Abstract
Stroke is an acute cerebrovascular disease that is now the most important cause of death due to brain problems in our country. CircRNAs are RNA circles that have been extensively involved in the disease. We aimed to investigate the mechanism of circ_0129657 in the pathogenesis of stroke. In this study, quantitative real-time polymerase chain reaction (RT-qPCR) and western blot assays were used to assess the expression of circ_0129657, miR-194-5p, and glia maturation factor beta (GMFB). Cell viability was measured by Cell Counting Kit-8 (CCK-8) assay. 5-Ethynyl-2′-Deoxyuridine (EdU) assay was used to detect cell proliferation. Flow cytometry was used to detect cell apoptosis. Dual-luciferase reporter, RNA pull-down, and RNA immunoprecipitation (RIP) assays were used to assess the relationship between miR-194-5p and circ_0129657 or GMFB. Mouse middle cerebral artery occlusion (MCAO) model was applied to mimic the cerebral ischemia/reperfusion injury. Our data showed that the levels of circ_0129657 and GMFB were significantly increased and the expression of miR-194-5p was significantly decreased in oxygen-glucose deprivation (OGD)-induced human brain microvascular endothelial cells (HBMECs). Silencing circ_0129657 expression in OGD-induced HBMECs could promote cell viability and cell proliferation. Moreover, circ_0129657 depletion also could inhibit apoptosis and inflammatory factor secretion. Circ_0129657 functioned as a sponge for miR-194-5p and could regulate GMFB expression via miR-194-5p competition. Furthermore, miR-194-5p downregulation or GMFB restoration could partially reverse the effects of circ_0129657 silencing on cell biological properties in OGD-induced HBMECs. Meanwhile, circ_0129657 knockdown decreased cerebral infarction volume and neurological impairment in MCAO mouse models. In conclusion, our findings suggest that circ_0129657 can inhibit cell proliferation and promote apoptosis and inflammatory factor secretion in HBMECs after oxygen-glucose deprivation via miR-194-5p/GMFB axis, providing evidence that circ_0129657 has the potential as a useful biological molecular marker in the diagnosis of stroke.
Keywords:
Introduction
Stroke, also known as cerebral stroke, is an acute cerebrovascular disease with a wide range of causes [Citation1–4]. It is mainly caused by the sudden rupture of blood vessels in the brain or blockage of blood vessels [Citation2], further causing brain tissue damage or brain blood circulation disorders of the disease [Citation2, Citation5]. Insufficient blood supply to the brain is followed by neurological dysfunction, which can lead to disability or loss of autonomic consciousness [Citation3, Citation6, Citation7]. Due to the rapid onset of the disease, the optimal treatment time is very short, resulting in a high mortality rate in addition to a high morbidity rate. Therefore, stroke has been a serious health problem for people all around the world [Citation7]. It also imposes a huge economic burden on families and countries [Citation8]. Therefore, it is of utmost importance to prevent the onset of the disease or to save the life of the patients within the best resuscitation time. Here, we aimed to investigate bio-diagnostic markers that could be used as valid markers for stroke and provide a theoretical basis for the early onset of the disease.
Circular RNAs (circRNAs) are an interesting class of RNA molecules due to their covalently closed structure and high stability [Citation9]. There is a large amount of literature reporting that circRNAs are important in the development of diseases. For example, Circ_0007059 expression was significantly reduced in nephritis tissues and cells, and circ_0007059 regulated proliferation and apoptosis in nephritis cell models [Citation10]. Recently, the importance of circRNAs in stroke pathogenesis has become increasingly clear [Citation11]. Interestingly, the expression of circ_0129657, formed by back-spliced exons of IQ motif containing GTPase activating protein 2 (IQGAP2), was significantly upregulated in oxygen-glucose deprivation (OGD)-induced human brain microvascular endothelial cells (HBMECs) [Citation12]. Nonetheless, the role of circ_0129657 in stroke pathogenesis is unclear.
MicroRNAs (miRNAs) are a large class of non-coding single-stranded RNA molecules that are mainly encoded by endogenous genes [Citation13, Citation14]. Recent studies have reported that miRNAs play an important regulatory role in human physiological and pathological responses [Citation14, Citation15]. Importantly, circRNAs could further play regulatory roles as sponges of miRNAs [Citation10, Citation12]. For instance, circ_0007919 was significantly dysregulated in ulcerative colitis and could function as a sponge for let-7a and miR-138, thereby regulating the expression of EPC1 and VIPR1 and the progression of ulcerative colitis [Citation16]. MiR-194-5p expression was decreased in neurological disorders [Citation17], deep hypothermic cycling process [Citation4], cerebral Infarction [Citation18] and ischemic cerebral infarction [Citation19]. However, the specific molecular mechanism of miR-194-5p in the development of stroke remains unclear.
Currently, more and more studies are focusing on circRNA-miRNA-mRNA regulatory mechanisms that circRNA might function as competing for endogenous RNAs (ceRNAs) to sequester miRNAs away from mRNAs [Citation20, Citation21]. In this study, circ_0129657 expression was significantly increased in OGD-induced HBMECs. Furthermore, bioinformatics analysis exhibited that miR-194-5p possessed some binding sites with circ_0129657 or glia maturation factor beta (GMFB) for the first time. It has been reported that plasma GMFB might perform as a convenient non-invasive adjunct to neuroimaging for stroke diagnosis and prognosis [Citation22]. Therefore, we aim to validate the role of circ_0129657 in OGD-induced HBMEC dysfunction, and to illuminate whether the involvement of circ_0129657 in OGD-induced HBMEC injury was mediated by miR-194-5p/GMFB axis.
Materials and methods
Cell line and OGD treatment
The cells used in this study were HBMECs (Sciencell, Carlsbad, CA, USA, cat. no. 1000). HBMECs were cultured in a 5% CO2 and 37 °C incubator. DMEM (Gibco, Carlsbad, CA, USA, cat. no. 11965-092) and 10% FBS (Gibco, cat. no. 07905) were used for cell culture. The cell culture conditions in the control group remained unchanged and those in the OGD group were replaced with serum-free medium and the cells were cultured under anaerobic conditions with 1% O2, 5% CO2 and 94% N2 for 24 h, as previously described [Citation23–25].
Constructs, plasmids and transfection of HBMECs
siRNA against circ_0129657 (si-circ_0129657) and siRNA control (si-NC) were from Geneseed (Guangzhou, China). Mature miR-194-5p mimic, mimic control (miR-NC), miR-194-5p inhibitor (anti-miR-194-5p) and inhibitor mock (anti-miR-NC) were from Ribobio (Guangzhou, China). The GMFB coding sequence expression plasmid pcDNA3.1 (+)-GMFB and nontarget plasmid control pcDNA were from VectorBuilder (Guangzhou, China). All transient transfections were done using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA, cat. no. L3000) as per the accompanying instructions.
Quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was extracted by Trizol (Invitrogen, cat. no. 15596026). First, HBMECs were washed with cold PBS. The cleaning solution was discarded, and then Trizol was added for cell lysis. The extracted total RNA was then reverse-transcribed into cDNA using a reverse transcription kit (TaRaKa, Dalian, China, cat. no. RR037A). The SYBR-Green method was further used for RT-qPCR using SYBR Premix ExTaq kit (TaKaRa, cat. no. RR820A) and specific primer sets (Supplement Table S1). Finally, U6 was used as an internal reference for miRNAs, and β-actin was used as an internal reference for circRNAs and genes. The relative expression levels of each gene were calculated using the 2−ΔΔct method. Each test should be repeated three times.
Treatment of Rnase R and actinomycin D
For Rnase R treatment, 1 µg of total RNA from OGD-induced HBMECs was exposed to 0 units (Mock) or 3 units of Rnase R (Lucigen, Middleton, WI, USA, cat. no. RNR07250) for 30 min at 37 °C. For actinomycin D treatment, OGD-induced HBMECs were stimulated with 2 mg/mL actinomycin D (Biofount, Beijing, China, cat. no. RASP-101) for 0, 6, 12 and 24 h. RNA was then measured by RT-qPCR to evaluate circ_0129657 and IQGAP2 mRNA levels.
Cell Counting Kit-8 (CCK-8) assay
HBMECs that had been transfected with plasmids were inoculated onto 96-well plates. After 24 h of cell wall incubation, 10 μL of CCK-8 reagent (MedChemExpress, Shanghai, China, cat. no. HY-K0301) was added to cells for a total of 3 h. The plates were placed on an enzyme-labeled analyzer (Mlbio, Shanghai, China) and the absorbance values were measured at 450 nm.
5-Ethynyl-2′-deoxyuridine (EdU) assay
The EdU Apollo 488 Cell Proliferation Kit (Ribobio, cat. no. C10310-3) was used to assess the proliferative capacity of HBMECs. First, HBMECs were inoculated in 24-well plates and incubated for 24 h. After incubation, EdU solution was added to each well, and cells were co-incubated for 2 h, followed by the staining with Apollo 488 reagent. Then, 10 μL 4′,6-diamidino-2-phenylindole (DAPI, Beyotime, Shanghai, China, cat. no. C1005) was added to each well to stain the cell nuclei. Finally, the images were photographed using an inverted fluorescence microscope, and then the images were counted and calculated for EdU proliferation-positive cells. Finally, the rate of EdU-positive cells was calculated to indicate the proliferative capacity of the cells.
Enzyme‑linked immunosorbent assay (ELISA)
The Caspase-3 Assay Kit (Abcam, Cambridge, UK, cat. no. ab39401) was used to detect the caspase-3 activity of OGD-induced HBMECs after the indicated transfection, as per the manufacturing protocols. The optical density (OD) values were detected at 405 nm.
Flow cytometry assay
Using the Annexin V-FITC Apoptosis Kit (Beyotime, Shanghai, China, cat. no. C1062S), we detected the apoptosis of HBMECs. HBMECs after the indicated transfection were inoculated into 6-well plates and subjected to OGD treatment. After cell culture for 36 h, Annexin V-FITC (5 μL) and PI (10 μL) were added into each well and incubated for 20 min without light at 4 °C. Finally, cell apoptosis rate was measured by flow cytometry (Geneunion, Guangzhou, China).
Western blot assay
HBMECs were then lysed using RIPA lysis buffer (Beyotime, cat. no. P0013C). Proteins were separated using SDS-polyacrylamide gel electrophoresis and transferred to the PVDF membranes (Millipore, Shanghai, China) at the end of electrophoresis. The membranes were blocked with skim milk at room temperature for 1.5 h. The PVDF membranes were then washed three times with 1 × TBST buffer and incubated overnight at 4 °C using primary antibodies. The PVDF membranes were washed 3 times with 1 × TBST buffer and then incubated with goat anti-rabbit or anti-mouse IgG secondary antibodies conjugated with HRP (ab205718 or ab205719, Abcam, 1:5,000) for 1 h in room temperature and then washed 3 times with 1 × TBST buffer. Finally, immunoreactive bands were developed using the ECL kit (Abcam, cat. no. ab65623). All primary antibodies (Abcam) used at 1:1,000 dilution were: proliferating cell nuclear antigen (PCNA, ab29), BCL2-associated X, apoptosis regulator (Bax, ab32503), BCL2 apoptosis regulator (Bcl2, ab32124), interleukin-1beta (IL-1β, ab216995), interleukin-6 (IL-6, ab233706), tumor necrosis factor-α (TNF-α, ab183218), GMFB (ab224322), and β-actin (ab8226) as a loading buffer.
Dual‑luciferase reporter assay
The segments of circ_0129657 and GMFB 3′UTR harboring the predicted miR-194-5p complementary sequence or miss-matched seed sites were subcloned into the pMIR-REPORT vector (Thermo Fisher Scientific, Uppsala, Sweden) to construct the luciferase reporters. The above plasmids along with the pRL-TK Renilla luciferase vector (Promega, Milan, Italy) were co-transfected with miR-194-5p mimic or mimic mock into HBMECs, and then the cells were assayed for luciferase activity after 48 h using the Dual-Luciferase Reporter System (Promega, cat. no. E1910). The luciferase activity was then calculated and normalized.
RNA pull‑down assay
HBMECs were lysed in RIPA lysis buffer, and then lysates were incubated with biotinylated probes (Bio-miR-NC or Bio-miR-194-5p, Ribobio) and Dynabeads M-280 Streptavidin beads (Invitrogen) overnight at 4 °C. The beads were eluted using a washing buffer, and then, the eluted material was lysed using Trizol lysis solution. And total RNA extraction was performed. Finally, RT-qPCR was performed to analyze the enrichment level of circ_0129657.
RNA immunoprecipitation (RIP) assay
The RIP-Assay Kit (Gzscbio, Guangzhou, China, cat. no. KT102-01) was used to perform the RIP assay. Lysates extracted with RIPA lysis buffer from HBMECs were incubated with Magnetic beads (Invitrogen) pre-coupled with an antibody against Ago2 (ab186733, Abcam, 1:30) and IgG (ab172730, Abcam, 1:100) at 4 °C for 6 h. After the incubation, 300 μL RIP wash buffer was added to each tube, followed by RNA extraction, and then, RT-qPCR was carried out to gauge circ_0129657 and miR-194-5p enrichment levels.
Middle cerebral artery occlusion (MCAO) model of ischemic stroke
This animal experiment was approved by the Affiliated Hangzhou First People’s Hospital, Zhejiang University School of Medicine. Twenty male mice (5–6 weeks old, n = 5 per group, Vital River Laboratory, Beijing, China) were randomly divided into 4 groups: sham group and MCAO group: mice were injected with the empty lentiviral vector; MCAO + sh-NC group: mice were injected with a lentiviral vector containing sh-NC (GeneChem, Shanghai, China); and MCAO + sh-circ_0129657 group: sh-circ_0129657 (GeneChem) were injected into the cerebral cortex of mice before preparing a model of MCAO ischemic stroke. Two weeks later, the mouse MACO model was established. Generally, these mice were anesthetized with 3% isoflurane and the anesthesia was maintained during surgery with 1.5% isoflurane. Then, a fishing thread (diameter of 0.26 mm) with a round tip was inserted from the left common carotid artery into the lumen of the internal carotid artery to occlude the origin of the middle cerebral artery. After occluding for 1 h in the mouse’s middle cerebral artery, the mice were allowed to reperfuse for 24 h. For the sham group, the mice went through the same procedure, only a monofilament was inserted. For other groups, 24 h after MCAO, these mice were given analgesia, sacrificed, and then the brains were quickly removed for follow-up analysis. MCAO surgery was carried out as previously described [Citation26].
2,3,5-Triphenyltetrazolium chloride (TTC) staining and neurological impairment assessment
Assessment of cerebral infarct volume was conducted as previously described [Citation27, Citation28]. In short, the obtained brains were rapidly cut into sections at 2 mm. After being incubated in 2% TTC solution (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C for 10 min, the tissues were fixed in 4% paraformaldehyde for 1-2 days. After scanning using a scanner, the infarct volume was calculated using Image J software. The dyed red was normal tissue, and the white was infarcted tissue. Meanwhile, the possible interference of brain edema on infarct volume was corrected (whole contralateral hemisphere volume – nonischemic ipsilateral hemisphere volume) and the infracted volume was expressed as a ratio of the whole contralateral hemisphere [Citation29].
As previous described [Citation30], the researchers assessed the nerve defects 24 h after MCAO surgery by bind evaluation. Neurological injury of the mice was assessed using the 6-point scale: 0, no obvious deficits; 1, failure to entirely stretch the left forepaw when the tail of mice was stimulated; 2, contralateral circling to the left when the tail of mice was stimulated; 3, walking or circling to the left; 4, walking only when stimulated; 5, no response to stimulation and low consciousness.
Statistical analysis
All experiments in the study were replicated at least three times. GraphPad Prism 8 software (GraphPad Inc., La Jolla, CA, USA) was used to perform statistics and analysis of all data. A Student’s t-test was used for the comparison of differences between the two groups, and one-way or two-way ANOVA was used to evaluate the comparison of differences between more than two groups. P value less than 0.05 was statistically significant.
Results
In OGD-induced HBMECs, cell proliferation was inhibited and apoptosis and inflammatory factor expression were promoted
We first treated HBMECs with OGD and assessed the changes in the biological properties compared to the control group. In OGD-induced HBMECs, cell viability (Figure S1A) and cell proliferation capacity (Figure S1B) were significantly reduced. At the same time, the activity of caspase-3 (Figure S1C) and cell apoptosis rate (Figure S1D) were increased. Moreover, the expression of pro-apoptotic protein Bax was upregulated and the levels of proliferating marker PCNA and anti-apoptotic protein Bcl2 were downregulated in OGD-induced HBMECs (Figure S1E–H). We also examined the expression of inflammatory factors, and the secretion levels of IL-1β, IL-6 and TNF-α were significantly increased in the OGD-induced cells compared to the control group cells (Figure S1I–L). These data indicated that in HBMECs, OGD suppressed cell proliferation and induced cell apoptosis and inflammatory cytokine production.
Figure 1. Circ_0129657 was upregulated in OGD-induced HBMECs. (A) The expression of circ_0129657 was assessed by RT-qPCR. (B and C) The stability of circ_0129657 was tested by Rnase R and actinomycin D treatments in OGD-induced HBMECs. (D) The formation and structure of circ_0129657 were shown. ***p < 0.001, n = 3.
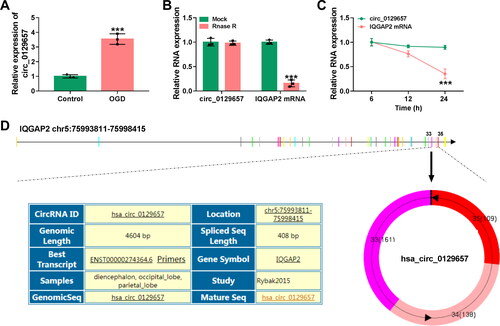
Circ_0129657 expression was upregulated in OGD-induced HBMECs
According to the previous literature [Citation12], we chose the top five overexpressed circRNAs in HBMECs after OGD. Then, these circRNAs were detected in the OGD group or the Control group using RT-qPCR. As a result, circ_0129657 presented the highest fold change in the OGD group among 5 circRNAs (Figure S2A). Therefore, circ_0129657 was selected for further study. We then examined the expression of circ_0129657 in OGD-induced HBMECs, and the result showed the expression of circ_0129657 was significantly increased (). Next, we examined the stability of circ_0129657 in OGD-induced HBMECs using Rnase R and actinomycin D treatments. The results showed that circ_0129657, rather than IQGAP2 linear mRNA, was resistant to Rnase R digestion (). Moreover, actinomycin D treatment inhibited the expression of IQGAP2 linear mRNA but not circ_0129657 (). As shown in , we found that circ_0129657 was a 408 bp long circRNA that was generated by back-splicing of exons of IQGAP2 pre-mRNA. In conclusion, these results suggested that the expression of circ_0129657 was upregulated in OGD-induced HBMECs.
Figure 2. Silencing circ_0129657 promoted cell proliferation and inhibited apoptosis and inflammatory factor expression in OGD-induced HBMECs. (A-M) HBMECs were introduced with si-circ_0129657 or si-NC before OGD treatment. (A) Circ_0129657 knockdown efficiency was detected by RT-qPCR. (B) Effect of silencing circ_0129657 on cell viability was assessed by CCK-8 assay. (C) Changes in cell proliferation were detected after silencing circ_0129657 by EdU assay. (D) The caspase-3 activity was investigated by ELISA kits. (E) The effect of silencing circ_0129657 on apoptosis was measured by flow cytometry assay in OGD-induced HBMECs. (F–I) The protein expression levels of PCNA, Bax and Bcl2 were assessed by western blot assay after silencing circ_0129657 expression. (J–M) The protein expression levels of IL-1β, IL-6 and TNF-α were investigated by western blot assay in OGD-induced HBMECs. ***p < 0.001, n = 3.
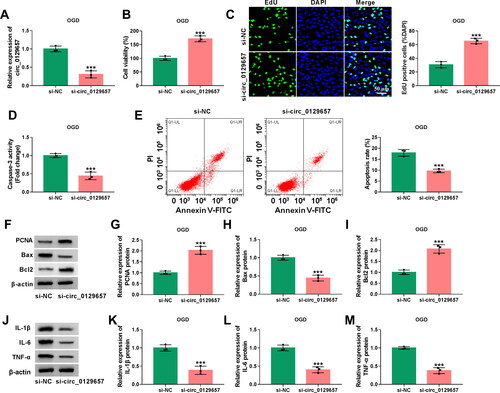
Effects of circ_0129657 silencing on the biological properties in OGD-induced HBMECs
We further explored the effects of circ_0129657 silencing on the biological properties in OGD-induced HBMECs. Firstly, we examined the efficiency of si-circ_0129657 transfection, and the results showed that circ_0129657 was significantly low expressed in the si-circ_0129657 group (). Then, we examined the effects of circ_0129657 silencing on cell viability and cell proliferation capacity in OGD-induced HBMECs, and the results showed that in the si-circ_0129657 group, cell viability and proliferation capacity were significantly upregulated (). Next, we found a significant decrease in caspase-3 activity and apoptosis rate after reducing the expression of circ_0129657 in OGD-induced HBMECs (). To verify the above results, we examined the marker proteins of proliferation and apoptosis, and the results were consistent with the above results (). Finally, we found that in OGD-induced HBMECs, the expression of inflammatory factors IL-1β, IL-6 and TNF-α was also significantly reduced after silencing circ_0129657 (). These results indicated that silencing circ_0129657 promoted cell proliferation and inhibited apoptosis and inflammatory factor secretion in OGD-induced HBMECs.
Circ_0129657 acted as a sponge for miR-194-5p
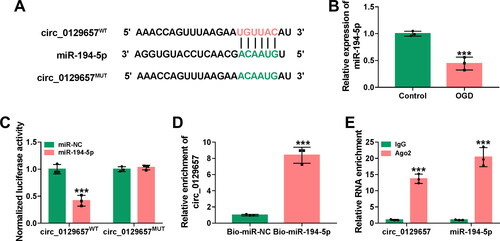
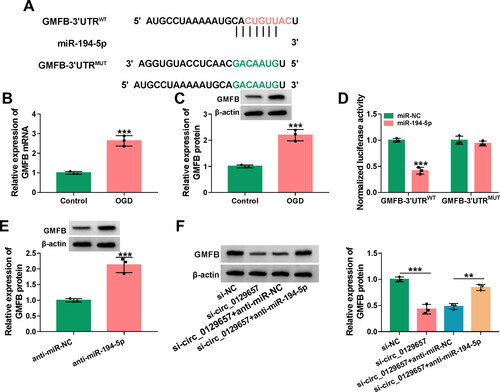
Further, we explored how circ_0129657 exerted the regulatory effect in OGD-induced HBMECs. We used the circInteractome prediction website (https://circinteractome.nia.nih.gov/) to search for many potential miRNAs that are potentially bound to circ_0129657. Among these miRNAs, miR-194-5p and miR-496 are lowly expressed in a stroke model and inhibit the progression of the disease [Citation4, Citation31]. Then, these two miRNAs were subjected to RT-qPCR analysis responding to circ_0129657 downregulation. As shown in Figure S2B, miR-194-5p was significantly increased in HBMECs. Hence, we chose miR-194-5p for further research. The binding sites for circ_0129657 and miR-194-5p were shown in . The expression of miR-194-5p was significantly reduced in OGD-induced HBMECs (). To validate the relationship between circ_0129657 and miR-194-5p, we performed luciferase reporter, RNA pull-down and RIP assays. Dual-luciferase reporter results showed that the luciferase activity of circ_0129657WT was significantly reduced after co-transfection with miR-194-5p mimic in OGD-induced HBMECs (). The results of RNA pull-down and RIP assays showed the interaction between circ_0129657 and miR-194-5p (). These results suggested that circ_0129657 could act as a sponge for miR-194-5p.
Figure 3. Circ_0129657 directly bound to miR-194-5p. (A) The putative miR-194-5p binding of circ_0129657 was shown. (B) The expression of miR-194-5p was measured by RT-qPCR in OGD-induced HBMECs. (C) The luciferase activity of circ_0129657WT and circ_0129657MUT was detected by dual-luciferase reporter assay. (D and E) RIP and RNA pull-down assays were used to assess the interaction between circ_0129657 and miR-194-5p. ***p < 0.001, n = 3.
The effects of circ_0129657 silencing on the biological properties of OGD-induced HBMECs could be partially reversed by miR-194-5p downregulation
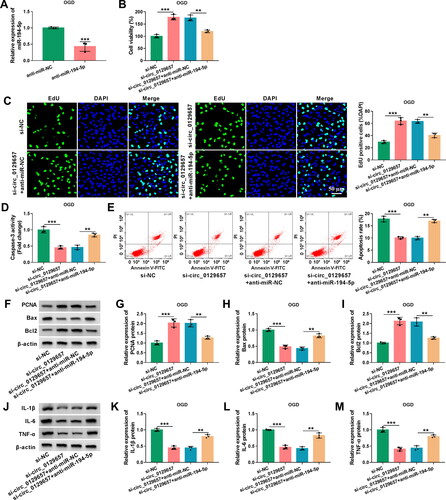
We further investigated whether miR-194-5p was involved in the regulation of circ_0129657 in OGD-induced HBMECs. We detected a significant reduction in miR-194-5p expression after transfection with miR-194-5p inhibitor in OGD-induced HBMECs (). Cell viability and EdU assays revealed that circ_0129657 silencing-driven viability () and proliferation () enhancement was reversed by transfection with miR-194-5p inhibitor in OGD-induced HBMECs. Knockdown of circ_0129657 inhibited caspase-3 activity and cell apoptosis rate, while these effects were reversed by transfection with miR-194-5p inhibitor (). Silencing circ_0129657 promoted the expression of PCNA and Bcl2 and inhibited the expression of Bax, while these effects were abated by miR-194-5p reduction (). Knockdown of circ_0129657 suppressed the expression of inflammatory factors, which was reverted by transfection with miR-194-5p inhibitor (). The above findings demonstrated that circ_0129657 silencing exerted regulatory effects on the biological properties of OGD-induced HBMECs through miR-194-5p.
Figure 4. MiR-194-5p inhibitor partially rescued the effects of circ_0129657 silencing on cell biological properties in OGD-induced HBMECs. (A) The knockdown efficiency of miR-194-5p was examined by RT-qPCR in HBMECs after transfection by anti-miR-194-5p or anti-miR-NC. (B-M) HBMECs were introduced with si-circ_0129657 + anti-miR-194-5p, si-circ_0129657 + anti-miR-NC, si-circ_0129657 or si-NC before OGD treatment. (B) Cell viability was assessed by CCK-8 assay. (C) EdU assay was used to measure cell proliferative capacity. (D and E) Caspase-3 activity and apoptosis of cells were assessed by ELISA kits and flow cytometry assay, respectively. (F-I) The western blot assay examined the protein expression of PCNA, Bax and Bcl2 in OGD-induced HBMECs after the indicated transfection. (J-M) Western blot assay was used to assess the expression of IL-1β, IL-6 and TNF-α. **p < 0.01, ***p < 0.001, n = 3.
GMFB was a target of miR-194-5p
We used the targetscan database (http://www.targetscan.org/vert_72/) to predict GMFB as a potential target of miR-194-5p. showed the binding sites of miR-194-5p and GMFB. In OGD-induced HBMECs, the mRNA and protein expression levels of GMFB were significantly elevated (). Reporter assays showed that the luciferase activity of GMFB-3′UTRWT was reduced after transfection with miR-194-5p mimic (). However, no reduction was observed in the luciferase activity of GMFB-3′UTRMUT with miR-194-5p overexpression (). In OGD-induced HBMECs after transfection with miR-194-5p inhibitor, the protein expression level of GMFB was significantly increased (), indicating that miR-194-5p regulated the expression of GMFB. In addition, functional analysis displayed that GMFB overexpression might overturn the repression of miR-194-5p on OGD-induced HBMEC apoptosis and inflammatory response (Figure S3). Furthermore, the protein expression level of GMFB was decreased by circ_0129657 silencing, while this effect was reversed after transfection with miR-194-5p inhibitor (). Briefly, circ_0129657 could regulate the expression of GMFB through miR-194-5p.
Figure 5. MiR-194-5p targeted GMFB. (A) The binding site of miR-194-5p and GMFB was shown. (B and C) The mRNA expression and protein expression of GMFB were assessed by RT-qPCR and western blot assay. (D) Dual-luciferase reporter assay was used to measure the luciferase activity of GMFB-3′UTRWT or GMFB-3′UTRMUT after co-transfection with miR-194-5p mimic or miR-NC mimic. (E) The protein expression of GMFB was tested after transfection with miR-194-5p inhibitor by western blot assay. (F) Western blot assay was used to check the protein expression of GMFB after transfection with si-NC, si-circ_0129657, si-circ_0129657 + anti-miR-NC or si-circ_0129657 + anti-miR-194-5p in OGD-induced HBMECs. **p < 0.01, ***p < 0.001, n = 3.
Overexpression of GMFB reversed the effects of circ_0129657 silencing on cell biological properties in OGD-induced HBMECs
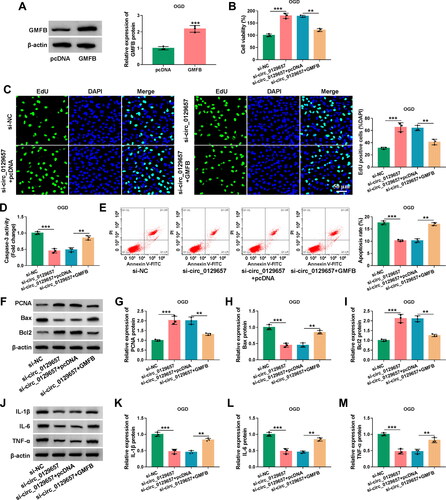
We had known that circ_0129657 could regulate the expression of GMFB through miR-194-5p. We then investigated the regulatory role of GMFB in circ_0129657 regulation of cellular functions. After transfection of GMFB expression plasmid in OGD-induced HBMECs, the protein expression level of GMFB was significantly upregulated (). Overexpression of GMFB in OGD-induced HBMECs could partially reverse cell viability and proliferation promotion induced by circ_0129657 silencing (). The activity of caspase-3 was significantly reduced by circ_0129657 silencing, and this was reversed by GMFB expression elevation in OGD-induced HBMECs (). Moreover, circ_0129657 silencing-mediated apoptosis inhibition was upregulated by GMFB expression elevation (). We examined the expression of PCNA, Bax and Bcl2 proteins, and the impact of circ_0129657 silencing on these marker proteins was reversed by GMFB expression elevation (). Additionally, reduced expression of circ_0129657 inhibited the expression of inflammatory factors, which was reversed by GMFB expression elevation (). In addition, our data displayed that the downregulation of circ_0129657 might relieve cerebral ischemia/reperfusion injury in MCAO mice (Figure S4). In conclusion, the effects of circ_0129657 silencing on cell biological properties were reversed by GMFB restoration.
Figure 6. The influence of circ_0129657 and GMFB on OGD-induced HBMECs progression. (A) The protein expression of GMFB was detected using western blot assay in OGD-induced HBMECs after transfection by GMFB expression plasmid or pcDNA control. (B-M) HBMECs were transfected with si-NC, si-circ_0129657, si-circ_0129657 + pcDNA or si-circ_0129657 + GMFB expression plasmid. (B) CCK-8 assay was used to analyze the viability of cells treated as indicated. (C) EdU assay was used to investigate cell proliferation. (D) ELISA kits were used to detect caspase-3 activity in treated HBMECs. (E) Flow cytometry assay was used to assess the apoptosis rate of treated HBMECs. (F–I) The expression levels of PCNA, Bax and Bcl2 were tested by western blot assay. (J–M) Western blot assay was used to measure the protein expression of IL-1β, IL-6 and TNF-α. **p < 0.01, ***p < 0.001, n = 3.
Discussion
Stroke has a very low chance of being cured [Citation32, Citation33]. The sequelae of the disease are numerous and can cause a range of problems such as disability, aphasia, and mental disorders in severe cases [Citation32, Citation34, Citation35]. In this study, we found by functional assays that silencing circ_0129657 promoted cell proliferation and inhibited apoptosis. Among the molecular mechanisms of circ_0129657, we found that circ_0129657 acted as a sponge for miR-194-5p and regulated GMFB expression in OGD-induced HBMECs through miR-194-5p. Furthermore, the effects of circ_0129657 silencing on the biological properties could be counteracted by the downregulation of miR-194-5p or overexpression of GMFB. In short, circ_0129657 could regulate the biological properties of OGD-induced HBMECs through the miR-194-5p/GMFB axis.
CircRNAs have important regulatory roles in many diseases, such as chronic liver diseases [Citation36], atherosclerosis [Citation37], and ischemic stroke [Citation12]. Xu et al. reported that circ_0129657 was highly expressed in OGD-induced HBMECs [Citation12]. Consistently, our data revealed that circ_0129657 expression was significantly upregulated in OGD-induced HBMECs. We also unveiled that silencing circ_0129657 promoted cell proliferation and inhibited cell apoptosis and inflammatory factor secretion in OGD-induced HBMECs.
We further investigated the specific regulatory mechanisms of circ_0129657 regulation in OGD-induced HBMECs. Some studies have reported that circRNAs can function as sponges for miRNAs, thereby being involved in the disease process [Citation10, Citation12]. We first confirmed that miR-194-5p was a downstream target of circ_0129657. MiR-194-5p has established a critical role in human diseases. For instance, miR-194-5p exerts a suppressive role in nephroblastoma and ovarian cancer [Citation38, Citation39]. MiR-194-5p can attenuate myocardial ischemia/reperfusion injury depending on the regulation of MAPK1 expression through the PTEN/AKT pathway [Citation40]. Moreover, dysregulation of miR-194-5p is associated with the pathogenesis of cerebral infarction [Citation18]. Our results first concluded that circ_0129657 could act as a sponge for miR-194-5p, and miR-194-5p could function as a mediator of the circ_0129657 function.
GMFB is a brain protein that is upregulated in many malignancies or brain diseases, predicting a poor prognosis in these diseases [Citation22, Citation41, Citation42]. Moreover, plasma GMFB may be a non-invasion adjunct to neuroimaging for the diagnosis and prognosis of stroke [Citation22]. We first confirmed that GMFB was a direct miR-194-5p target. In the molecular mechanism study, circ_0129657 could regulate the expression of GMFB through miR-194-5p. Furthermore, overexpression of GMFB could counteract the effects of circ_0129657 silencing on the biological properties of OGD-induced HBMECs.
To sum up, our results showed the increased expression of circ_0129657 in OGD-induced HBMECs. Our findings suggested that silencing of circ_0129657 promoted cell proliferation and inhibited apoptosis and inflammatory factor secretion of OGD-induced HBMECs through the miR-194-5p/GMFB axis. Based on these results, circ_0129657 might be an effective molecular marker and target for stroke diagnosis and treatment.
Supplemental Material
Download MS Word (1.5 MB)Data availability statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Zhang H, Lin S, McElroy CL, et al. Circulating pro-Inflammatory exosomes worsen stroke outcomes in aging. Circ Res. 2021;129(7):1–11.
- Han B, Jiang W, Cui P, et al. Microglial PGC-1alpha protects against ischemic brain injury by suppressing neuroinflammation. Genome Med. 2021;13(1):47.
- Duran-Laforet V, Pena-Martinez C, Garcia-Culebras A, et al. Pathophysiological and pharmacological relevance of TLR4 in peripheral immune cells after stroke. Pharmacol Ther. 2021;228:107933.
- Wang X, You Z, Zhao G, et al. MicroRNA-194-5p levels decrease during deep hypothermic circulatory arrest. Sci Rep. 2018;8(1):14044.
- Lyu Z, Park J, Kim KM, et al. A neurovascular-unit-on-a-chip for the evaluation of the restorative potential of stem cell therapies for ischaemic stroke. Nat Biomed Eng. 2021;5(8):847–863.
- Koch PJ, Park CH, Girard G, et al. The structural connectome and motor recovery after stroke: predicting natural recovery. Brain. 2021;144(7):2107–2119.
- Zeiger WA, Marosi M, Saggi S, et al. Barrel cortex plasticity after photothrombotic stroke involves potentiating responses of pre-existing circuits but not functional remapping to new circuits. Nat Commun. 2021;12(1):3972.
- Bonkhoff AK, Schirmer MD, Bretzner M, et al. Outcome after acute ischemic stroke is linked to sex-specific lesion patterns. Nat Commun. 2021;12(1):3289.
- Chen L, Shan G. CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49–57.
- Guo PW, Huang HT, Ma J, et al. Circular RNA-0007059 protects cell viability and reduces inflammation in a nephritis cell model by inhibiting microRNA-1278/SHP-1/STAT3 signaling. Mol Med. 2021;27(1):113.
- Wang YY, Wang YZ, Zhang HY, et al. The role of circular RNAs in brain and stroke. Front Biosci (Landmark Ed). 2021;26(5):36–50.
- Xu X, Wu Z, Qiu H, et al. Circular RNA circPHC3 promotes cell death and apoptosis in human BMECs after oxygen glucose deprivation via miR-455-5p/TRAF3 axis in vitro. Neuropsychiatr Dis Treat. 2021;17:147–156.
- Kapoor P, Chowdhry A, Bagga DK, et al. MicroRNAs in oral fluids (saliva and gingival crevicular fluid) as biomarkers in orthodontics: systematic review and integrated bioinformatic analysis. Prog Orthod. 2021;22(1):31.
- Homayoonfal M, Asemi Z, Yousefi B. Targeting microRNAs with thymoquinone: a new approach for cancer therapy. Cell Mol Biol Lett. 2021;26(1):43.
- Rajool Dezfuly A, Safaee A, Salehi H. Therapeutic effects of mesenchymal stem cells-derived extracellular vesicles’ miRNAs on retinal regeneration: a review. Stem Cell Res Ther. 2021;12(1):530.
- Wang T, Chen N, Ren W, et al. Integrated analysis of circRNAs and mRNAs expression profile revealed the involvement of hsa_circ_0007919 in the pathogenesis of ulcerative colitis. J Gastroenterol. 2019;54(9):804–818.
- Osei J, Kelly W, Toffolo K, et al. Thymosin beta 4 induces significant changes in the plasma miRNA profile following severe traumatic brain injury in the rat lateral fluid percussion injury model. Expert Opin Biol Ther. 2018;18(sup1):159–164.
- Zou JB, Chai HB, Zhang XF, et al. Reconstruction of the lncRNA-miRNA-mRNA network based on competitive endogenous RNA reveal functional lncRNAs in cerebral infarction. Sci Rep. 2019;9(1):12176.
- Takuma A, Abe A, Saito Y, et al. Gene expression analysis of the effect of ischemic infarction in whole blood. IJMS. 2017;18(11):2335.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Yuan Z, Yu Z, Zhang Y, et al. Analysis of the clinical diagnostic value of GMFB in cerebral infarction. Curr Pharm Biotechnol. 2020;21(10):955–963.
- Jiang S, Zhao G, Lu J, et al. Silencing of circular RNA ANRIL attenuates oxygen-glucose deprivation and reoxygenation-induced injury in human brain microvascular endothelial cells by sponging miR-622. Biol Res. 2020;53(1):27.
- Yang X, Li X, Zhong C, et al. Circular RNA circPHKA2 relieves OGD-Induced human brain microvascular endothelial cell injuries through competitively binding miR-574-5p to modulate SOD2. Oxid Med Cell Longev. 2021;2021:3823122.
- Li J, Wang J, Wang Z. Circ_0006768 upregulation attenuates oxygen-glucose deprivation/reoxygenation-induced human brain microvascular endothelial cell injuries by upregulating VEZF1 via miR-222-3p inhibition. Metab Brain Dis. 2021;36(8):2521–2534.
- Wu L, Chen C, Li Y, et al. UPLC-Q-TOF/MS-based serum metabolomics reveals the anti-ischemic stroke mechanism of nuciferine in MCAO rats. ACS Omega. 2020;5(51):33433–33444.
- Kaundal RK, Sharma SS. Ameliorative effects of GW1929, a nonthiazolidinedione PPARγ agonist, on inflammation and apoptosis in focal cerebral ischemic-reperfusion injury. Curr Neurovasc Res. 2011;8(3):236–245.
- Kaundal RK, Shah KK, Sharma SS. Neuroprotective effects of NU1025, a PARP inhibitor in cerebral ischemia are mediated through reduction in NAD depletion and DNA fragmentation. Life Sci. 2006;79(24):2293–2302.
- Zuo X, Lu J, Manaenko A, et al. MicroRNA-132 attenuates cerebral injury by protecting blood-brain-barrier in MCAO mice. Exp Neurol. 2019;316:12–19.
- Hu Q, Manaenko A, Bian H, et al. Hyperbaric oxygen reduces infarction volume and hemorrhagic transformation through ATP/NAD(+)/Sirt1 pathway in hyperglycemic Middle cerebral artery occlusion rats. Stroke. 2017;48(6):1655–1664.
- Wang F, Liu J, Wang D, et al. Knockdown of circ_0007290 alleviates oxygen-glucose deprivation-induced neuronal injury by regulating miR-496/PDCD4 axis. Metab Brain Dis. 2022;37(3):807–818.
- Ajoolabady A, Wang S, Kroemer G, et al. Targeting autophagy in ischemic stroke: from molecular mechanisms to clinical therapeutics. Pharmacol Ther. 2021;225:107848.
- Zhang SR, Phan TG, Sobey CG. Targeting the immune system for ischemic stroke. Trends Pharmacol Sci. 2021;42(2):96–105.
- Camilleri M. Gastrointestinal motility disorders in neurologic disease. J Clin Invest. 2021;131:e143771.
- D’Souza A, Dave KM, Stetler RA, et al. Targeting the blood-brain barrier for the delivery of stroke therapies. Adv Drug Deliv Rev. 2021;171:332–351.
- Zeng X, Yuan X, Cai Q, et al. Circular RNA as an epigenetic regulator in chronic liver diseases. Cells. 2021;10(8):1945.
- Sun X, Deng K, Zang Y, et al. Exploring the regulatory roles of circular RNAs in the pathogenesis of atherosclerosis. Vascul Pharmacol. 2021;141:106898.
- Liu H, Ren SY, Qu Y, et al. MiR-194-5p inhibited metastasis and EMT of nephroblastoma cells through targeting crk. Kaohsiung J Med Sci. 2020;36(4):265–273.
- Bai R, Dou K, Wu Y, et al. The NF-κB modulated miR-194-5p/IGF1R/PPFIBP axis is crucial for the tumorigenesis of ovarian cancer. J Cancer. 2020;11(12):3433–3445.
- Zhang Q, Wu X, Yang J. miR-194-5p protects against myocardial ischemia/reperfusion injury via MAPK1/PTEN/AKT pathway. Ann Transl Med. 2021;9(8):654.
- Xu FF, Zhang ZB, Wang YY, et al. Brain-derived glia maturation factor beta participates in lung injury induced by acute cerebral ischemia by increasing ROS in endothelial cells. Neurosci Bull. 2018;34(6):1077–1090.
- Yamazaki H, Tateyama H, Asai K, et al. Glia maturation factor-beta is produced by thymoma and may promote intratumoral T-cell differentiation. Histopathology. 2005;47(3):292–302.

