Abstract
Aim: The intestinal flora may be washed out after bowel cleansing and may be beneficially influenced by probiotics during recolonization. In this randomized placebo-controlled double-blind study, the effect of bowel cleansing with or without subsequent inoculation by Lactobacillus plantarum 299v was assessed. Patients and methods: Patients who underwent bowel cleansing were included. After colonoscopy, patients consumed a drink with or without L. plantarum 299v (1011 cfu/day). Faecal samples were collected: before bowel cleansing (week 0), the first sample after colonoscopy (week 1), after 2 and 4 weeks of consumption (week 2 and week 4), and 4 weeks after cessation of the drinks (week 8). Total colony forming units (cfu) of bacteria per gram faeces were counted. Denaturing gradient gel electrophoresis (DGGE) was used to analyse the dominant flora, expressed as ‘similarity values’. Results: Twenty-two patients completed the study (12 probiotic, 10 placebo). The mean concentration of lactobacilli increased (p<0.05) from 4.7±1.9 at week 0 to 6.5±0.8 at week 2 in the probiotic group and returned back to 4.6±1.4 at week 8 (p<0.05). DGGE profiles demonstrated high similarity values within subjects over time: mean value 86.5%±2.6. No significant differences in similarity values were seen after bowel cleansing (week 2) or between probiotic and placebo groups. Conclusions: Although counts of faecal lactobacilli increased during probiotic consumption, no further changes in culture and DGGE results were observed. Therefore, the faecal flora can be considered as relatively stable over time: bowel cleansing or probiotic consumption had no major influence on the composition of the faecal flora.
Introduction
The human gastrointestinal tract comprises a complex ecosystem with a myriad of bacteria that probably have substantial impact on the physiology and health of the host. The highest numbers of bacteria are found in the colon, which contains about 1011 mainly anaerobic (99.9%) bacteria per gram intestinal content. The predominant isolates found in the colon are Bacteroides spp., bifidobacteria, Clostridium spp. and eubacteria Citation[1], Citation[2]. The intestinal flora contains enzymes, and produces vitamins and short chain fatty acids from non-absorbed carbohydrates and plays a role in the defence against pathogens by influencing the barrier function of the gut, the immune system and the colonization resistance of the intestinal tract Citation[2]. The intestinal flora is suspected to play a role in the pathogenesis of gastrointestinal diseases such as ulcerative colitis, Crohn's disease and irritable bowel syndrome Citation[3], Citation[4].
Each individual is thought to develop a unique, stable and complex intestinal flora (i.e. ‘bacterial fingerprint’) after birth by ingestion of environmental microbes colonizing the previously sterile colon Citation[5]. After 2 years of age, the intestinal flora has stabilized and adapted to an ‘adult-like’ composition influenced by genotype and environmental factors Citation[5]. High similarity values between the bacterial fingerprints of monozygotic twins have been found compared with the similarity values found in marital partners or genetically unrelated individuals. This demonstrates the important role of genetic factors in influencing the composition of the intestinal flora Citation[6]. Furthermore, environmental factors like diet and antibiotic use have been shown to influence the intestinal flora Citation[7]. Adequate knowledge of factors influencing the composition of the intestinal flora may give further insight into the development and pathogenesis of gastrointestinal diseases and may provide opportunities to modulate the flora with the aim of therapeutic intervention.
Bowel cleansing, required as preparation for colonoscopic examinations, could wash out the luminal bacteria from the gastrointestinal tract. In patients with Crohn's disease bowel cleansing has been found to decrease disease activity and to induce clinical improvement Citation[8–10]. Van den Bogaard et al. demonstrated a reduction in aerobic as well as in anaerobic faecal bacteria and found that the intestinal flora had to recolonize after bowel cleansing Citation[11]. Probiotics may have a beneficial effect in this recolonization phase.
Probiotics are defined as ‘mono- or mixed cultures of live micro-organisms which, when applied to animal or man, beneficially affect the host by improving the properties of the indigenous flora’ Citation[12]. Intake of the probiotic strain Lactobacillus plantarum 299v has been demonstrated to increase the numbers of faecal lactobacilli Citation[13], Citation[14]. Moreover, L. plantarum 299v was shown to adhere to human mucosa cells in vitro, competing with the colonization of E. coli, and is able to survive passage through the gastrointestinal tract. This bacterium may be a suitable candidate to influence the recolonization of the intestinal flora after bowel cleansing Citation[15], Citation[16].
Changes in the composition of the intestinal flora can be studied by culture techniques. Due to insufficiently selective media, it is estimated that over 50% of the bacteria in the gastrointestinal tract have not yet been cultured and/or are not yet identified Citation[17], Citation[18]. For these reasons the intestinal flora should preferably be studied with molecular techniques. Polymerase chain reaction (PCR) in combination with denaturing gradient gel electrophoresis (DGGE) is a useful technique to study changes in the dominant individual faecal flora over time by producing bacterial fingerprints Citation[19].
In this study the effect of bowel cleansing on the intestinal bacterial composition with and without subsequent treatment by the probiotic L. plantarum 299v was investigated using both culture techniques and PCR/DGGE analyses.
Patients and methods
Subjects
Twenty-three consecutive patients undergoing a bowel cleansing procedure prior to colonoscopy were enrolled in the study. Patients were not allowed to consume probiotics, prebiotics or antibiotics during the 2 weeks before the study or during the period of the study. The use of medication was registered and, if clinically possible, dose and type of the medication were kept stable during the study period. Participants were asked to continue their normal dietary habits. The study was approved by the Medical Ethical Committee of the University Hospital Maastricht and was performed in accordance with the declaration of Helsinki. All participants gave written informed consent.
Study design
The study was double-blind and placebo-controlled. The patients were randomized to consume probiotic or placebo drinks. The total duration of the study was 9 weeks: 1 week before bowel cleansing and colonoscopy (week 0), 4 weeks probiotic or placebo consumption after colonoscopy (weeks 1–4) and 4 weeks after cessation of the probiotic or placebo consumption (weeks 5–8).
Patients started with the consumption of the probiotic and placebo drinks in the evening after the colonoscopy. During week 1 until week 4, patients consumed 200 ml (100 ml in the morning and 100 ml in the evening) of a fermented oatmeal drink with L. plantarum 299v (109 cfu/ml; probiotic) or without L. plantarum 299v (placebo) daily. Probiotic and placebo drinks were indistinguishable as regards smell, taste and colour.
At 24 h before colonoscopy, the patients took the macrogol/electrolytes laxative Kleanprep® to clean the gastrointestinal lumen. After colonoscopy, the endoscopist completed a questionnaire about the effectiveness of the bowel cleansing in colon ascendens, transversum, descendens and rectum (‘very good’: no faeces visible, not even in the diverticula; ‘good’: some faeces visible without hampering the colonoscopy; ‘poor’: faeces visible, hampering the colonoscopy).
Five faecal samples were collected: one before bowel cleansing (week 0), the first faecal sample which could be produced after the colonoscopy (week 1), after consumption of probiotic or placebo for 2 and 4 weeks (weeks 2 and 4) and 4 weeks after cessation of the drinks (week 8). Together with the faecal sampling, the patients completed a questionnaire on medication, bowel habits (defecation frequency and faeces consistency scores from hard lumps (score 1) until watery diarrhoea (score 7) according to the Bristol scale Citation[20]), compliance, side effects and changes in dietary habits. Participants were excluded from analyses if <85% of the faecal samples were collected and/or <85% of the probiotic/placebo drinks were consumed.
Microbiological analyses
Faecal samples were collected and processed as described previously Citation[13]. Tenfold serial dilutions of the faecal samples were made in physiological saline and 40 µl were inoculated on agar plates to culture and count total (facultative) aerobic bacteria, enterobacteriaceae, enterococci, total (facultative) anaerobic bacteria, Bacteroides spp., clostridia, lactic acid bacteria and lactobacilli Citation[13].
DNA extraction
DNA from faecal samples was isolated with the fast DNA SPIN® Kit for soil (Q-biogene, Heidelberg, Germany) with disruption of bacterial cells by highly energetic mechanical means using the FastPrep® Instrument (Q-biogene, Heidelberg, Germany). Genomic DNA was purified with a proprietary silica matrix, eliminating contaminants that inhibit reactions.
PCR
PCR was performed with a Taq polymerase kit. The primers F-0968-GC and R-1401 used in this study amplify the V6–V8 region of 16S rRNA. The PCR mixture contained 5 µl of 10× PCR buffer containing 15 mM MgCl2, 3 µl of 50 mM MgCl2, 1 µl of deoxynucleoside triphosphate preparation (10 mM), 1 µl of each primer (10 µM), 0.25 µl of Taq DNA polymerase (5 U/µl), 37.75 µl sterile Milli-Q water and 1 µl of a 10-fold diluted DNA solution.
The following PCR program was used: pre-denaturation for 5 min at 94°C, 35 cycles of denaturation at 94°C for 30 s, annealing at 56°C for 20 s and elongation at 68°C for 45 s. The program ended with a post-elongation step at 68°C for 7 min followed by cooling down to 4°C. The amplification of the amplicons was confirmed on ethidium-stained 1% agarose gels for 20 min at 100 V.
DGGE analysis
PCR products were analysed on DGGE gels according to the method described by Zoetendal et al. Citation[17] with the following modifications. The 8% polyacrylamide gels contained a urea/formamide gradient from 30% to 60%. Electrophoresis was performed in 0.5× Tris acetic acid EDTA (TAE) buffer (2.42 g Tris, 570 µl acetic acid, 0.37 g EDTA) at 85 V for 16 h using the DCode System apparatus (Biorad, Hercules, CA, USA). Amplicons of isolates of L. plantarum 299v were electrophoresed next to the PCR products of the faecal samples as well as a marker. After electrophoresis, the gels were stained as described by Sanguinetti et al. using the rapid silver staining procedure Citation[21] and scanned at 400 DPI. DGGE profiles were compared using the Bionumerics software version 3.0 (Applied-Maths, Sint-Martens-Latem, Belgium). The similarity between the five faecal samples of each individual was determined by calculating the similarity value using the Pearson's product-moment correlation coefficient Citation[22].
Similarity values were called ‘deviant’ when the value was below the mean similarity value of the group minus one standard deviation. Similarity values at different time points were correlated with changes in type or dose of medication.
Statistical analyses
Statistical evaluation of changes in culture results between groups and within groups during the study period was carried out using the linear mixed model analysis with SPSS version 11.0. Non-parametric Wilcoxon signed-rank test and Mann-Whitney U test with a Bonferroni-Holm correction were used for comparison of DGGE results within and between groups, respectively. A p value of 0.05 was considered to be statistically significant.
Results
Twenty-two patients completed the study: 12 in the probiotic group (4 male, 8 female, mean age 46.5±8.7 years) and 10 in the placebo group (3 male, 7 female, mean age 55.0±16.1 years). No significant differences in gender and age were seen between the probiotic and placebo groups. One patient (probiotic group) was excluded because he collected only three of five faecal samples. Clinical diagnoses of patients at colonoscopy included: inflammatory bowel diseases (IBD) (n=5), irritable bowel syndrome (n=4), colorectal adenomas (n=5), diverticular disease (n=2), rectal ulcer (n=1) and ileocaecal resection (n=1). Four patients had normal findings at colonoscopy and had no diagnoses of gastrointestinal diseases. Patients used different types of medication such as corticosteroids (n=3), 5-ASA medication (n=4), proton pump inhibitors (n=3), laxatives or anti-diarrhoea medication (n=7), and other non-gastrointestinal medication such as antihistamines, analgesis and cholesterol-lowering medication (n=17). Ten patients changed the type and two patients the dose of their gastrointestinal medication during the study period.
No significant differences were observed in defecation frequency and faeces consistency scores between the probiotic and placebo groups or within both groups over time (). The mean period of time between colonoscopy and collection of the first faecal sample produced after colonoscopy (i.e. sample of week 1) was 82 h (range 20–146) in the probiotic group and 60 h (range 23–120) in the placebo group and did not differ significantly (p=0.46).
Table I. Defecation frequency (per day) and faeces consistency scores according to the Bristol scale (1, hard lumps to 7, watery diarrhoea) in week 0 (before bowel cleansing), week 1 (first faecal sample after colonoscopy), week 2 (after consumption of probiotic/placebo for 2 weeks), week 4 (after consumption of probiotic/placebo for 4 weeks) and week 8 (four weeks after cessation of the probiotic/placebo drinks) in 22 patients (mean±SD).
In 13 patients the effectiveness of the bowel cleansing was scored: 2 patients had a ‘poor’ bowel cleansing in the whole colon and rectum and 5 patients in some parts of the colon and rectum. The other six patients had a ‘very good’ or ‘good’ bowel cleansing procedure. Furthermore, no side effects were reported after the treatment with L. plantarum 299v or the placebo drink. The compliance was 96% for the probiotic drinks and 90% for the placebo drinks.
Culture
During the consumption of the drinks (weeks 2 and 4), a significant increase in the number of lactic acid bacteria (p=0.008) and lactobacilli (p<0.005) was observed in the probiotic group but not in the placebo group (). The number of lactic acid bacteria (p=0.006) and lactobacilli (p<0.005) decreased significantly after cessation of the drinks (week 8) compared with the number during the consumption of the probiotic drinks (week 2 and 4). No significant differences could be observed in other aerobic and anaerobic bacterial counts before (week 0) and after bowel cleansing (week 1) and during probiotic intake (week 2 and 4), with the exception of the total number of aerobic bacteria, which increased significantly (p=0.001) in the probiotic group in week 1 compared with week 0.
Table II. Number of bacterial strains (log cfu/g faeces) in week 0 (before bowel cleansing), week 1 (first faecal sample after colonoscopy), week 2 (after consumption of probiotic/placebo for 2 weeks), week 4 (after consumption of probiotic/placebo for 4 weeks) and week 8 (4 weeks after cessation of the probiotic/placebo drinks) in 22 patients (mean±SD).
DGGE analyses
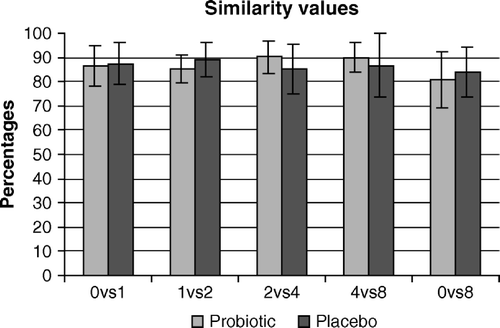
Faecal samples for DGGE analyses and calculations of similarity values were available for 20 patients (11 probiotic and 9 placebo group). Mean similarity values over time were relatively high (probiotic group = 86.8±3.1 and placebo group = 86.3±2.2) (). No major differences could be observed between probiotic and placebo groups or within the groups.
Figure 1. Mean similarity values (in percentages) in the probiotic and placebo group using PCR/DGGE. Comparisons were made between weeks 0 and 1 (0vs1), weeks 1 and 4 (1vs4), weeks 4 and 8 (4vs8) and weeks 0 and 8 (0vs8).
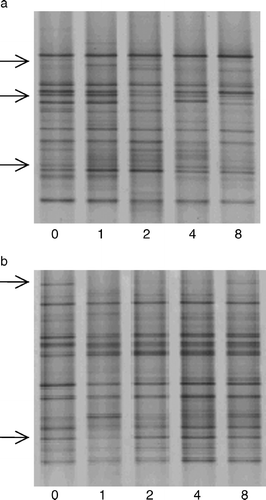
Eighteen individual similarity values were lower than the mean of the total group minus the standard deviation and therefore defined as ‘deviant’. These time points were checked for possible changes in type or dose of medication. No correlation could be seen between these time points and changes in medication except for one patient who stopped the use of loperamide in week 4 resulting in a ‘deviant’ similarity value when comparing week 2 to week 4.
shows the DGGE pattern of two individual patients during the study period: one from the probiotic and one from the placebo group. Changes in individual banding patterns could be observed in both probiotic and placebo patients mainly after bowel cleansing (n=11), and returned within 2 weeks. For example, in the probiotic patient (b), some bands disappeared after the bowel cleansing procedure (week 1 compared with week 0). However, in week 8 these bands were again present.
Discussion
In this study, pre-colonoscopic bowel cleansing procedure and subsequent consumption of L. plantarum 299v had no significant influence on the composition of the colonic flora based on both culture results and bacterial fingerprints of the dominant faecal flora, indicating that the composition of the faecal flora remains relatively stable over time despite these major interventions.
No changes in the faecal bacterial numbers due to bowel cleansing could be demonstrated by culture with the exception of the number of total aerobic bacteria. The effectiveness of the bowel cleansing (very good, good and poor) did not influence the culture results significantly (data not shown) and was successful in the majority of patients. The number of total aerobic bacteria increased significantly after bowel cleansing in all patients in the probiotic group. In contrast, van den Bogaard et al. demonstrated a reduction of log 2–3 in the faecal number of total aerobic bacteria and of log 4–5 in the number of total anaerobic bacteria after bowel cleansing Citation[11]. These inconsistent results might be explained by the use of different bowel cleansing procedures (saline versus Kleanprep). Moreover, van den Bogaard et al. collected clear rectal effluent immediately after bowel cleansing, while in the present study the first faecal sample after colonoscopy was collected with a mean period of time of 82 h in the probiotic and 60 h in the placebo group. This difference in faeces collection time comparing the probiotic to the placebo group could not be explained by differences in defecation frequency or faeces consistency.
The increase in the number of total aerobic bacteria might also be caused by the consumption of the probiotic drinks. However, the increase in aerobic bacteria did not persist during the probiotic consumption and disappeared in week 4. In addition, previous studies performed with healthy volunteers consuming L. plantarum 299v for 4 and 2 weeks, respectively, could not observe an increase in the number of total aerobic bacteria Citation[13], Citation[16].
Furthermore, the consumption of the probiotic, L. plantarum 299v, increased the amount of lactic acid bacteria and lactobacilli significantly with log 1.5 cfu/g faeces. These results were in accordance with the results of a previous study in which 20 healthy volunteers consumed L. plantarum 299v for 4 weeks Citation[13]. However, the number of lactobacilli recovered in faeces during probiotic consumption was lower in this study (log 6.8 cfu/g faeces) compared with the previous one (log 8.2 cfu/g faeces), while the daily dose of L. plantarum 299v was the same (1011 cfu/day). Comparison of the pretreatment faecal bacterial concentrations to the results found in a previous study with healthy volunteers Citation[13], demonstrated lower bacterial concentrations for total aerobic bacteria, enterococci, total anaerobic bacteria, Bacteroides spp. and lactic acid bacteria in the present study, although similar culture methods were used in both studies. These differences could be explained by higher defecation frequency and faeces consistency scores found in the present study, which can result in dilution of the faecal samples and lower numbers of bacteria counted per gram faeces. These looser stool samples could be due to the inclusion of various types of patients such as IBD patients. For example, decreased numbers of lactobacilli and aerobic bacteria have been reported in IBD patients in the literature Citation[3], Citation[23]. However, exclusion of the bacterial concentrations of the IBD patients did not result in an increase of bacterial concentrations.
In this study, PCR combined with DGGE analyses was found to be a useful additive tool to study the effects of bowel cleansing and probiotic consumption on the dominant faecal flora over time. The intra-individual similarity values calculated between the faecal samples obtained before and after bowel cleansing were relatively high with a mean of 86.5%. Zoetendal et al. calculated similarity values ranging from 66 to 88% comparing two faecal samples of four individuals over time in a 4 month period Citation[6]. In this study, bowel cleansing did not significantly influence the overall banding patterns of the dominant flora. Although bowel cleansing was successful, it cannot be ruled out that even very small amounts of remaining bacteria can result in a recolonization of the original flora.
The increase in the number of lactobacilli during probiotic consumption was not reflected in one single band of the molecular fingerprint, as the dominant flora consists of <1% of lactobacilli and universal primers used for the analyses of the dominant faecal flora detect the most prevalent species, representing 90–99% of the total faecal flora Citation[24].
Factors affecting the intestinal flora, except for bowel cleansing and L. plantarum 299v intake, were kept as stable as possible during the study period. However, medication taken in this study (such as sulfasalazine and proton pump inhibitors) has been demonstrated to influence the faecal bacterial composition Citation[25], Citation[26]. Changes in dose and type of these compounds could not be correlated to the ‘deviant’ similarity values found in this study except in one patient. This patient stopped the use of loperamide in week 4, resulting in changes in the banding patterns of the dominant flora and in a similarity value of 76%. Loperamide has been found to change the number of streptococci, but did not cause bacterial overgrowth in 14 children receiving the drug because of severe diarrhoea Citation[27].
Similarity values did not change due to bowel cleansing but some changes in individual bands occurred, mainly in the first week after bowel cleansing, suggesting that bowel cleansing could have an influence on specific bacterial species. Moreover, repeated bowel cleansing procedures might result in a more prominent elimination of the bacterial mass. To obtain further insight into bowel cleansing as a therapeutic modality in gastrointestinal diseases like IBD, the influences of bowel cleansing on specific bacterial subspecies have to be studied in defined patient groups using PCR/DGGE with specific primers instead of universal primers.
This study was performed with consecutive patients undergoing a bowel cleansing procedure instead of healthy volunteers, which could have influenced culture results and DGGE banding patterns of the dominant faecal flora. However, even in this heterogeneous population, the intestinal flora remained stable over time. Unfortunately, the numbers of patients with specific clinical diagnoses were too small to compare these groups statistically and in a future study more patients should be included.
Seven patients in this study were older than 60 years. As the faecal flora might be changed by the physiological conditions of the elderly, their flora might be different compared with the younger participants Citation[28]. However, no significant differences were found in the faecal samples collected before bowel cleansing of the younger (<60 years) compared to the older patients (>60 years) (data not shown).
In conclusion, drastic environmental intervention and changes in dietary factors such as bowel cleansing and probiotic consumption did not result in major changes in the dominant faecal flora. Also, changes in type or dose of medication did not significantly alter the bacterial fingerprint of the flora, supporting the results of previous studies indicating that the individual faecal flora is relatively stable over time. However, some changes in single species of the dominant flora of individuals could be noticed, particularly in the early phase of bowel recolonization.
References
- Marteau P, Pochart P, Dore J, Bera-Maillet C, Bernalier A, Corthier G. Comparative study of bacterial groups within the human cecal and fecal microbiota. Appl Environ Microbiol 2001; 67: 4939–42
- Simon GL, Gorbach SL. The human intestinal microflora. Dig Dis Sci 1986; 31(9 Suppl): 147S–162S
- Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002; 122: 44–54
- Bradley HK, Wyatt GM, Bayliss CE, Hunter JO. Instability in the faecal flora of a patient suffering from food-related irritable bowel syndrome. J Med Microbiol 1987; 23: 29–32
- Hopkins MJ, Sharp R, Macfarlane GT. Variation in human intestinal microbiota with age. Dig Liver Dis 2002; 34(Suppl 2)S12–S18
- Zoetendal EG, Akkermans ADL, Akkermans-van Vliet WM, Visser de AGM, Vos de WM. The host genotype affects the bacterial community in the human gastrointestinal tract. Microb Ecol Health Dis 2001; 13: 129–34
- Finegold SM, Sutter VL. Fecal flora in different populations, with special reference to diet. Am J Clin Nutr 1978; 31(10 Suppl): S116–S122
- Wellmann W, Fink PC, Schmidt FW. Whole-gut irrigation as antiendotoxinaemic therapy in inflammatory bowel disease. Hepatogastroenterology 1984; 31: 91–3
- Wellmann W, Schmidt FW. Intestinal lavage in the treatment of Crohn's disease: a pilot study. Klin Wochenschr 1982; 60: 371–3
- Wellmann W, Fink PC, Benner F, Schmidt FW. Endotoxaemia in active Crohn's disease. Treatment with whole gut irrigation and 5-aminosalicylic acid. Gut 1986; 27: 814–20
- van den Bogaard AE, Weidema WF, van Boven CP, van der Waay D. Recolonization and colonization resistance of the large bowel after three methods of preoperative preparation of the gastrointestinal tract for elective colorectal surgery. J Hyg (Lond) 1986; 97: 49–59
- Havenaar R, Ten Brink B, Huis in't Veld J. Selection of strains for probiotic use. Probiotics, the scientific basis, R Fuller. Chapman & Hall, London 1992; 209–24
- Goossens D, Jonkers D, Russel M, Stobberingh E, Van Den Bogaard A, Stockbrugger R. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: a placebo-controlled study on the onset and duration of effects. Aliment Pharmacol Ther 2003; 18: 495–505
- Johansson M, Nobaek S, Berggren A, Nyman M, Bjorck I, Arhne S, et al. Survival of Lactobacillus plantarum DSM 9843 (299v), and effect on the short-chain fatty acid content of faeces after ingestion of a rose-hip drink with fermented oats. Int J Food Microbiol 1998; 42: 29–38
- Adlerberth I, Ahrne S, Johansson M, Molin G, Hanson L, Wold A. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl Environ Microbiol 1996; 62: 2244–51
- Goossens D, Jonkers D, Russel M, Thijs A, van den Bogaard A, Stobberingh E, et al. Survival of the probiotic, L. plantarum 299v and its effects on the faecal bacterial flora, with and without gastric acid inhibition. Dig Liver Dis 2005; 37: 44–50
- Zoetendal EG, Akkermans AD, de Vos WM. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 1998; 64: 3854–9
- Suau A, Bonnet R, Sutren M, Godon JJ, Gibson GR, Collins MD, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 1999; 65: 4799–807
- Muyzer G, Smalla K. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 1998; 73: 127–41
- O'Donnell L, Virjee J, Heaton K. Detection of pseudodiarrhoea by simple clinical assessment of intestinal transit rate. BMJ 1990; 300: 439–40
- Sanguinetti CJ, Dias Neto E, Simpson AJ. Rapid silver staining and recovery of PCR products separated on polyacrylamide gels. Biotechniques 1994; 17: 914–21
- Hane BG, Jager K, Drexler HG. The Pearson product-moment correlation coefficient is better suited for identification of DNA fingerprint profiles than band matching algorithms. Electrophoresis 1993; 14: 967–72
- Ott S, Musfeldt M, Wenderoth D, Hampe J, Brant O, Folsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004; 53: 685–93
- Walter J, Hertel C, Tannock GW, Lis CM, Munro K, Hammes WP. Detection of Lactobacillus, Pediococcus, Leuconostoc, and Weissella species in human feces by using group-specific PCR primers and denaturing gradient gel electrophoresis. Appl Environ Microbiol 2001; 67: 2578–85
- Danielsson D, Kjellander J, Jarnerot G. The effect of metronidazole and sulfasalazine on the fecal flora in patients with Crohn's disease. Scand J Gastroenterol 1981; 16: 183–92
- Jonkers DM, Stobberingh EE, Houben GMP, Stockbrugger RW. The influence of long-term treatment with omeprazole on changes in faecal aerobic flora. Gastroenterology 1994; 106(Suppl 4)A101
- Lambert-Zechovsky N, Bingen E, Cezard JP, Mashako L, Marinier E, Navarro J. Effects of loperamide on the fecal flora in children in severe diarrheas. Pathol Biol (Paris) 1987; 35: 656–60
- Mitsuoka T. Intestinal flora and aging. Nutr Rev 1992; 50: 438–46