Abstract
Increased proliferation and migration of abnormal airway smooth muscle cells (ASMCs) are significantly associated with asthma. Recently, the function of long non-coding RNAs (lncRNAs) in ASMCs has been identified. Our research attempted to resolve the function of the lncRNA DGCR5 in ASMCs and focus on its in-depth mechanisms of proliferation and migration. Quantitative real-time PCR and western blotting were used to evaluate the expressions of RNA and proteins, respectively. The CCK-8 assay was used to detect cell proliferation. Transwell assays were used to assess migration. Luciferase and RNA pull-down assays were used to verify the targeting between miR-204-5p and lncRNA DGCR5 or SRSF7. LncRNA DGCR5 was upregulated in ASMCs stimulated with PDGF-BB. Knockdown of DGCR5 reversed the effect of platelet-derived growth factor BB (PDGF-BB) on the proliferation and migration of ASMCs. The results of this study showed that lncRNA DGCR5 binds to miR-204-5p, and SRSF7 is the target of miR-204-5p. Rescue experiments verified the interaction between miR-204-5p and DGCR5, as well as that between miR-204-5p and SRSF7. Overall, this study revealed that the lncRNA DGCR5/miR-204-5p/SRSF7 signalling axis has key regulatory effects on the proliferation and migration of ASMCs.
Keywords:
Introduction
Asthma is a chronic inflammatory respiratory disease, which is characterised by chronic airway hyperreaction, airway inflammation, and airway remodelling [Citation1]. The proliferation of airway smooth muscle cells (ASMCs) has a key effect on airway remodelling [Citation2]. Although asthma is a chronic disease, its reversibility is different from that of other chronic obstructive pulmonary diseases (COPDs). Asthma is a risk factor for persistent airflow restriction, which may lead to the exacerbation of clinical symptoms in patients with asthma. In asthma, there is a relationship between the inflammatory state of the airways and the severity of hyperresponsiveness [Citation3].
Airway smooth muscle remodelling plays a pivotal role in the occurrence of chronic respiratory diseases such as asthma [Citation4, Citation5]. Under normal circumstances, ASMCs remain static and non-migrating, but inflammatory cytokines, growth factors, contractile agonists, extracellular matrix proteins, and other stimuli can induce ASMC proliferation and migration, which is related to the development of severe asthma [Citation6]. PDGF-BB is significantly upregulated in asthmatic tissues and can enhance ASMC proliferation and migration ability and aggravate the airway remodelling process [Citation7]. PGE2 plays a protective role in airway smooth muscle and promotes its proliferation [Citation8]. Endothelin-1 (ET-1) has been indirectly implicated in the pathophysiology of asthma and induces airway inflammation and remodelling [Citation9]. Transforming growth factor-β1(TGF-β1) can induce airway smooth muscle cell proliferation [Citation10]. However, further clarity is needed on the molecular mechanisms of ASMC proliferation and migration.
Long non-coding RNAs (lncRNAs) are non-protein encoding transcripts with a length of at least 200 nucleotides. It is usually divided into antisense, pseudogene, intron, and intergenic lncRNAs [Citation11]. With their classic mechanism, they can regulate gene expression by translational inhibition, splicing modification, and mRNA degradation by binding to genes directly, and can also act as competitive endogenous RNA to microRNAs to stabilise mRNA [Citation12, Citation13]. At present, a number of studies have revealed the effect of lncRNA on asthma, for example, lncRNA BCYRN1 promoted the proliferation and migration of ASMCs [Citation14], and lncRNA TUG1 promoted proliferation and migration, and inhibited the apoptosis of AMSCs [Citation15]. DiGeorge Syndrome Critical Region Gene 5 (DGCR5) has a tumour suppressing or carcinogenic effect in a variety of cancers [Citation16]. LncRNA DGCR5, also known as lncRNA0037, was first discovered in Huntington’s disease [Citation17]. Interestingly, it was found that DGCR5 is downregulated in most tumour cells, and it reduced the proliferation and invasion of cervical cancer by activating Wnt signalling [Citation18]. The low expression level of DGCR5 in hepatocellular carcinoma is associated with poor prognosis. However, DGCR5 is upregulated in gallbladder cancer, and inhibition of lncRNA impairs cell proliferation and migration through the JNK and p38/MAPK pathways [Citation19]. These results indicate that lncRNAs play different roles in the pathogenesis of different cancers. Nevertheless, the specific function of lnc-DGCR5 in the process of ASMCs is worth exploring.
More evidence suggests that lncRNA and target genes jointly and competitively bind to the same miRNA response element, thereby avoiding degradation of the mRNA. MicroRNAs are non-coding, single-stranded RNAs that are less than 21 nucleotides in length. They can regulate both transcriptional and post-transcriptional levels, and bind to the 3′-untranslated region (3’UTR) of mRNA with completely complementary sequences to degrade mRNA [Citation20]. MiR-204-5p functions as a tumour suppressor that inhibits tumour cell proliferation, autophagy, and metastasis in multiple cancer types [Citation21]. However, its role in asthma has not well documented.
SRSF7 encodes a splicing factor that regulates the different steps of RNA processing. As recently reported, it plays a key role in lung cancer cell survival by controlling the splicing of the apoptotic protein Fas and knocking down SRSF7-induced lung cancer cell apoptosis [Citation22]. MicroRNAs regulate the expression of SRSF7 in cancer cells by binding to the 3’UTR of the target gene and downregulate its expression [Citation23]. However, the relationship between miR-204-5p and SRSF7 and the role of SRSF7 in asthma are still largely unknown.
In the present study, we aimed to explore the role of lnc-DGCR5 in platelet derived growth factor (PDGF)-BB-induced ASMCs, as well as the targeted association between miR-204-5p and SRSF7. We hypothesised that lnc-DGCR5 regulated cell proliferation and migration via the miR-204-5p/SRSF7 axis. Our findings may provide new evidence in terms of asthma therapy.
Materials and methods
ASMC isolation
The hospital ethics committee approved this study and acquired the support of all patients participating in the study, who signed an informed consent form. Smooth muscle samples were obtained from the healthy parts of the main trachea of patients with lung laceration or pulmonary tuberculosis undergoing pneumonectomy. These subjects underwent fiberoptic bronchoscopy to confirm the health of the trachea before pneumonectomy. The patients with the following conditions were excluded from this study: upper respiratory tract infection or acute attack within 3 months prior to bronchoscopy, bronchial carcinoid tumour, lung cancer. The specific operation is briefly described as follows. After separating the ASMCs from the surrounding tissues, these bundles were cut into small pieces and cultured in DMEM (10567014, Gibco, Grand Island, NY, USA) supplemented with 15% foetal bovine serum (FBS; 10099, Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin (15070063, Gibco) at 37 °C and 5% CO2. The medium was replaced every three days until confluence was formed, and a trypsin-EDTA (0.25% trypsin, 1 mM EDTA in HBSS; 25200072, Gibco) solution was used for passage. The cells at the passage 4–6 were used in this study.
Cell treatment
To induce asthma cell model, ASMCs were treated with 10 μM PGE2 (HY-101952, MedChemExpress, Monmouth Junction, NJ, USA), 10 ng/mL ET-1 (HY-P71446, MedChemExpress), 10 ng/mL TGF-β1 (7754-BH-100/CF, R&D Systems, Minneapolis, MN, USA), and 25 ng/mL PDGF-BB (HY-P7087, MedChemExpress) for 24 h. Additionally, ASMCs were treated with 25 ng/mL PDGF-BB for 0, 1, 3, 6, 12, and 24 h. Vehicle (dimethyl sulphoxide, DMSO, D2650, Sigma-Aldrich, St. Louis, MO, USA) at the concentration of 0.2%, was used to treat ASMCs for 24 h as the negative control group.
Cell transfection
Lnc-DGCR5 siRNAs (si-lnc-DGCR5 1#: 5′-CGTGGAGACACAGATGTGTGCACAT-3′ and si-lnc-DGCR5 2#: 5′-GGAGACACAGATGTGTGCACATGCA-3′), siRNA negative control (si-nc), empty vector, and SRSF7 overexpressing vector were synthesised from Genepharma (Shanghai, China). MiR-204-5p inhibitor (miR20000265-1-5), nc inhibitor (miR2N0000001-1-5), mimic (miR10000265-1-5), and nc mimic (miR1N0000001-1-5) were purchased from Ribobio (Guangzhou, China). ASMCs were transfected with the vectors mentioned above using Lipofectamine 2000 (11668027, Invitrogen). Following 48 h, transfected cells were harvested.
Cell counting kit-8 (CCK-8)
A CCK-8 (C0037, Beyotime, Shanghai, China) was used to detect cell viability. Briefly, transfected ASMCs at the concentration of 2000 cells/well were treated with PDGF-BB for 24 h. Then, CCK-8 reagent (10 μL) was added to incubate with ASMCs at 37 °C for 2 h. The optical value at 450 nm was recorded using a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Colony formation assay
Transfected ASMCs were incubated at 37 °C with 5% CO2 for 2 weeks. Then, ASMCs were fixed with 4% paraformaldehyde (158127, Sigma-Aldrich) and stained with 0.1% crystal violet (C0775, Sigma-Aldrich). The number of colonies was counted under a microscope (Olympus, Tokyo, Japan).
Transwell assay
Cell migration was determined by Transwell assay using 24-well chambers (8 μm pore; 3428, Corning, Corning, NY, USA). Serum-free DMEM containing 1 × 105 ASMCs were added in the upper chambers, and complete DMEM was added in the low chambers. After incubation for 24 h, the migrated ASMCs were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Finally, the stained cells were visualised under an inverted microscope at five randomly fields.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from ASMCs using Trizol reagent (15596026, Invitrogen). RNA concentration and integrity were analysed. For miR-204-5p detection, reverse transcription (RT) was performed using miRcute Plus miRNA First-Strand cDNA Kit (4992909, TIANGEN, Beijing, China), followed by qPCR was carried out using miRcute Plus miRNA qPCR Kit (SYBR Green) (4992779, TIANGEN). For lnc-DGCR5 and SRSF7 detection, RT and qPCR was conducted using FastKing One Step RT-qPCR kit (SYBR Green) (4993103, TIANGEN). The expression levels were calculated by the 2−ΔΔCt method. U6 was the internal control of miR-204-5p, and GAPDH was the internal control of lnc-DGCR5 and SRSF7. The primers were listed as follows: lnc-DGCR5 F 5′-CCTGCGAGCACATAGGCATA-3′ and R 5′-AGGGCCTCTGCTCCAGATTA-3′, SRSF7 F 5′-GATTGTCATCGTTACAGCCG-3′ and R 5′-CTTGATCGTCGAGGAGATGC-3′, GAPDH F 5′-GAAGGTCGGAGTCAACGGAT-3′ and R 5′-CCTGGAAGATGGTGATGGG-3′, miR-204-5p F 5′-TTCCCTTTGTCATCCTATGCCT-3′ and R 5′- CTCAACTGGTGTCGTGGA −3′, and U6 F 5′-CCTGCGCAAGGATGAC-3′ and R 5′-GTGCAGGGTCCGAGGT-3′.
Western blot
Proteins were extracted by lysing the ASMCs using RIPA buffer, and the concentration was measured using a BCA kit (P0012, Beyotime). A total of 30 μg protein was separated on SDS-PAGE and transferred to PVDF membranes (FFP26, Beyotime). The membranes were blocked using 5% skim milk for 1 h, incubated with primary antibodies at 4 °C overnight, and incubated with the secondary antibody at room temperature for 1 h. The bands were visualised using a ECL reagent (P0018S, Beyotime) and quantified using the Image J software. GAPDH was the internal control.
The antibodies used in western blot were all purchased from Abcam (Cambridge, MA, USA) and listed below: anti-PCNA (ab92552, 1:5000), anti-MMP9 (ab76003, 1:1000), anti-MMP3 (ab52915, 1/1000), anti-SRSF7 (ab138022, 1;1000), anti-GAPDH (ab9485, 1:2500) and goat anti-rabbit H&L HRP (ab6721, 1:3000).
Bioinformatic analysis
The targeting relationship between miR-204-5p and lnc-DGCR5 was predicted using the LncBase database. The targeting relationship between miR-204-5p and SRSF7 was predicted using the starBase database. The enrichment analysis of the biological pathway related to miR-204-5p targets (top 50 targets) were performed using the DAVID software.
Dual luciferase reporter assay
The Luciferase reporter assay was carried out according to the method of the Unal study [Citation24]. The 3′-UTR of lnc-DGCR5 and SRSF7 containing potential binding sites within miR-204-5p were inserted into the pGL3-Basic vector (Promega, Madison, WI, USA). The pGL3-Basic vector with pRL-TK vector (Promega) was co-transfected with mimic or nc mimic into HEK293T cells using Lipofectamine 2000 (Invitrogen). After 48 h of the transfection, the luciferase activity was measured using the dual-luciferase reporter assay system (E1980, Promega).
RNA pull-down assay
As previously described [Citation25]. in vitro transcription and biotin labelling of miR-204-5p were conducted using Bioco (Tianjin, China) with the antisense sequence as the negative control. The labelled product was purified using a RNeasy Mini Kit (Qiagen, USA). The ASMCs were lysed and the lysate was incubated with the biotin-labelled product for 1 h at 4 °C. Streptavidin Sepharose beads (65306, Invitrogen) were then added and incubated for 1 h. After elution the beads, the levels of lnc-DGCR5 and SRSF7 in the precipitated complex was evaluated using qRT-PCR.
Statistics analysis
An independent sample t-test was used for two-group comparisons. One-way analysis of variance (ANOVA) was used for multiple group comparisons. Statistical significance was set at p < 0.05.
Results
This study aimed to explore the role of lnc-DGCR5 in asthma. We demonstrated that lnc-DGCR5 promoted proliferation and migration of ASMCs via the miR-204-5p/SRSF7 axis.
LncRNA DGCR5 is up-regulated in the ASMCs stimulated with PDGF-BB
In order to know the expression level of lnc-DGCR5 in ASMCs of asthma, ASMCs were derived from the healthy part of the patient’s trachea and cultured in vitro. We used PGE2, ET-1, TGF-β1 and PDGF-BB to establish the cell model of asthma. We found that the expression level of lnc-DGCR5 was upregulated when the cells were treated with ET-1, TGF-β1, and PDGF-BB, but not affected by PGE2, compared with the vehicle group ().
Figure 1. Lnc-DGCR5 is up-regulated in ASMCs treated by PDGF-BB. (a) The expression level of lnc-DGCR5 increased in ASMCs treated with PGE2, ET-1, TGF-β1, and PDGF-BB. (b) The expression level of lnc-DGCR5 increased in PDGF-BB-treated ASMCs in a time-dependent manner. *p < 0.05 vs vehicle or 0 groups, **p < 0.01 vs vehicle or 0 groups.
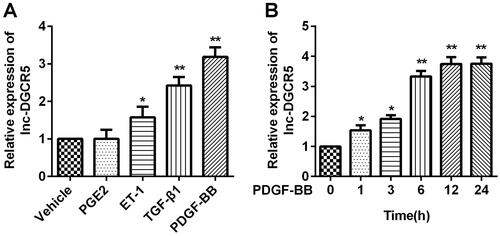
It is worth knowing whether the expression of lnc-DGCR5 is affected by PDGF-BB stimulation in ASMCs. ASMCs were cultured in a complete medium containing 25 ng/ml of PDGF-BB for 0, 1, 3, 6, 12, and 24 h. In particular, with the increase in PDGF-BB treatment time, the expression level of lnc-DGCR5 also increased (). This indicates that the cell model was successfully established, and the expression of lnc-DGCR5 is abnormally upregulated. These findings suggest that lnc-DGCR5 is involved in ASMCs treated with PDGF-BB.
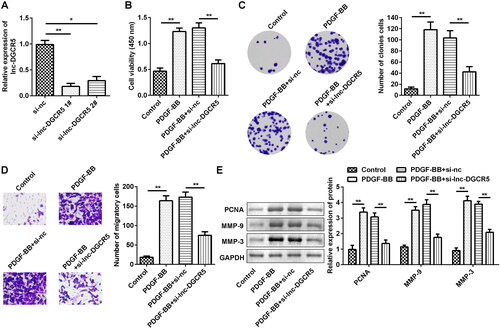
LncRNA DGCR5 promotes the PDGF-treated ASMCs proliferation and migration
To explore the role of lnc-DGCR5 in ASMCs, si-lnc-DGCR5 1#, si-lnc-DGCR5 2#, or si-nc were transfected in ASMCs to inhibit lnc-DGCR5 expression in ASMCs (). The inhibition of lnc-DGCR5 has no significant difference on cell viability, proliferation and migration compared to control (Supplementary Figure S1). Thus, we then performed the experiment in cells treated with PDGF-BB. The CCK-8 assay confirmed that cell viability was increased in PDGF-BB-treated AMSCs, but it was significantly inhibited by reduced lnc-DGCR5 expression (). Colony formation and Transwell assays showed that PDGF-BB treatment notably promoted the proliferation and migration of ASMCs, while silencing of lnc-DGCR5 significantly reduced the effect of PDGF-BB treatment on the proliferation and migration of ASMCs (). Western blot results indicated that PDGF-BB promoted the expression levels of proliferation marker PCNA and migration-related proteins MMP-9 and MMP-3, while knockdown of lnc-DGCR5 significantly reversed the effects ().
Figure 2. Lnc-DCR5 promotes the proliferation and migration of ASMCs. (a) Inhibiting the expression of lnc-DGCR5 was obtained through the siRNAs. (b) CCK-8 assay indicated that the cell viability of ASMCs treated by PDGF-BB is weakened when the expression of lnc-DGCR5 was inhibited. (c) Colony formation assay showed the significantly decreased proliferation ability of ASMCs when the expression of lnc-DGCR5 was inhibited. (d) Migrated ability of ASMCs, as observed using the Transwell assay, showed the migration of ASMCs after transfection. (e) Western blotting confirmed the effect of lnc-DGCR5 on PCNA, MMP-3, and MMP-9 expression in PDGF-BB-treated ASMCs. *p < 0.05, **p < 0.01.
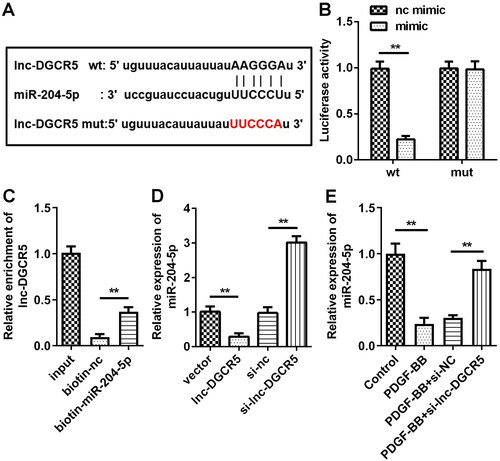
miR-204-5p acts as the target of lnc-DGCR5 in the ASMCs
LncRNA can competitively bind miRNAs, which can reduce its ability to affect the protein encoded by the target gene, thereby dynamically regulating gene expression. To explore whether lnc-DGCR5 plays a role in ASMCs through regulating miRNAs, bioinformatic analysis showed that the potential miRNAs interact with lnc-DGCR5. In addition, there are binding sites between miR-204-5p and lnc-DGCR5 (). Compared with the mimic-NC transfection, miR-204-5p mimic transfection significantly inhibited the luciferase activity of the reporter vector containing lnc-DGCR5-wt, while the luciferase activity of the reporter vector containing lnc-DGCR5-mut did not affect luciferase activity (). The results of RNA pull-down assay confirmed that miR-204-5p can directly target lnc-DGCR5 (). In ASMCs, the transfection of lnc-DGCR5 siRNA enhanced the expression of miR-204-5p, while the overexpression of lnc-DGCR5 decreased the expression of miR-204-5p (). Moreover, qRT-PCR revealed that PDGF-BB treatment downregulated miR-204-5p expression in ASMCs as compared to that in the controls, while knockdown of lnc-DGCR5 increase the expression of miR-204-5p in PDGF-BB-induced ASMCs (). Overall, these data indicate that miR-204-5p is the target of lnc-DGCR5, and lnc-DGCR5 can directly regulate the expression of miR-204-5p.
Figure 3. miR-204-5p acts as the target of lnc-DGCR5 in the ASMCs. (a) Bioinformatic analysis for the binding sites between miR-210-5p and lnc-DGCR5. (b) Luciferase assay was used to detect the direct interaction between miR-204-5p and lnc-DGCR5. (c) RNA pull-down assay confirmed that miR-204-5p can directly target lnc-DGCR5. (d) The expression level of miR-204-5p in the ASMCs was downregulated when lnc-DGCR5 was overexpressed, and its expression level was increased when lnc-DGCR5 was inhibited. (e) The expression level of miR-204-5p was downregulated in ASMCs treated by PDGF-BB, and knockdown of lnc-DGCR5 reversed the downregulation. **p < 0.01.
Lnc-DGCR5 promotes the proliferation and migration of ASMCs treated with PDGF-BB by sponging miR-204-5p
To confirm the function of miR-204-5p in ASMCs, an inhibitor was designed to downregulate miR-204-5p level (). When the expression of lnc-DGCR5 was inhibited, the results of CCK-8 and colony formation assays showed that cell viability in the miR-204-5p inhibitor group was more active than that in the control group (nc inhibitor), confirming that miR-204-5p can suppress the proliferation of cells when lnc-DGCR5 is inhibited (). In addition, downregulation of lnc-DGCR5 significantly reduced the migrated cell number and downregulated the levels of PCNA, MMP-9, and MMP-3 in PDGF-BB-treated ASMCs, while downregulation of miR-204-5p abrogated the reduction induced by si-lnc-DGCR5, as determined by Transwell assay and western blot (). Together, these results demonstrate that miR-204-5p can regulate proliferation and migration of AMSCs under PDGF-BB-stimulated conditions.
Figure 4. MiR-204-5p inhibition reversed the effect of lnc-DGCR5 knockdown. (a) The inhibitor can effectively inhibit the expression of miR-204-5p. (b) CCK-8 analysis showed that miR-204-5p inhibitor can rescue the cell viability in PDGF-BB-treated AMSC when lnc-DGCR5 was inhibited. (c) The results of the colony formation assay showed the miR-204-5p inhibitor can rescue proliferation ability in PDGF-BB-treated AMSC when lnc-DGCR5 was inhibited. (d) The results of the Transwell assay showed that the miR-204-5p inhibitor can rescue the migration ability of PDGF-BB-treated AMSCs when lnc-DGCR5 was inhibited. (e) Western blotting results showed that miR-204-5p inhibitor can rescue the expression level of PCNA, MMP-9, and MMP-3 in PDGF-BB-treated AMSCs when lnc-DGCR5 was inhibited. *p < 0.05, **p < 0.01.
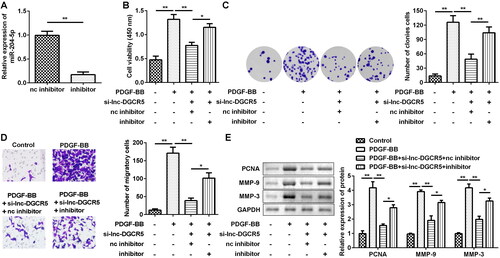
SRSF7 acts as the target of miR-204-5p in the ASMCs
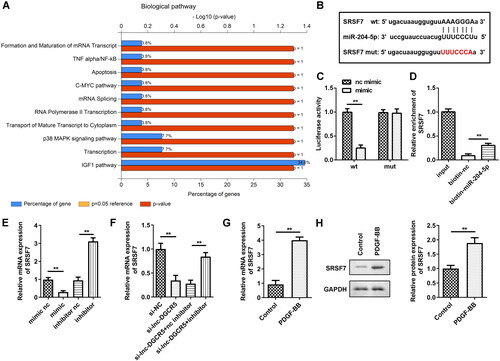
To understand the potential molecular mechanisms that mediate miR-204-5p-induced ASMC proliferation inhibition, bioinformatic analysis was used to screen the target mRNAs of miR-204-5p. The enrichment analysis results indicated that the top 50 miR-204-5p targets are associated with several pathways, such as formation and maturation of mRNA transcript pathway (). SRSF7 is a splicing factor, and variable mRNA splicing can regulate smooth muscle cell proliferation and migration [Citation26]. Thus, we focussed on whether miR-204-5p targets SRSF7 to mediate the biological behaviours of ASMCs. The TargetScan results predicted that SRSF7 has the binding sites in miR-204-5p () as it can shuttle continuously between the nucleus and cytoplasm, with additional involvement in mRNA transport and translation [Citation8] and may have an active role in cancer cells. To confirm whether miR-204-5p directly binds to the 3′UTR of SRSF7 mRNA, the wt and mutated binding sites on the SRSF7 mRNA were used to construct the luciferase reporter vector. The luciferase reporter assay showed that co-transfection of miR-204-5p and SRSF7-wt decreased luciferase activity (). In addition, the RNA-pull down assay also showed that miR-204-5p can directly bind to SRSF7 (). In ASMCs, transfection of miR-204-5p inhibitor increased the expression of SRSF7, but the miR-204-5p mimics decreased the expression of SRSF7 (). Additionally, silencing of lnc-DGCR5 downregulated SRSF7, while inhibition of miR-204-5p reversed the downregulation (). The expression of SRSF7 was upregulated in ASMCs treated with PDGF-BB (). Meanwhile, protein expression of SRSF7 was also increased in ASMCs treated with PDGF-BB (). Thus, these data suggest that SRSF7 acts as a target of miR-204-5p in ASMCs.
Figure 5. miR-204-5p targets SRSF7 directly. (a) The enrichment analysis of miR-204-5p targets-related biological pathway. (b) The binding site of miR-204-5p in SRSF7 3’-UTR. (c) The luciferase assay showed that miR-204-5p could directly targets SRSF7. (d) RNA pull-down assay confirmed that miR-204-5p can directly target SRSF7. (e) The expression level of SRSF7 in the ASMCs was downregulated when miR-204-5p was overexpressed, and its expression level increased when miR-204-5p was inhibited. (f) knockdown of lnc-DGCR5 decreased SRSF7 expression, and inhibition of miR-204-5p reversed the decrease. (g) The expression level of SRSF7 was upregulated in ASMCs treated by PDGF-BB. (h) Protein level of SRSF7 increased in ASMCs treated with PDGF-BB. **p < 0.01.
miR-204-5p inhibits ASMCs proliferation and migration through inhibiting SRSF7 expression
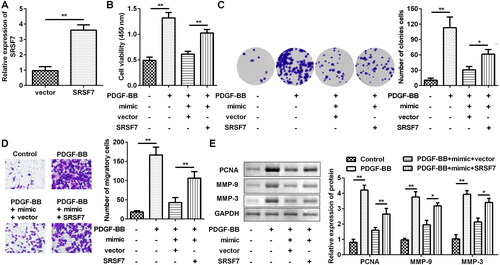
Previous data have shown that SRSF7 is a direct target of miR-204-5p in ASMCs, and several studies have shown that SRSF7 can suppress proliferation and increase apoptosis in some carcinomas [Citation22, Citation27]. However, little is currently known about the role of SRSF7 in ASMCs. After transfection of SRSF7 overexpressing vector, the expression level of SRSF7 was increased by about 4 times as compared to that in the control group (). The results of the CCK-8 and colony formation assays have revealed that overexpression of SRSF7 can improve the cell viability and proliferation ability of ASMCs transfected with miR-204-5p mimics (). To further determine the role of SRSF7 in ASMC migration, the SRSF7 overexpression vector reversed the inhibition of cell migration induced by miR-204-5p (). The protein levels of PCNA, MMP-9, and MMP-3 were increased by SRSF7 overexpression (), as shown by western blotting. Taken together, these results indicate that miR-204-5p inhibits ASMC proliferation and migration, at least in part, by inhibiting the expression of SRSF7 in ASMCs.
Figure 6. Overexpression of SRSF7 can rescue the proliferation and migration of ASMC treated by PDGF-BB, which is attenuated by overexpression of miR-204-5p. (a) qRT-PCR confirmed the overexpression efficiency of SRSF7. (b) CCK-8 analysis showed that overexpression of SRSF7 can rescue the cell viability in PDGF-BB-treated AMSC when miR-204-5p was overexpressed. (c) The results of the colony formation assay showed that overexpression of SRSF7 can rescue the proliferation ability in PDGF-BB-treated AMSC when miR-204-5p was overexpressed. (d) The results of Transwell assay showed that overexpression of SRSF7 can rescue migration ability in PDGF-BB-treated AMSC when miR-204-5p was overexpressed. (e) Western blotting showed that overexpression of SRSF7 can rescue the expression level of PCNA, MMP-9, and MMP-3 in PDGF-BB-treated AMSC when miR-204-5p was overexpressed. *p < 0.05, **p < 0.01.
Discussion
We investigated the effects of a novel lncRNA (lnc-DGCR5) on the proliferation and migration of ASMCs. In PDGF-BB-treated ASMCs, the expression level of lnc-DGCR5 was upregulated. In addition, the loss-of-function assay indicated that lnc-DGCR5 promoted the proliferation and migration of PDGF-BB-induced ASMCs. These findings support the important role of lnc-DGCR5 in the pathogenesis of asthma.
It is well known that lncRNAs can bind with miRNAs to block their inhibitory effects on the expression of their targets. Several studies have indicated that lnc-DGCR5 sponges miRNAs to participate in the development of human diseases. For example, lnc-DGCR5 downregulates miR-21 to inhibit the proliferation of colorectal cancer cells migration and invasion [Citation28]. Additionally, lnc-DGCR5 inhibits apoptosis and promotes tumour growth in lung adenocarcinoma via sponging miR-22-3p [Citation29]. Our results showed that lnc-DGCR5 binds with miR-204-5p. Our study provides convincing evidence that miR-204-5p is a key regulator of ASMC function in humans. miR-204-5p usually acts as a tumour suppressor in head and neck squamous cell carcinoma (HNSCC) and can inhibit tumour growth, metastasis, and stemness [Citation30]. In addition, the overexpression of miR-204-5p caused significant changes in the metabolic properties of cancer cells in oral squamous cell carcinoma and inhibited tumour growth and metastasis [Citation31]. The correlation between the expression of miR-204-5p and the clinical outcome of breast cancer patients showed a non-linear pattern, which was reproduced in the experimental detection of cancer cell behaviour and metastatic ability [Citation32]. Although miR-204-5p is highly expressed in ASMCs treated with PDGF-BB, the function of miR-204-5p in the regulation of ASMC phenotype remains to be elucidated. Here, we demonstrated that miR-204-5p is an important regulator that inhibits the proliferation and migration of ASMCs.
MiR-204-5p targets multiple mRNAs to regulate the progression of diseases. For instance, miR-204-5p inhibits inflammation of synovial fibroblasts by targeting FOXC1 in osteoarthritis [Citation33]. In addition, miR-204-5p targets PRR11 to induce cell cycle arrest and inhibit migration and invasion of breast cancer cells [Citation34]. Moreover, miR-204-5p has been revealed to participate in the development of asthma. A previous study has reported that miR-204-5p inhibited the proliferation and extracellular matrix production by targeting Six1 in asthma [Citation35]. Our research validated a novel direct target of miR-204-5p, SRSF7, is upregulated in ASMCs treated with PDGF-BB, and its overexpression can improve cell viability and migration.
Conclusion
Overall, this study reveals the crucial regulatory effect of the lnc-DGCG5/miR-204-5p/SRSF7 axis on ASMC proliferation and migration. This discovery reveals the mechanism by which lnc-DGCG5 regulates the progression of asthma. These findings suggest that the targeted overexpression of lnc-DGCG5 and SRSF7 in ASMCs may be a potential new therapy to inhibit the proliferation of ASMCs associated with the onset of asthma.
Ethical approval
This study protocol was approved by the Ethics Committee of Shenzheng Shiyan People’s Hospital.
Supplemental Material
Download MS Word (535.9 KB)Disclosure statement
The authors declared that they have no conflict interest
Additional information
Funding
References
- Samitas K, Delimpoura V, Zervas E, et al. Anti-IgE treatment, airway inflammation and remodelling in severe allergic asthma: current knowledge and future perspectives. Eur Respir Rev. 2015;24(138):1–10.
- Olafsdottir TA, Theodors F, Bjarnadottir K, et al. Eighty-eight variants highlight the role of T cell regulation and airway remodeling in asthma pathogenesis. Nat Commun. 2020;11(1):393.
- Postma DS, Kerstjens HA. Characteristics of airway hyperresponsiveness in asthma and chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1998;158(5 Pt 3):S187–S92.
- Kudo M, Ishigatsubo Y, Aoki I. Pathology of asthma. Front Microbiol. 2013;4:263.
- Makinde T, Murphy RF, Agrawal DK. The regulatory role of TGF-beta in airway remodeling in asthma. Immunol Cell Biol. 2007;85(5):348–356.
- Fixman ED, Stewart A, Martin JG. Basic mechanisms of development of airway structural changes in asthma. Eur Respir J. 2007;29(2):379–389.
- Dai Y, Li Y, Cheng R, et al. TRIM37 inhibits PDGF-BB-induced proliferation and migration of airway smooth muscle cells. Biomed Pharmacother. 2018;101:24–29.
- Sastre B, del PV. Role of PGE2 in asthma and nonasthmatic eosinophilic bronchitis. Mediators Inflamm. 2012;2012:645383.
- Labram B, Namvar S, Hussell T, et al. Endothelin-1 mediates Aspergillus fumigatus-induced airway inflammation and remodelling. Clin Exp Allergy. 2019;49(6):861–873.
- Chen M, Zhang W, Shi J, et al. TGF-beta1-Induced airway smooth muscle cell proliferation involves TRPM7-dependent calcium influx via TGFbetaR/SMAD3. Mol Immunol. 2018;103:173–181.
- Bridges MC, Daulagala AC, Kourtidis A. LNCcation: lncRNA localization and function. J Cell Biol. 2021;220:e202009045.
- Booton R, Lindsay MA. Emerging role of MicroRNAs and long noncoding RNAs in respiratory disease. Chest. 2014;146(1):193–204.
- Sun W, Lv J, Duan L, et al. Long noncoding RNA H19 promotes vascular remodeling by sponging let-7a to upregulate the expression of cyclin D1. Biochem Biophys Res Commun. 2019;508(4):1038–1042.
- Zhang XY, Zhang LX, Tian CJ, et al. LncRNAs BCYRN1 promoted the proliferation and migration of rat airway smooth muscle cells in asthma via upregulating the expression of transient receptor potential 1. Am J Transl Res. 2016;8(8):3409–3418.
- Lin J, Feng X, Zhang J, et al. Long noncoding RNA TUG1 promotes airway smooth muscle cells proliferation and migration via sponging miR-590-5p/FGF1 in asthma. Am J Transl Res. 2019;11:3159–3166.
- Xue C, Chen C, Gu X, et al. Progress and assessment of lncRNA DGCR5 in malignant phenotype and immune infiltration of human cancers. Am J Cancer Res. 2021;11:1–13.
- Johnson R. Long non-coding RNAs in Huntington’s disease neurodegeneration. Neurobiol Dis. 2012;46(2):245–254.
- Liu Y, Chang Y, Lu S, et al. Downregulation of long noncoding RNA DGCR5 contributes to the proliferation, migration, and invasion of cervical cancer by activating Wnt signaling pathway. J Cell Physiol. 2019;234(7):11662–11669.
- Liu S, Chu B, Cai C, et al. DGCR5 promotes gallbladder cancer by sponging MiR-3619-5p via MEK/ERK1/2 and JNK/p38 MAPK pathways. J Cancer. 2020;11(18):5466–5477.
- Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. 2021;14:dmm047662.
- Yang F, Bian Z, Xu P, et al. MicroRNA-204-5p: a pivotal tumor suppressor. Cancer Med. 2023;12(3):3185–3200. Epub ahead of print.
- Fu Y, Wang Y. SRSF7 knockdown promotes apoptosis of colon and lung cancer cells. Oncol Lett. 2018;15(4):5545–5552.
- Boguslawska J, Sokol E, Rybicka B, et al. microRNAs target SRSF7 splicing factor to modulate the expression of osteopontin splice variants in renal cancer cells. Gene. 2016;595(2):142–149.
- Unal H. Luciferase reporter assay for unlocking ligand-mediated signaling of GPCRs. Methods Cell Biol. 2019;149:19–30.
- Torres M, Becquet D, Guillen S, et al. RNA pull-down procedure to identify RNA targets of a long non-coding RNA. J Vis Exp. 2018;(134):57379.
- Xie N, Chen M, Dai R, et al. SRSF1 promotes vascular smooth muscle cell proliferation through a Δ133p53/EGR1/KLF5 pathway. Nat Commun. 2017;8:16016.
- Liu Z, Ma L, Gu Y, et al. Long non-coding RNA LINC01123 promotes cell proliferation, migration and invasion via interacting with SRSF7 in colorectal cancer. Pathol Res Pract. 2022;232:153843.
- Huang H, Yang X, Chen J, et al. lncRNA DGCR5 inhibits the proliferation of colorectal cancer cells by downregulating miR-21. Oncol Lett. 2019;18(3):3331–3336.
- Dong HX, Wang R, Jin XY, et al. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J Cell Physiol. 2018;233(5):4126–4136.
- Zhuang Z, Yu P, Xie N, et al. MicroRNA-204-5p is a tumor suppressor and potential therapeutic target in head and neck squamous cell carcinoma. Theranostics. 2020;10(3):1433–1453.
- Rajthala S, Dongre H, Parajuli H, et al. Combined in situ hybridization and immunohistochemistry on archival tissues reveals stromal microRNA-204 as prognostic biomarker for oral squamous cell carcinoma. Cancers (Basel). 2021;13(6):1307.
- Hong BS, Ryu HS, Kim N, et al. Tumor suppressor miRNA-204-5p regulates growth, metastasis, and immune microenvironment remodeling in breast cancer. Cancer Res. 2019;79(7):1520–1534.
- He X, Deng L. miR-204-5p inhibits inflammation of synovial fibroblasts in osteoarthritis by suppressing FOXC1. J Orthop Sci. 2022;27(4):921–928.
- Su Q, Shen H, Gu B, et al. miR-204-5p hampers breast cancer malignancy and affects the cell cycle by targeting PRR11. Comput Math Methods Med. 2022;2022:4010947.
- Yang Z, Qu Z, Yi M, et al. MiR-204-5p inhibits transforming growth factor-β1-induced proliferation and extracellular matrix production of airway smooth muscle cells by regulating Six1 in asthma. Int Arch Allergy Immunol. 2020;181(4):239–248.