Abstract
As an autoimmune disease, systemic lupus erythematosus (SLE) can affect multiple organs and systems. Whether SLE can increase the risk of coeliac disease (CeD) was not evaluated until now. We performed a two-sample Mendelian randomisation study to evaluate the relationship between SLE and CeD, and found that SLE can significantly increase the risk of CeD, suggesting the association between SLE and abnormal intestinal immune microenvironment.
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disease characterised by the production of autoantibody. The diseases predominantly affects females and results in progressive damage to multiple organs over time [Citation1,Citation2]. The association between SLE and other autoimmune diseases was examined in previous studies. Coeliac disease (CeD) is an autoimmune genetic disorder that primarily affects the digestive tract and is caused by intolerance to dietary gluten. The pattern of inheritance is unknown. When gluten protein is consumed, it triggers an immune response that damages the small intestinal villi and prevents adequate absorption of nutrients [Citation3,Citation4]. Jun Inamo found that CeD was associated with significantly increased risk of SLE by Mendelian randomisation (MR) analysis [Citation5]. However, whether SLE can also increase the risk of CeD was not evaluated until now. A case study reported that a patient developed CeD after the diagnosis of SLE for 8 years [Citation6]. In addition, about 70% of patients with SLE showed abdominal injury in the autopsy and endoscopic study although only 10% of these patients exhibited gastrointestinal symptoms [Citation7,Citation8], suggesting SLE might increase the risk of gastrointestinal diseases, including CeD.
Mendelian randomisation (MR) analysis is an effective method that can infer the causal relationship between exposure and outcome by constructing the single nucleotide polymorphism (SNP) as an instrument variable (IV) based on genome-wide association studies [Citation9]. Utilising the Mendelian law of heredity, which assumes that parental alleles are randomly assigned to offspring, MR simulates the causal relationships between exposures and outcomes. In addition, MR draws on the experiences gained from randomised trials. Therefore, MR provides a powerful tool for investigating causality [Citation10]. To examine the causal relationship between SLE and CeD, we performed a two-sample MR study with SLE as the exposure and CeD as the outcome.
Methods
The summary data from open access genome-wide association studies data were obtained (https://gwas.mrcieu.ac.uk/). We extracted summary statistics for SLE involving 5,201 cases and 9,066 controls from European ancestry [Citation11]. For CeD, a GWAS set involving 1,973 cases and 210,964 controls from European ancestry was obtained (https://gwas.mrcieu.ac.uk/datasets/finn-b-K11_COELIAC/). A threshold of linkage disequilibrium for the selection of significant verified SNPs (p < 5.00 × 10−08) associated with SLE as IVs was set at r2 <0.001 to improve inference, and clumping distance was set at 10,000 kb.
The inverse variance weighting (IVW) method was used as the primary approach, with supplementary methods being employed utilising other algorithms. The effect size estimated the causal relationships between SLE and the risk of CeD was exhibited by the β-coefficient. Gene names of rsID were obtained through NCBI (https://www.ncbi.nlm.nih.gov/snp/). The data were analysed with R, version 4.2.2, and package ‘‘TwoSampleMR”. P values of less than 0.05 were considered statistically significant.
Results
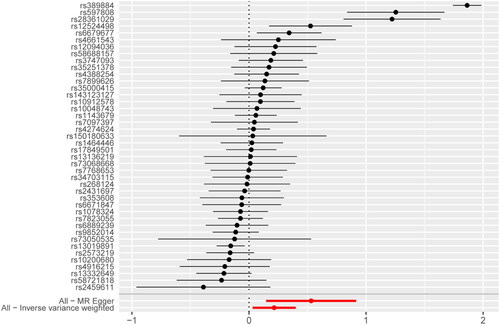
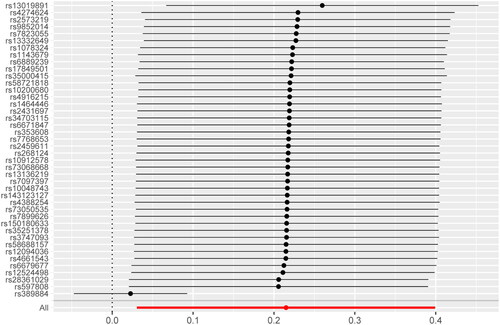
After removing linkage disequilibrium and SNPs being palindromic with intermediate allele frequencies, 42 SNPs were incorporated into the final analysis. IVW method showed that the genetic risk of SLE was associated with the increased risk of CeD (βIVW = 0.21, p = 0.022), and odds ratio (OR) = 1.24 (95% confidence interval (CI) = 1.03–1.49), as indicated in . The MR-Egger method also supported the result (βMR-Egger = 0.53, p = 0.010), and OR = 1.70 (95%CI = 1.16–2.50). A series of sensitivity analyses were conducted to examine the association. Cochran’s Q test suggested possible heterogeneity between IVs estimates (p < 0.05). However, the MR-PRESSO indicated that the association still existed after removing the outlier. MR-Egger regression showed no directional horizontal pleiotropy (intercept = −0.126, p = 0.076). MR leave-one-out sensitivity analysis showed stable results, except for rs389884 (). The top SNPs identified were as follows: (1) rs389884, a single nucleotide variant (SNV) of STK19, encoding a serine/threonine kinase; (2) rs597808, a SNV of ATXN2 intron, the protein encoded by this gene can involve in endocytosis and modulate mTOR signals; (3) rs28361029, a SNV not well described in NCBI; (4) rs12524498, a SNV not well described in NCBI; (5) rs6679677, a SNV of PHTF1.
Discussion
Our study indicated that SLE was associated with the increased risk of CeD. The possibility of CeD should be taken into consideration in the patients with SLE accompanied by gastrointestinal symptoms. CeD is a T-cell mediated disease, previous studies reported that abnormal T cell homeostasis in the intestine in patients with CeD [Citation12,Citation13]. Gut microbiota dysbiosis are commonly associated with abnormal gut immune microenvironment [Citation14]. Our team reported that SLE patients have significantly different gut microbiota composition compared with healthy controls, and faecal microbiota transplantation could serve as a feasible, safe, and potentially effective therapy for patients with SLE through modifying the gut microbiome and its metabolic profile [Citation15]. Our study results in combination with that from Jun Inamo [Citation5], suggesting that SLE and CeD exist bidirectional casual association, abnormal intestinal immune microenvironment might play an important role in the development of SLE.
Authors’ contributions
Q.W., Q.L. and M.Z. designed the study, were responsible for the analysis and interpretation of data. Q.W. and S.J. drafted and revised the article. All authors approved the final version of the article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data is available on individual request by contacting the corresponding author.
Additional information
Funding
References
- Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):1–3.
- Caielli S, Wan Z, Pascual V. Systemic lupus erythematosus pathogenesis: interferon and Beyond. Annu Rev Immunol. 2023;41:533–560.
- Catassi C, Verdu EF, Bai JC, et al. Coeliac disease. Lancet. 2022;399(10344):2413–2426.
- Iversen R, Sollid LM. The immunobiology and pathogenesis of celiac disease. Annu Rev Pathol. 2023;18:47–70.
- Inamo J. Association between celiac disease and systemic lupus erythematosus: a mendelian randomization study. Rheumatology (Oxford). 2020;59(9):2642–2644.
- Mirza N, Bonilla E, Phillips PE. Celiac disease in a patient with systemic lupus erythematosus: a case report and review of literature. Clin Rheumatol. 2007;26(5):827–828.
- Takeno M, Ishigatsubo Y. Intestinal manifestations in systemic lupus erythematosus. Intern Med. 2006;45(2):41–42.
- Endo H, Kondo Y, Kawagoe K, et al. Lupus enteritis detected by capsule endoscopy. Intern Med. 2007;46(18):1621–1622.
- Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. Jama. 2017;318(19):1925–1926.
- Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of Hypothesis-Free causality. Annu Rev Genomics Hum Genet. 2015;16:327–350.
- Bentham J, Morris DL, Graham DSC, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47(12):1457–1464.
- Sollid LM, Jabri B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat Rev Immunol. 2013;13(4):294–302.
- Meresse B, Malamut G, Cerf-Bensussan N. Celiac disease: an immunological jigsaw. Immunity. 2012;36(6):907–919.
- Sollid LM, Iversen R. Tango of B cells with T cells in the making of secretory antibodies to gut bacteria. Nat Rev Gastroenterol Hepatol. 2023;20(2):120–128.
- Huang C, Yi P, Zhu M, et al. Safety and efficacy of fecal microbiota transplantation for treatment of systemic lupus erythematosus: an EXPLORER trial. J Autoimmun. 2022;130:102844.