100 μl of Schiff’s reagent (Servicebio) was added to each sample, and they were incubated in a humid chamber at 37 °C in the dark for 30 min to 1 h. Then, 100 μl of hematoxylin staining solution was added to each sample and left to stain for 30 s. Finally, the samples were dehydrated using an alcohol gradient, cleared with xylene twice, and mounted using neutral resin (Biosharp, Anhui, China). The morphology of the kidney was observed using an automatic digital slice scanner (Panoramic MIDI II, 3Dhitech, Hungary).
Sirius Red staining analysis
The kidney tissue was fixed in 4% paraformaldehyde and dehydrated using graded ethanol, then embedded in paraffin to produce 7 μm-thick tissue sections. Prior to pathological staining, the sections were baked in a 65 °C oven for 10 min and then deparaffinized with xylene and rehydrated with graded ethanol. After that, the tissue sections were stained with Sirius Red (Solarbio, Beijing, China) for 1 h, rinsed with running water to remove excess dye from the surface, and stained with hematoxylin (Servicebio) for 20–30 s to stain the nuclei. Excess dye was then removed with running water, and the sections were dehydrated with graded ethanol and made transparent using xylene twice before being fixed with neutral resin (Biosharp). Collagen fiber deposition of the kidney tissues in each group was observed under a light microscope (Panoramic MIDI II).
Transmission electron microscopy analysis
For transmission electron microscopy (TEM) analysis, we isolated kidney samples of size 1 mm3. These samples were fixed in 2.5% glutaraldehyde at room temperature for 2–3 h and then stored at 4 °C. Subsequently, the samples were rinsed five times for 15 min each, using a buffer solution consisting of sodium dihydrogen phosphate and disodium hydrogen phosphate (0.1 M, pH = 7.4). After this, the samples were fixed in 1% osmium tetroxide for 1.5 h in the dark. We then dehydrated the samples using ethanol and acetone, penetrated them with resin, and embedded them. Ultrathin tissue sections of 65 nm were prepared and stained with 2% uranyl acetate for 25 min and lead citrate for 7 min. Finally, the sections were washed and baked under infrared light for 10 min before being observed using a transmission electron microscope (Hitachi Limited, Tokyo, Japan). Each group comprised at least three mice.
Immunohistochemistry analysis
For immunohistochemistry analysis, the tissue sections were initially dewaxed and rehydrated using xylol and a graded alcohol series with decreasing concentrations of ethanol. To avoid interference from endogenous peroxidase activity, the sections were treated with 3% H2O2 for 15 min, followed by blocking with primary antibody diluent (Servicebio) for 1 h. Subsequently, the sections were incubated overnight at 4 °C with rabbit anti-γH2AX antibody (1:200, BIOSS, Beijing, China) and rabbit anti-CASPASE-3 antibody (1:200, HUABIO, Hangzhou, China). After washing the sections three times with PBS, goat anti-rabbit secondary antibody (1:200, ZSGB-BIO, Beijing, China) was applied for 1 h at room temperature. After three additional washes with PBS, a DAB kit (ZSGB-BIO) was used to generate positive signals, followed by counterstaining with hematoxylin. Dehydration, transparentization, and mounting were performed as outlined earlier in this section. Images were captured using an automated digital slide scanner (Panoramic MIDI II), and Image J 1.4.3.67 software (Media Cybernetics, Silver Spring, MD, USA) was used to analyze and express staining results as the average optical density (AOD).
Quantitative polymerase chain reaction (qPCR) analysis
Total RNA was extracted using TRIzol reagent (Invitrogen, California, USA) and reverse transcribed with a cDNA synthesis kit (YEASEN, Shanghai, China) following the manufacturer’s instructions. The resulting cDNAs were then amplified through qPCR with the addition of SYBR Green Master Mix (YEASEN) in a Light Cycler qPCR instrument (Roche LC96, Basel, Switzerland) following the specifications provided by the manufacturer. qPCR was conducted under the following conditions: a 5 min at 95 °C, followed by 35 cycles of 30 s at 95 °C, 30 s at 58 °C, 30 s at 72 °C, and a 10 min extension at 72 °C. Gene expression changes were normalized to Actb and analyzed by the 2−ΔΔCt method [Citation23]. The primer sequences used for qPCR were as follows:
P53 F: GAAGTCCTTTGCCCTGAAC;
P53 R: CTAGCAGTTTGGGCTTTCC.
Caspase-3 F: TGGTGATGAAGGGGTCATTTATG;
Caspase-3 R: TTCGGCTTTCCAGTCAGACTC.
Actb F: CATATTGCGATCCTGGAATGAG;
Actb R: GATGAACCTGGCTGACTATCA.
Western blot
The total proteins extracted from the tissue were obtained using RIPA buffer (Beyotime, Shanghai, China), which contained PMSF (Beyotime) to inhibit protease degradation. Next, the samples were separated via electrophoresis on an SDS-PAGE gel (Solarbio) and then transferred onto polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). After that, the membranes were incubated overnight at 4 °C with one of the primary antibodies: γH2AX (BIOSS, Beijing, China) at a 1:500 dilution, CASPASE-3 (HUABIO, Hangzhou, China) at a 1:500 dilution, and β-actin (HUABIO) at a 1:5000 dilution. Next, a horseradish peroxidase-labeled secondary antibody (Thermo Fisher, Waltham, MA USA) was added and incubated at room temperature for 1 h. Finally, the protein bands were developed using an ECL Western Blot Kit (Millipore, Bedford, MA, USA), and the relative intensities of the bands were determined using Quantiscan software (Biosoft, Cambridge, United Kingdom).
Statistical analyses
GraphPad Prism 7.0 software was used to analyze all the experimental data obtained in this study. The results are presented as the mean ± SEM values. The differences between the groups were assessed with a Student’s t-test. One-way ANOVA was used to compare multiple groups. p < 0.05 was considered statistically significant.
Results
Nsun5 knockout leads to significantly reduced survival rate in mice
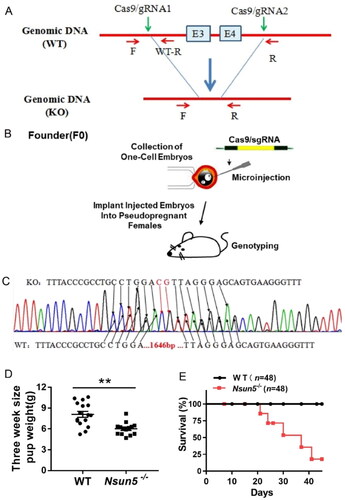
To generate Nsun5 knockout mice, we chose exons 3–4 of Nsun5 for mutation. The mutation was transferred from foreign donor DNA to the recipient chromosome through nonhomologous recombination at the target site using either Cas9 or gRNA (). Founder mice were prepared using single-cell embryo microinjection (). Subsequently, heterozygous hybridization was used to produce homozygous Nsun5-/- mice, and the direct sequencing results are shown in . Studies showed that the body weight of Nsun5-/- mice at PD 21 was significantly reduced (). Compared to WT mice, Nsun5 deficiency resulted in a gradual decrease in the survival rate. The survival rate gradually decreased from 100% at PD 21 to 20% at 40 days after birth. However, the mice that managed to survive appeared to live normally (for each genotype, n = 48) (). These results indicate that deletion of Nsun5 poses a threat to the life of mice, resulting in a significant reduction in survival rate.
Figure 1. Nsun5 knockout mice resulted in a significant reduction in survival.
(A) Strategy for generating Nsun5 deficiency mice involved selecting exon 3–4 of Nsun5 for mutation, and using Cas9 or gRNA to transfer mutations from exogenous donor DNA into the recipient chromosomes by nonhomologous recombination at the target site. (B) The pooled system of single-cell embryo microinjection was utilized to generate founder mice. (C) Homozygote Nsun5 deficiency mice were directly sequenced and the results were obtained. (D) The weight of PD21 individual pups was observed to be reduced in Nsun5-/- mice compared to WT mice. For each genotype, n = 6. (E) The survival of Nsun5-/- mice was measured. For each genotype, n = 48. The data, represented as mean ± SEM analyzed using the Student’s t-test, *p < 0.05, **p < 0.01.
Nsun5-/-mice suffered from multiple organ damage
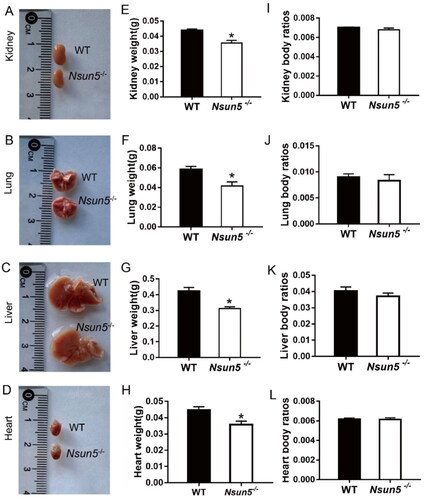
In order to investigate the cause of death of Nsun5-/- mice, we collected kidneys, lungs, livers, and hearts of PD 21 mice for biological observation. Representative images of each organ are shown in . We found that the weight of the kidneys (), lungs (), livers (), and hearts () of PD 21 Nsun5-/- mice were significantly reduced. However, there was no significant difference in kidney body ratio (), lung body ratio (), liver body ratio (), and heart body ratio () of PD 21 mice.
Figure 2. Nsun5-/- mice suffered multiple organ damages.
(A–D) Representative photos of the kidney (A), lung (B), liver (C), and heart (D) sizes at PD21 pups are shown. (E–H) The weight of kidney (E), lung (F), liver (G), and heart (H) in PD21 pups were reduced in Nsun5-/- mice compared to WT mice. (I–L) There were no significant difference in kidney body ratios (I), lung body ratios (J), liver body ratios (K), and heart body ratios (L) of PD21 pups. For each genotype, n = 5–7. The data presented are the mean ± SEM and were analyzed using the Student’s t-test, *p < 0.05, **p < 0.01.
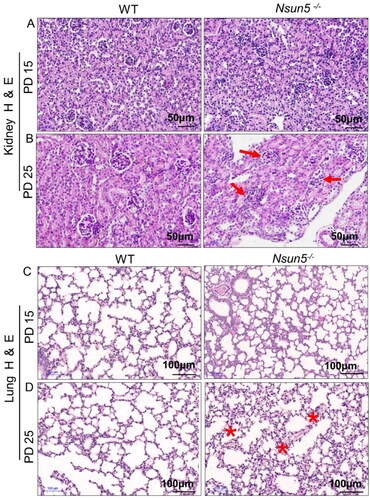
To further investigate the effect of Nsun5 deficiency on the organs of mice, we observed the morphological changes of the organs through H&E staining. Compared with the kidneys of WT mice, the kidney tissue of Nsun5-/-mice showed no significant difference on PD 15 (). However, PD 25, the kidney tissue of Nsun5-/- mice showed severe defects and glomerular degeneration (). Similarly, the lung tissue of Nsun5-/- mice showed no significant difference compared to WT mice on PD 15 (), but on PD 25, the lung alveolar walls of Nsun5-/- mice appeared thicker (). These results suggest that Nsun5 deficiency not only reduces organ weight in mice but also causes damage to kidney and lung tissue, with the kidneys being more severely affected.
Figure 3. Analysis of kidney and lung tissue in Nsun5-/- mice.
(A) Representative images of H&E-stained kidney sections of Nsun5-/- and WT mice at postnatal day (PD) 15 showed normal morphology. (B) Representative images of H&E-stained kidney sections of Nsun5-/- and WT mice at PD 25 exhibited glomerular degeneration (red arrows). Scale bars: 50μm. (C) Representative images of H&E-stained lung sections of Nsun5-/- and WT mice at PD 15 showed normal tissue structure. (D) Representative images of H&E-stained lung sections of Nsun5-/- and WT mice at PD 25 displayed incrassated thickening of alveolar walls (red stars). Scale bars: 100μm.
Nsun5-/- mouse lethal damage of kidney
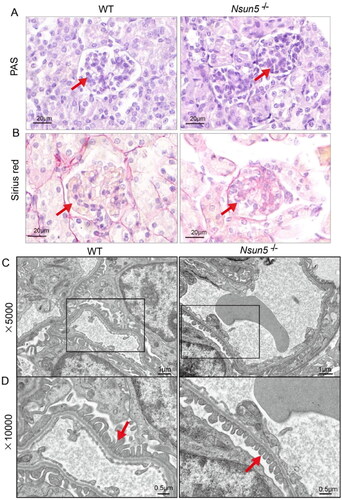
Based on our previous research, we have discovered that Nsun5-/-mice experience the most severe kidney damage and tissue defects on PD 25. This led us to speculate that this may be the underlying cause of their mortality. To test our hypothesis, we continued further investigate into the condition of the Nsun5-/-mice’s kidneys on PD 25. PAS staining revealed significant glycogen deposition in the glomeruli of the Nsun5-/-mice (indicated by the red arrow in ). Additionally, Sirius Red staining showed notable collagen fiber hyperplasia in the glomeruli of the Nsun5-/-mice compared to the control group (also indicated by the red arrow in ). Furthermore, electron microscopy revealed shortened foot processes of the glomeruli in the Nsun5-/-mice (red arrow in ). Based on these findings, it can be inferred that Nsun5 deficiency leads to lethal damage of kidney in mice, which ultimately results in their demise.
Figure 4. Kidney analysis of Nsun5-/- mice.
(A) Representative images of kidney sections stained with periodic acid-Schiff (PAS) from Nsun5-/- and WT mice at PD 25 show significant glomerular glycogen deposition (red arrows). Scale bars: 20μm. (B) Representative images of kidney sections stained with Sirius red from Nsun5-/- and WT mice at PD 25 show significant glomerular collagen fiber hyperplasia (red arrows). Scale bars: 20μm. (C–D) Under electron microscopy, shortened podocyte foot processes (red arrows) can be observed in Nsun5-/- mice compared to WT mice at PD 25. The magnifications in (C) and (D) are 5000 × and 10,000×, respectively.
Nsun5 deficiency induces the accumulation of DNA damage and apoptosis
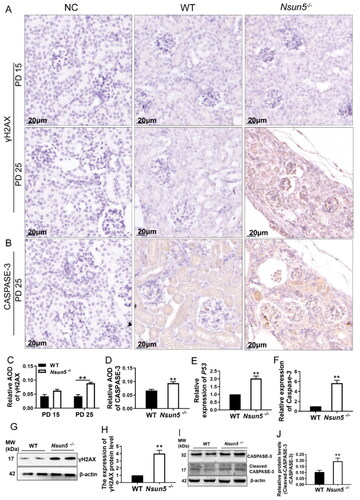
In order to further investigate the mechanism of kidney damage, we measured the expression levels of kidney γH2AX in mice on PD 15 and PD 25. Previous studies have indicated that γH2AX serves as a marker for DNA damage. Therefore, we detected its expression in the kidney through immunohistochemical staining. We observed elevated expression level of kidney γH2AX () and CASPASE-3 () in Nsun5-/- mice. Additionally, we quantified the expression levels of γH2AX () and CASPASE-3 (). The results indicated that the expression levels of both γH2AX and CASPASE-3 were significantly higher on PD 25 compared to those on PD 15 in the kidneys of Nsun5-/- mice. Moreover, we also conducted qPCR to analyze the mRNA expression levels of P53 () and Caspase-3 (), which revealed significantly increases in both. Further detection of the expression levels of γH2AX and CASPASE-3 via Western Blot showed that the expression levels of γH2AX () and Cleaved-CASPASE-3 () were significantly increased, consistent with earlier findings. Hence, we can conclude that Nsun5-/- mice experience severe kidney damage and even death due to heightened DNA damage and increased apoptotic accumulation.
Figure 5. Nsun5-/- mice exhibited high expression of apoptosis signal and DNA damage in the kidneys.
(A) Immunohistochemistry showed that the staining intensity of the γH2AX positive cells is increased in the kidney Nsun5-/- mice compared to WT at PD 25 not PD 15. Scale bar: 20μm. (B) Immunohistochemistry showed that the staining intensity of the CASPASE-3 positive signal is increased in the kidney of Nsun5-/- mice compared to WT at PD 25. (C-D) Average optical density (AOD) was used for the quantitative analysis of immunohistochemistry staining, and the data are shown in bar graphs. (E-F) The mRNA expression levels of P53 (E) and Caspase-3 (F) in the kidney of Nsun5-/- mice were higher than those in WT mice (n = 6-8 per genotype). (G) Results of Western blotting showed that the protein level of γH2AX was increased in the kidney Nsun5-/- mice compared to those in WT. (H) The results of Western blotting were quantified and displayed in bar graphs. The values are given as mean ± SEM analyzed using Student’s t-test. * P < 0.05, ** P < 0.01 (I) Results of Western blotting showed that the protein level of CASPASE-3 was increased in the kidney of Nsun5-/- mice compared to those in WT. (J) The results of Western blotting were quantified and displayed in bar graphs. The values are given as mean ± SEM analyzed using Student’s t-test. * p < 0.05, ** p < 0.01.
Discussion
In this study, we aimed to investigate the role of NSUN5 in mice. To achieve this, we utilized the CRISPR/Cas9 system to generate an Nsun5 knockout mouse model. Our findings demonstrate that Nsun5-/- mice exhibit reduced body weight and a gradual decrease in survival rate, reaching only 20% after PD 21. These mice also displayed multiple organ damage, with the kidney suffering the most severe damage. Further analysis revealed lethal damage of kidney, including kidney glycogen deposition and fibrosis. Additionally, there was a notable shortening in the primary foot processes of glomeruli in Nsun5-/- mice. Our mechanistic studies uncovered a significant increase in γH2AX and CASPASE-3 in the kidneys of PD 21 Nsun5-/- mice, indicating severe DNA damage and apoptosis. Taken together, our research provides evidence that NSUN5 plays a pivotal role in kidney development, and its absence can have detrimental effects on the kidneys.
NSUN5 belongs to the NSUN family. Some NSUN family proteins are associated with the regulation of protein translation, RNA processing, metabolism, and cell differentiation [Citation24–26]. For example, NSUN2 and NSUN4 have been shown to participate in cell proliferation, differentiation, and protein biosynthesis [Citation27]. Additionally, NSUN5 deficiency has been linked to a reduction in protein synthesis during mammalian development [Citation28]. In this study, we reported for the first time that the lack of NSUN5 contributes to kidney damage, ultimately putting individuals at risk of life-threatening complications. Kidney disease is rapidly becoming one of the leading global causes of death and poses a serious threat to human health [Citation29]. Therefore, it is imperative that we urgently investigate the underlying mechanisms of kidney disease.
Kidney is one of the vital organs in the body and is indispensable for maintaining the normal operation of the urinary system. The population with kidney injuries manifest both the structural and functional changes [Citation30]. Kidney injury is characterized by a decline in kidney function and is a global public health concern with high morbidity and mortality [Citation31]. The etiology of acute kidney injury and chronic kidney disease is heterogeneous [Citation32]. Apoptosis, the major form of cell death, contributes to the decline in kidney function. According to report, in the kidney, apoptosis contributes to the loss of parenchymal cell during acute and chronic kidney injury [Citation33]. Several types of apoptosis have recently been identified as regulated processes, mediated by diverse molecules [Citation34]. However, the factors triggering kidney injury still need to be investigated in detail. Damage to the kidneys can lead to a series of adverse consequences, posing a threat to the patient’s physical health. In this study, we not only found a significant reduction in the weight of mouse kidneys after Nsun5 knockout but also observed lethal damage to the kidneys.
Cell survival is closely related to DNA damage, especially in tissues and organs that undergo extensive cell proliferation and differentiation events. DNA damage can lead to genetic mutations, which can delay cell cycle progression and trigger cell death [Citation35]. Various types of mutations can arise from DNA damage, eventually leading to diseases such as cancer [Citation36] and aging [Citation37]. In the course of conducting pathway analyses, it was determined that the DNA damage response is implicated in kidney injury. Specifically, gene mutations associated with DNA damage reactions that interfere with normal kidney function have the potential to result in embryonic mortality [Citation38]. γH2AX has been widely used by many researchers as a tool to detect DNA damage [Citation39]. P53 plays a key role in response to DNA damage, enabling a variety of cellular responses including cell cycle arrest, apoptosis, and DNA repair [Citation40]. Caspase-3, an effector caspase, plays a crucial role in initiating apoptosis [Citation41]. In the course of apoptosis, caspase-3 becomes activated and subsequently cleaves various downstream substrates, resulting in significant morphological alterations in cells undergoing apoptosis [Citation42]. In our study, we observed elevated expression levels of kidney γH2AX, P53, and cleaved Caspase-3 in Nsun5-/- mice. Our findings suggest that Nsun5 deficiency leads to severe DNA damage and cell apoptosis, resulting in lethal kidney damage and death before puberty in mice. In summary, the excessive DNA damage and cell apoptosis during kidney development are important factors contributing to the death of Nsun5-/- mice. However, since the analysis was carried out at PD 21 when Nsun5-/- mice started to die, the observed phenotype (increase in apoptotic signals) might be a phenomenon in the dying process.
This research has several limitations. Firstly, the 7-micrometer-thick slices potentially had biases in their evaluation toward degeneration or accumulation of PAS or Sirius Red-positive substances. A thinner slice would be beneficial for improving the accuracy of results in tissue analysis. This needs further improvement. Secondly, it remains unclear how the absence of NSUN5 in the kidney is related to epigenetic events and epigenetic transcription events. It is not yet known whether this phenomenon is caused by insufficient m5C methylation of DNA damage and apoptosis genes. Last, it has yet to be examined if the mechanism of NSUN5 deficiency affecting other organ systems is the same as that observed in the kidney. If these findings can be corroborated in larger prospective clinical studies, they may prove helpful in managing patients with kidney disease.
Conclusion
In conclusion, we found that mice lacking NSUN5 died before puberty due to kidney fatal damage caused by DNA damage and cell apoptosis. Taken together, these results suggest that NSUN5 is associated with reduced DNA damage and cell apoptosis in the kidney, which is essential for preventing fatal damage to the kidney.
Ethical approval
All animal experiments were carried out in compliance with the Guidelines for the Care and Use of Laboratory Animals of Shandong Normal University and were authorized by the Animal Ethics Committee of Shandong Normal University.
Informed consent statement
Not applicable.
Authors’ contributions
CZ and HYZ conceived and designed the study. HYZ performed the experiments and drafted the manuscript. XHL acquired and analyzed data. CZ and HYZ revised the manuscript. All authors read and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
This manuscript contains all of the data produced during this research.
Additional information
Funding
References
- Gao Y, Fang J. RNA 5-methylcytosine modification and its emerging role as an epitranscriptomic mark. RNA Biol. 2021;18(sup1):1–13. doi: 10.1080/15476286.2021.1950993.
- Trixl L, Lusser A. The dynamic RNA modification 5-methylcytosine and its emerging role as an epitranscriptomic mark. WIREs RNA. 2019;10(1):e1510. doi: 10.1002/wrna.1510.
- Breiling A, Lyko F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin. 2015;8:24.
- Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m(5)C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes . 2019;10(2):102. doi: 10.3390/genes10020102.
- Liu Y, Siejka-Zielińska P, Velikova G, et al. Bisulfite-free direct detection of 5-methylcytosine and 5-hydroxymethylcytosine at base resolution. Nat Biotechnol. 2019;37(4):424–429. doi: 10.1038/s41587-019-0041-2.
- Chen YS, et al. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip Rev RNA. 2021;12(4):e1639.
- Campbell EM, et al. Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with tat for TAR binding and methylation. PLOS Pathog. 2020;16(3):e1008430. doi: 10.1371/journal.ppat.1008430.
- Flores JV, Cordero-Espinoza L, Oeztuerk-Winder F, et al. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Reports. 2017;8(1):112–124. doi: 10.1016/j.stemcr.2016.11.014.
- Squires JE, Patel HR, Nousch M, et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40(11):5023–5033. doi: 10.1093/nar/gks144.
- Chi L, Delgado-Olguin P. Expression of NOL1/NOP2/sun domain (nsun) RNA methyltransferase family genes in early mouse embryogenesis. Gene Expr Patterns. 2013;13(8):319–327. doi: 10.1016/j.gep.2013.06.003.
- Bourgeois G, Ney M, Gaspar I, et al. Eukaryotic rRNA modification by yeast 5-methylcytosine-methyltransferases and human proliferation-associated antigen p120. PLOS One. 2015;10(7):e0133321. doi: 10.1371/journal.pone.0133321.
- Sajini AA, Choudhury NR, Wagner RE, et al. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun. 2019;10(1):2550. doi: 10.1038/s41467-019-10020-7.
- Haag S, Sloan KE, Ranjan N, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. Embo J. 2016;35(19):2104–2119. doi: 10.15252/embj.201694885.
- Trixl L, Amort T, Wille A, et al. RNA cytosine methyltransferase Nsun3 regulates embryonic stem cell differentiation by promoting mitochondrial activity. Cell Mol Life Sci. 2018;75(8):1483–1497. doi: 10.1007/s00018-017-2700-0.
- Metodiev MD, Spåhr H, Loguercio Polosa P, et al. NSUN4 is a dual function mitochondrial protein required for both methylation of 12S rRNA and coordination of mitoribosomal assembly. PLOS Genet. 2014;10(2):e1004110. doi: 10.1371/journal.pgen.1004110.
- Li C, Wang S, Xing Z, et al. A ROR1-HER3-lncRNA signalling axis modulates the Hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19(2):106–119. doi: 10.1038/ncb3464.
- Ortiz-Barahona V, Soler M, Davalos V, et al. Epigenetic inactivation of the 5-methylcytosine RNA methyltransferase NSUN7 is associated with clinical outcome and therapeutic vulnerability in liver cancer. Mol Cancer. 2023;22(1):83. doi: 10.1186/s12943-023-01785-z.
- Schosserer M, Minois N, Angerer TB, et al. Methylation of ribosomal RNA by NSUN5 is a conserved mechanism modulating organismal lifespan. Nat Commun. 2015;6(1):6158. doi: 10.1038/ncomms7158.
- Sharma S, Yang J, Watzinger P, et al. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013;41(19):9062–9076. doi: 10.1093/nar/gkt679.
- Dominissini D, Rechavi G. 5-methylcytosine mediates nuclear export of mRNA. Cell Res. 2017;27(6):717–719. doi: 10.1038/cr.2017.73.
- Zhang T, Chen P, Li W, et al. Cognitive deficits in mice lacking Nsun5, a cytosine-5 RNA methyltransferase, with impairment of oligodendrocyte precursor cells. Glia. 2019;67(4):688–702. doi: 10.1002/glia.23565.
- Liu J, Ren Z, Yang L, et al. The NSUN5-FTH1/FTL pathway mediates ferroptosis in bone marrow-derived mesenchymal stem cells. Cell Death Discov. 2022;8(1):99. doi: 10.1038/s41420-022-00902-z.
- Green MR, Sambrook J. Analysis and normalization of real-time polymerase chain reaction (PCR) experimental data. Cold Spring Harb Protoc. 2018;2018(10):769–777. doi: 10.1101/pdb.top095000.
- Yang X, Yang Y, Sun B-F, et al. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017;27(5):606–625. doi: 10.1038/cr.2017.55.
- Hussain S, Sajini AA, Blanco S, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4(2):255–261. doi: 10.1016/j.celrep.2013.06.029.
- Harris T, Marquez B, Suarez S, et al. Sperm motility defects and infertility in male mice with a mutation in Nsun7, a member of the sun domain-containing family of putative RNA methyltransferases. Biol Reprod. 2007;77(2):376–382. doi: 10.1095/biolreprod.106.058669.
- Sun Z, Xue S, Zhang M, et al. Aberrant NSUN2-mediated m(5)C modification of H19 lncRNA is associated with poor differentiation of hepatocellular carcinoma. Oncogene. 2020;39(45):6906–6919. doi: 10.1038/s41388-020-01475-w.
- Heissenberger C, Liendl L, Nagelreiter F, et al. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 2019;47(22):11807–11825. doi: 10.1093/nar/gkz1043.
- Sanz AB, Sanchez-Niño MD, Ramos AM, et al. Regulated cell death pathways in kidney disease. Nat Rev Nephrol. 2023;19(5):281–299. doi: 10.1038/s41581-023-00694-0.
- Wang W-J, Cai G-Y, Chen X-M. Cellular senescence, senescence-associated secretory phenotype, and chronic kidney disease. Oncotarget. 2017;8(38):64520–64533. doi: 10.18632/oncotarget.17327.
- Xia W, Li Y, Wu M, et al. Gasdermin E deficiency attenuates acute kidney injury by inhibiting pyroptosis and inflammation. Cell Death Dis. 2021;12(2):139. doi: 10.1038/s41419-021-03431-2.
- Scholz H, Boivin FJ, Schmidt-Ott KM, et al. Kidney physiology and susceptibility to acute kidney injury: implications for renoprotection. Nat Rev Nephrol. 2021;17(5):335–349. doi: 10.1038/s41581-021-00394-7.
- Priante G, et al. Cell death in the kidney. Int J Mol Sci. 2019;20(14):3598.
- Linkermann A, Chen G, Dong G, et al. Regulated cell death in AKI. J Am Soc Nephrol. 2014;25(12):2689–2701. doi: 10.1681/ASN.2014030262.
- Rich T, Allen RL, Wyllie AH. Defying death after DNA damage. Nature. 2000;407(6805):777–783. doi: 10.1038/35037717.
- Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168(4):644–656. doi: 10.1016/j.cell.2017.01.002.
- Tian X, Firsanov D, Zhang Z, et al. SIRT6 is responsible for more efficient DNA double-strand break repair in Long-Lived species. Cell. 2019;177(3):622–638 e22. doi: 10.1016/j.cell.2019.03.043.
- Gupta N, Matsumoto T, Hiratsuka K, et al. Modeling injury and repair in kidney organoids reveals that homologous recombination governs tubular intrinsic repair. Sci Transl Med. 2022;14(634):eabj4772. doi: 10.1126/scitranslmed.abj4772.
- Yan L-X. γ-H2AX – a novel biomarker for DNA double-strand breaks. In Vivo. 2008;22:305–310.
- Rodriguez-Pastrana I, Birli E, Coutts AS. p53-dependent DNA repair during the DNA damage response requires actin nucleation by JMY. Cell Death Differ. 2023;30(7):1636–1647. doi: 10.1038/s41418-023-01170-9.
- Lan S, Yang B, Migneault F, et al. Caspase-3-dependent peritubular capillary dysfunction is pivotal for the transition from acute to chronic kidney disease after acute ischemia-reperfusion injury. Am J Physiol Renal Physiol. 2021;321(3):F335–F351. doi: 10.1152/ajprenal.00690.2020.
- Eskandari E, Eaves CJ. Paradoxical roles of caspase-3 in regulating cell survival, proliferation, and tumorigenesis. J Cell Biol. 2022;221(6):e202201159. doi: 10.1083/jcb.202201159.

