Abstract
Circular RNAs (circRNAs) has been manifested to be involved in the development of human diseases, including sepsis-associated acute kidney injury (SA-AKI). However, the function and mechanism of circ_0006944 in SA-AKI has not been validated. Lipopolysaccharide (LPS) was utilised to induce AKI cell model. Levels of genes and proteins were monitored by quantitative real-time polymerase chain reaction (qRT-PCR) and western blot. Cell counting kit 8 assay, EdU assay and flow cytometry were exploited to estimate cell proliferation and apoptosis. The concentrations of inflammation factors were measured via using ELISA assay. The levels of MDA and SOD were tested by the corresponding kits. The relationship between miR-205-5p and circ_0006944 or UBL4A was verified by dual-luciferase reporter assay and RIP assay. Circ_0006944 was overexpressed in SA-AKI patients, and interference of circ_0006944 restrained LPS-stimulated HK2 cell proliferation repression, apoptosis, inflammation and oxidative stress. Mechanistically, circ_0006944 could sponge miR-205-5p, and miR-205-5p interference counteracted circ_0006944 inhibition-mediated impact on the biological functions in LPS-induced HK2 cell. Additionally, UBL4A was targeted by miR-205-5p, and UBL4A overexpression also partially abolished the repressive impacts of miR-205-5p on LPS-triggered HK2 cell damage. Importantly, circ_0006944 sponged miR-205-5p to mediate the expression of UBL4A. Our outcomes identified that circ_0006944 exacerbated SA-AKI development via miR-205-5p/UBL4A axis, which might be a potential treatment and diagnosis biomarker for SA-AKI.
Keywords:
Introduction
Sepsis is a disease characterised by systemic inflammation, which usually leads to organ failure and seriously endangers human life and health [Citation1]. In recent years, the incidence of sepsis has been high and increasing [Citation2]. Accumulating studies have shown that sepsis could enhance the apoptosis of renal cells and the release of inflammatory factors, which led to acute kidney injury (AKI) [Citation3]. Approximately half of AKI patients are associated with sepsis and are a common complication of sepsis [Citation4]. Although great progress has been made in the treatment of sepsis-associated AKI (SA-AKI), the therapeutic effect is still unsatisfactory [Citation5]. In addition, SA-AKI has a complex characteristic pathophysiological mechanism. Therefore, the study of complex modulation network is urgently needed to investigate more effective treatment strategies for SA-AKI. A deep understanding of the underlying mechanism of SA-AKI dysfunction may provide new therapeutic targets for the prevention of SA-AKI.
More and more studies show that non-coding RNA plays an important role in the occurrence and development of SA-AKI. As a new type of non-coding RNA, circular RNAs (circRNAs) are involved in the regulation of a variety of cell biological process. CircRNAs are a class of non-coding RNAs with covalently closed-loop structures, which lack poly(A) tails and 5’caps, and can regulate gene expression by targeting microRNAs (miRNAs) [Citation6]. At present, numerous circRNAs exhibited pivotal regulatory roles in many human diseases, including SA-AKI [Citation7]. For example, Lu et al. manifested that the abundance of circHIPK3 was increased in SA-AKI patients and human tubule epithelial cells (HK2) upon lipopolysaccharide (LPS) treatment. Besides that, the circHIPK3 deficiency promoted the survival of HK2 cells, and reduced cell apoptosis, inflammation and oxidative damage after LPS stimulation by mediating miR-338-3p/FOXA1 pathway axis [Citation8]. Wang et al. showed that interference of circVMA21 resulted in the inhibition of cell damage in HK2 cells by functioning as a miR-7-5p decoy and mediating PPARA level under LPS stimulation [Citation9]. Zhou et al. declared that circ-BNIP3L knockdown attenuated LPS-evoked HK2 damage via regulating miR-370-3p/MYD88 axis [Citation10]. These findings confirmed that circRNAs were implicated in the modulation of SA-AKI morbidity by mediating the miRNA/mRNA pathway, and may be a potential therapeutic target for this disease. Circ_0006944 is derived from its host gene GTF2I, and has a genomic length of 294 bp, which is locates at chr7:74119495-74131270. Studies have shown that the expression of circ_0006944 is increased in the serum of sepsis patients compared with controls [Citation11]. However, the biological function of circ_0006944 in SA-AKI is indistinct.
Herein, this work aimed to disclose the potential mechanisms underlying circ_0006944 in HK2 growth, inflammation, and oxidative stress under LPS stimuli, which may provide a new insight into the pathogenesis of SA-AKI.
Materials and methods
Serum samples
The human serum specimens were acquired from 19 SA-AKI patients and 17 healthy volunteers recruited from Huangshi Central Hospital from August 2019 to June 2021. These serum samples were centrifuged and stored at −80 °C. This project was ratified by the Ethics Committee of Huangshi Central Hospital. All participants have signed written informed consents.
Cell culture, treatment and cell transfection
HK2 cells were procured from Procell (Wuhan, China), and then cultivated in Dulbecco’s modified Eagle medium (DMEM, Solarbio, Beijing, China) supplemented with 1% Penicillin-Streptomycin (Solarbio) and 10% foetal bovine serum (Solarbio) at 37 °C in an incubator with 5% CO2. After cells were passaged 3 generations, different doses of LPS (0, 4, 8 and 12 μg/mL) was adopted to induce HK2 cell damage in vitro. LPS (8 μg/mL) was used to stimulate HK2 cells in subsequent functional assay. Small interfering RNA to circ_0006944 (si-circ_0006944), miR-205-5p mimic and inhibitor, UBL4A overexpression plasmid, and their control were acquired from GenePharma (Shanghai, China). When cells reached 70% confluence, Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) was adopted to execute cell transfection.
The outcomes analysed with the quantitative real-time polymerase chain reaction (qRT-PCR)
TRIzol reagent (Invitrogen) was adopted to obtain total RNA. The concentration and purity of RNA were analysed by NanoDrop-1000 apparatus. Cytoplasmic and nuclear RNA isolation was implemented by employing the PARISTM Kit (Life Technologies, Scotland, UK) to disclose the localisation of circ_0006944. Meanwhile, 3 U/μg of RNase R (Epicentre, Madison, WI, USA) was used to incubate total RNA at 37 °C for 20 min. Additionally, the complementary DNA (cDNA) was formed from 2 μg RNA by the application of Reverse Transcription Kit (Invitrogen). Subsequently, SYBR Green (Invitrogen) was utilised to perform qRT-PCR. Primer sequences were exhibited in . The relative level was normalised to GADPH or U6 via utilising the 2-ΔΔCt strategy.
Table 1. The Primer sequences for qRT-PCR.
Cell viability assessment
Cell Counting Kit-8 (CCK-8) was exploited for the detection of cell viability. Shortly, HK2 cells were cultivated in 96-well plates for 48 h. After that, CCK-8 solution (Solarbio) was supplemented into cell well. 4 h later, the optical density value was monitored via microplate reader (Bio-Rad, Laboratories Inc., Hercules, CA, USA).
5-ethynyl-2’-deoxyuridine (EdU) assay
HK2 cells were labelled with EdU solution (RiboBio, Guangzhou, China) for 2 h, After that cells were fixed and permeabilized, Apollo staining was used to incubate cells for 30 min. Followed by cell nucleus was stained using DAPI, the EdU-positive cells were gauged through using fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry
HK2 cells was collected and resuspended, and then incubated with Annexin V-fluorescein isothiocyanate (Annexin V-FITC) and propiduim iodide (Solarbio) for 15 min at room temperature. Afterwards, flow cytometry (BD Biosciences, San Jose, CA, USA) was applied for cell apoptosis analysis.
Western blot assay
Total proteins were isolated from HK2 cell by RIPA lysis buffer (Beyotime, Shanghai, China). Subsequently, the protein was separated by loading on 10% SDS-PAGE and then transferred to polyvinylidene fluoride (PVDF) membrane (Beyotime, Shanghai, China). After blocking for 1 h, primary antibodies against Bax (ab32503, 1:1000), Cleaved-caspase 3 (ab32042, 1:500), UBL4A (ab241427, 1:1000) and GAPDH (ab181602, 1:10000) were utilised to incubate the membranes, followed by incubation with Goat anti-Rabbit-HRP-conjugated second antibody (ab205718, 1:5000). All antibodies were acquired from Abcam (Cambridge, UK). The bands were visualised by using BeyoECL Star Kit (Beyotime).
Enzyme-linked immunosorbent assay (ELISA)
ELISA experiments are used to gauge the levels of inflammatory factors in cells. Shortly, HK2 cell culture medium was collected, and based on the operation manual of corresponding ELISA detection kits (Elabscience, Wuhan, China), the levels of IL-6, IL-1β and TNF-α were subsequently evaluated.
Evaluation of malondialdehyde (MDA) and superoxide dismutase (SOD) levels
Oxidative stress damage was detected by assessing the expression of oxidative stress-associated factors (MDA, SOD). In brief, the supernatant of HK2 cells were used to estimate the levels of MDA and SOD by using MDA (BC0025, Solarbio) or SOD detection kit (BC0170, Solarbio) according to the manufacturer’s protocols.
Dual-luciferase reporter assays
Binding sites among miR-205-5p and circ_0006944 or UBL4A were predicted by starBase v3.0. The wild-type (WT) and mutant-type (MUT) fragment of circ_0006944 or UBL4A 3’UTR comprising the binding sites and mutation site of miR-205-5p were inserted into pmirGLO (Promega, Fitchburg, WI, USA). Later on, vectors and miR-205-5p/miR-NC were transfected into HK2 cells. Lastly, luciferase activities were measured to evaluate targeting relationship.
RNA immunoprecipitation (RIP) assays
Targeting relationship among miR-205-5p and circ_0006944 or UBL4A was validated by using RIP assay. Shortly, cells were lysed and incubated with anti-Argonaute2 (Ago2) or immunoglobulin G (IgG). Thereafter, immunoprecipitated RNAs were harvested and then analysed via using qRT-qPCR.
Statistical analysis
All experiments were performed by three independent repetitions. All data were presented as the mean ± standard deviation (SD). Statistical analyses were conducted by Student’s t-test or analysis of variance (ANOVA). Pearson correlation analysis was applied for correlations analysis. GraphPad Prism 7 was applied for experimental results analysis. p < 0.05 indicated statistically significant.
Results
The level of circ_0006944 was elevated in the serum samples of SA-AKI patients and LPS-induced HK2 cells
Firstly, we investigated circ_0006944 level in the serum of SA-AKI patients and healthy volunteers, and we found that the expression of circ_0006944 was aberrantly increased in SA-AKI patients than that in healthy volunteers (). ROC curve revealed that circ_0006944 had better clinical diagnostic value (). Furthermore, HK2 cells were stimulated with LPS to trigger cell injury, and circ_0006944 was manifested to be increased in a dose-dependent manner (). Later on, we identified the stability and subcellular localisation of circ_0006944. RNase R treatment failed to degrade circ_0006944 but not linear RNA GAPDH in HK2 cells, (). Additionally, reverse transcription experiments was performed by using random and oligo (dT) 18 primers, and the results suggested the level of circ_0006944 was decreased in oligo (dT) 18 primers group than that in random primers group (). Besides that, we also demonstrated circ_0006944 mainly localised in the cytoplasmic fraction of HK2 cells (). Collectively, circ_0006944 was up-regulated in SA-AKI.
Figure 1. Circ_0006944 level was up-regulated in SA-AKI patients and LPS-exposed HK2 cells. (A) The abundance of circ_0006944 was estimated in SA-AKI patients and healthy volunteers by qRT-PCR. (B) The diagnostic value of circ_0006944 assessed by the ROC curve. (C) Circ_0006944 level was explored in HK2 cells after stimulation by different doses of LPS. (D) The circular characteristic of circ_0006944 was determined by RNase R treatment. (E) Random and Oligo(dT)18 primers were applied to explore the level of circ_0006944 and GAPDH in reverse transcription. (F) Subcellular localisation assays was executed to uncover the distribution of circ_0006944 in HK2 cells. *p < 0.05, ***p < 0.001, ****p < 0.0001.
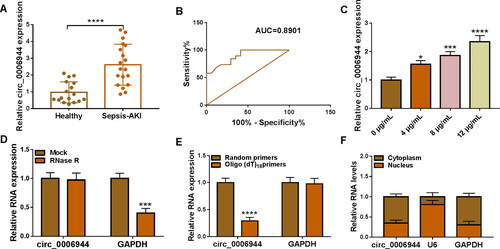
LPS treatment suppressed proliferation and expedited apoptosis, inflammatory response and oxidative stress in HK2 cells
HK2 cells were treated with different doses of LPS (0, 4, 8 and 12 μg/mL) for 24 h to disclose the impacts of LPS. It was verified that LPS exposure led to the inhibition of cell viability and proliferation (), enhancement of cell apoptosis (), Western blot analysis showed LPS treatment up-regulated Bax and Cleaved-caspase-3 protein level in HK2 cells (). Besides that, LPS exposure increased the levels of IL-6, IL-1β and TNF-α (). We also confirmed that the generation of MDA was increased while SOD was declined in HK2 cells with the increased doses of LPS (Figure 21,G). Overall, LPS stimulation constrained cell proliferation and induced cell apoptosis, inflammatory response and oxidative stress in HK2 cells.
Figure 2. LPS treatment caused HK2 cell injury. (A-J) HK2 cells were evoked to different concentrations of LPS (0, 4, 8 and 12 µg/mL) for 24 h. (A-B) Cell proliferation was checked by using CCK-8 and EdU assays. (C,D) Flow cytometry was implemented for cell apoptosis analysis. (E) The protein levels of Bax and Cleaved-caspase-3 was gauged by western blot analysis. (F–H) ELISA analysis were performed to measure the levels of IL-6, IL-1β and TNF-α. (I–J) The levels of MDA and SOD was analysed via using commercial kits. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
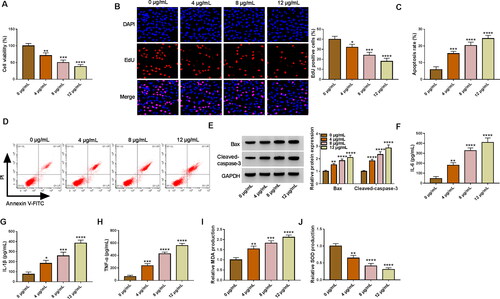
Silencing of circ_0006944 mitigated LPS-induced cell injury in HK2 cells
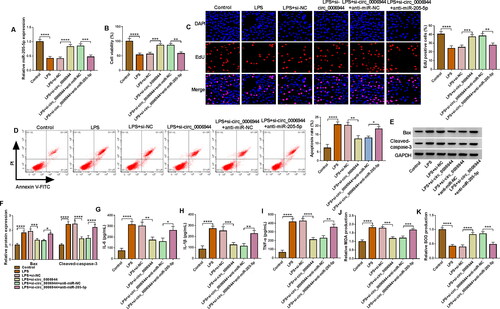
To assess the influence of circ_0006944 on LPS-induced HK2 cells, we implemented loss-of-function analyses via silencing circ_0006944. Circ_0006944 inhibition markedly impeded the elevation of LPS on circ_0006944 expression (). Functional experiments data manifested that the interference of circ_0006944 rescued the repression of LPS on cell proliferation (). Moreover, circ_0006944 knockdown distinctly counteracted the increase of LPS on cell apoptosis () and IL-6, IL-1β, TNF-α production (). Meanwhile, we demonstrate that the effects of LPS treatment on MDA and SOD levels in HK2 cells were overturned by the circ_0006944 deficiency (Figure 31,J). Collectively, these results suggested that circ_0006944 knockdown could partially reverse LPS-induced HK2 cell damage.
Figure 3. Circ_0006944 knockdown weakened the damage of HK2 cells caused by LPS treatment. HK2 cells were assigned to 4 groups: Control, LPS, LPS + si-NC and LPS + si-circ_0006944. (A) Circ_0006944 expression was tested by qRT-PCR assay. (B-D) CCK-8 assay, EdU assay and flow cytometry were implemented to investigate the viability, proliferation and apoptosis in HK2 cells, respectively. (E) Western blot assay was used to monitor the protein levels of Bax and Cleaved-caspase-3. (F-H) The concentrations of IL-6, IL-1β and TNF-α were counted via ELISA kits. (I-J) The production of MDA and SOD was checked by relevant kits. **p < 0.01, ***p < 0.001, ****p < 0.0001.
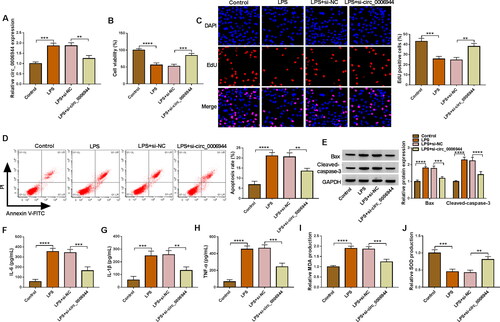
Circ_0006944 functioned as a sponge of miR-205-5p
Overlapping results of starbase 3.0 and circBank databases were represented by Venn diagrams, and three miRNAs could be targeted by circ_0006944. Further qRT-PCR analysis showed that circ_0006944 knockdown could conspicuously affect the level of miR-205-5p (). The binding sites between miR-205-5p and circ_0006944 were displayed in . MiR-205-5p mimic notably elevated miR-205-5p level in HK2 cells contrasted with miR-NC transfection (). Ulterior, the miR-205-5p introduction restrained the luciferase activity of WT-circ_0006944 group but not MUT-circ_0006944 group, affirming that circ_0006944 could bind to miR-205-5p (). Moreover, the enrichment of circ_0006944 and miR-205-5p by Ago2 protein confirmed the targeting relationship between circ_0006944 and miR-205-5p (). Furthermore, qRT-PCR results verified that miR-205-5p was apparently underexpressed in SA-AKI patient blood and LPS-treated HK2 cells (). Interestingly, a strong negatively related between circ_0006944 and miR-205-5p level was identified in SA-AKI patient blood (Figure 41). These outcomes affirmed that circ_0006944 had the sponge impact on miR-205-5p.
Figure 4. Circ_0006944 acted as a sponge of miR-205-5p. (A) Three miRNAs targeted by circ_0006944 were predicted by starbase and circBank databases. (B) The abundance of miRNA in si-NC and si-circ_0006944-transfected HK2 cells was monitored by qRT-PCR. (C) The predicted binding sites between circ_0006944 and miR-205-5p were shown. (D) MiR-205-5p level in HK2 cells transfected with miR-NC or miR-205-5p was assessed by qRT-PCR. (E-F) Dual-luciferase reporter assay and RIP assay were utilised to investigate the targeted relationship between miR-205-5p and circ_0006944. (G) MiR-205-5p abundance was obtained in SA-AKI patients and healthy volunteers by using qRT-PCR assay. (H) QRT-PCR was adopted to prove the effect of LPS (0, 5, 10 and 15 µg/mL) on miR-205-5p level. (I) Pearson correlation analysis was applied to estimate the correlation between circ_0006944 and miR-205-5p in SA-AKI patients. **p < 0.01, ***p < 0.001, ****p < 0.0001.
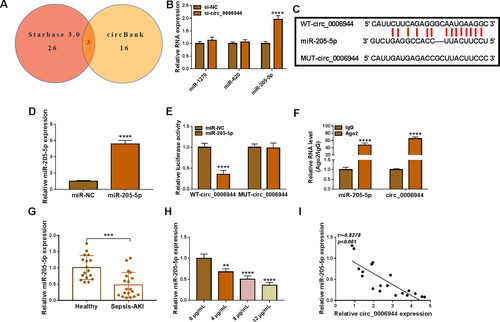
MiR-205-5p was a downstream effect factor of circ_0006944 in mediating LPS-triggered cell injury in HK2 cells
To uncover whether miR-205-5p was involved in circ_0006944-mediated modulation in LPS-stimulated HK2 cells injury, miR-205-5p inhibitor (anti-miR-205-5p) was used to decline the level of miR-205-5p in depletion of circ_0006944. As expected, the addition of anti-miR-205-5p evidently neutralised the increase of si-circ_0006944 on miR-205-5p level in LPS-treated HK2 cells (). ‘Rescue’ experiments validated that the anti-miR-205-5p introduction obviously overturned circ_0006944 inhibition-mediated enhancement on cell proliferation (), suppression on cell apoptosis () and IL-6, IL-1β and TNF-α production (), as well as facilitation on oxidative stress by detecting the level of MDA and SOD () in LPS-induced HK2 cells. In a word, we proved that miR-205-5p depletion could partially abolish the repression role of circ_0006944 silencing on LPS-triggered cell damage.
Figure 5. Effects of circ_0006944 downregulation and miR-205-5p inhibiter on LPS-induced HK2 cell dysfunction. HK2 cells were transfected with si-NC, si-circ_0006944, si-circ_0006944 + anti-miR-NC or si-circ_0006944+ anti-miR-205-5p, and then evoked with 10 μg/mL LPS for 24 h. (A) The abundance of miR-205-5p was counted by qRT-PCR. (B-E) The viability, proliferation and apoptosis was evaluated by CCK-8 assay, EdU assay and flow cytometry in HK2 cells. (F) The protein levels of Bax and Cleaved-caspase-3 were investigated with western blot assay. (G-I) The concentrations of IL-6, IL-1β and TNF-α were determined using ELISA assay. (J-K) The production of SOD and MDA levels were gauged using commercial kits. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
UBL4A served as a target gene for miR-205-5p
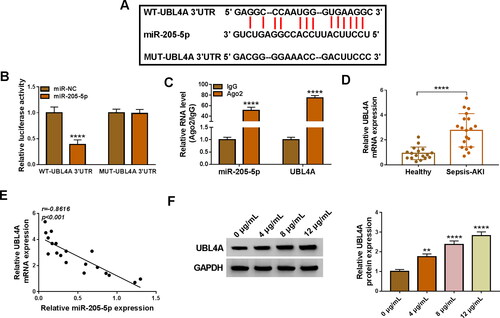
The starbase databases (http://starbase.sysu.edu.cn) has shown that miR-205-5p contained the binding sites of 3’UTR of UBL4A (). Dual-luciferase reporter assay and RIP assay () confirmed that miR-205-5p could bind to UBL4A in HK2 cells. Furthermore, the mRNA level of UBL4A was higher in serum samples from SA-AKI patients than that in control samples (). Interestingly, we disclosed a negative relation between the expression of miR-205-5p and UBL4A in SA-AKI samples (). Likewise, the elevation of UBL4A level was also observed in LPS-treated HK2 cells by western blot (). To sum up, miR-205-5p could directly interact with UBL4A.
Figure 6. UBL4A was a target for miR-205-5p. (A) The interaction sites between UBL4A 3’UTR and miR-205-5p were observed by starbase software. (B-C) The targeting relationship was demonstrated by dual-luciferase reporter and RIP assays. (D) The mRNA abundance of UBL4A was obtained by using qRT-qPCR in SA-AKI patients and and healthy controls. (E) Pearson’s correlation analysis was employed to disclose the relationship between the expressions of miR-205-5p and UBL4A. (F) UBL4A protein level was measured via western blot in LPS-triggered HK2 cells. **p < 0.01, ****p < 0.0001.
UBL4A overexpression abolished the effects of miR-205-5p on proliferation, apoptosis, inflammatory response and oxidative stress in LPS-treated HK2 cells
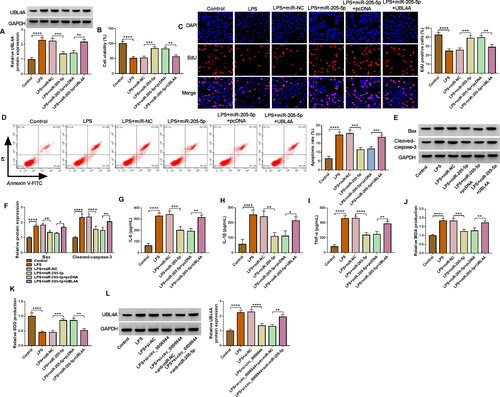
Next, rescue assays were implemented to reveal the correlation between miR-205-5p and UBL4A in LPS-stimulated HK2 cells. As exhibited in , the miR-205-5p-mediated reduction in UBL4A level was notably abated after co-transfection with pcDNA-UBL4A in LPS-induced HK2 cells. Afterwards, the cell proliferation was effectively increased in LPS-mediated HK2 cells following the enforced expression of miR-205-5p, whereas the effects were receded by the UBL4A introduction (). In addition, cell apoptosis rate impaired by miR-205-5p was abolished by addition of UBL4A in LPS-stimulated HK2 cells (), which was further validated by Bax and Cleaved-caspase-3 levels (). ELISA results manifested that the UBL4A overexpression could alleviate the repressive impact of miR-205-5p on IL-6, IL-1β and TNF-α levels in LPS-induced HK2 cells (). Besides that, miR-205-5p elevated MDA level and increased the level of SOD, which was also rescued by UBL4A overexpression in LPS-treated HK2 cells (). More importantly, western blot assay verified that circ_0006944 inhibition could hinder the protein level of UBL4A in LPS-treated HK2 cells, and the transfection of anti-miR-205-5p could substantially recede this impact (), demonstrating the circ_0006944/miR-205-5p/UBL4A axis in LPS-induced HK2 cells. In conclusion, these findings revealed that miR-205-5p apparently mitigated LPS-triggered cell dysfunction by mediating the expression of UBL4A.
Figure 7. UBL4A was a functional target of miR-205-5p in regulating LPS-evoked HK2 cells damage. HK2 cells were transfected with or without miR-NC mimic, miR-205-5p mimic, miR-205-5p mimic + pcDNA or miR-205-5p mimic + UBL4A and then treated with 10 μg/mL LPS for 24 h. (A) The protein level of UBL4A was revealed through using western blot assay. (B-C) The proliferation ability of HK2 cells was demonstrated by CCK-8 assay and EdU assay. (D-E) HK2 cells apoptosis rate was analysed by flow cytometry assay. (F) Protein levels of Bax, Cleaved-caspase-3 were assessed by western blot assay. (G-I) IL-6, IL-1β and TNF-α levels were gauged by ELISA kit. (J,K) Assay kits measured relative MDA level and SOD activity. (L) Western blot analysis for the abundance of UBL4A in LPS-induced HK2 cells transfected with si-NC, si-circ_0006944, si-circ_0006944 + anti-miR-NC or si-circ_0006944 + anti-miR-205-5p. *p < 0.05,**p < 0.01, ***p < 0.001 or ****p < 0.0001.
Discussion
Sepsis-induced AKI can lead to shock and death, and has become a global health concern [Citation5]. Although the study of this disease is increasing, the pathogenesis of this disease remains to be further explored [Citation12]. In recent years, the biological function and mechanism of circRNAs have been gradually recognised by bioinformatics analysis and RNA sequencing technology [Citation13]. CircRNA is a new characteristic marker for the development of many human diseases [Citation14], which also provides a new perspective for understanding the pathogenesis of SA-AKI [Citation15]. Whereas, the mechanisms of many circRNAs in the development of SA-AKI remain unclear. Our present study manifested the potential role of circ_0006944 in SA-AKI and found that circ_0006944 was higher in serum samples of SA-AKI patients than that in control samples, which were in line with previous studies [Citation11]. In addition, Cell function experiments manifested that circ_0006944 deficiency remarkably accelerated proliferation, while evidently retarded the apoptosis, inflammation and oxidative stress in LPS-treated HK2 cells. These results proved that circ_0006944 depletion might constrain the progress of SA-AKI.
CircRNAs can act as natural sponges for miRNAs and induce their inhibition of miRNA activity [Citation16]. CircRNA-miRNA interactions have been implicated in the pathological progression of SA-AKI [Citation15]. MiRNAs are short RNAs composed of 18-25 nucleotides, which can affect many biological processes via modulating the expression of target genes [Citation17]. Many studies have validated that miRNAs were involved in the progression of SA-AKI. Herein, our study affirmed that miR-205-5p could interact with circ_0006944 in HK2 cells. Also, circ_0006944 was identified as a miR-205-5p decoy via dual-luciferase reporter assay. Studies have found that miR-205-5p played a vital role in a variety of human diseases, including colorectal cancer [Citation18], atherosclerosis [Citation19], osteoporosis [Citation20] and SA-AKI [Citation21]. Wang et al. claimed that resveratrol attenuated SA-AKI by elevating the abundance of miR-205 and then regulating the viability of HK2 cells and the release of inflammatory cytokines stimulated by LPS [Citation22]. Han et al. indicated that depletion of lncRNA TapSAKI abrogated LPS-induced cytotoxicity in HK-2 cells by sponging miR-205-5p [Citation23]. Additionally, miR-205-5p could improve the pathomorphology and renal cell apoptosis of rats with septic kidney injury by mediating HMGB1-PTEN signalling pathway, and ultimately alleviated the progress SA-AKI [Citation21]. Our data disclosed that the expression of miR-205-5p was reduced in SA-AKI patients and LPS-stimulated HK2 cells, and downregulation of miR-205-5p abolished circ_0006944 deficiency-mediated trends on LPS-stimulated HK2 proliferation, apoptosis, inflammation and oxidative stress, suggesting that circ_0006944 modulated LPS-triggered cell dysfunction by serving as a sponge of miR-205-5p. Our findings were also consistent with a previous report that miR-205-5p was decreased in SA-AKI and inhibited SA-AKI progression [Citation21].
UBL4A was demonstrated to interact with miR-205-5p in HK2 cells. Ubiquitin-like protein 4 A (UBL4A) is located on the X chromosome (Xq28) and composed of 157 amino acid [Citation24]. Literature studies have shown that UBL4A played a pivotal role in the protein processing, and was involved in regulating the development of cancer and other diseases [Citation25]. Recent studies have found that UBL4A was also participated in the progress of SA-AKI. He et al. reported that miR-34b-3p protects septic mice from AKI by targeting UBL4A [Citation26]. In this study, we verified that UBL4A was elevated in SA-AKI patients and LPS-stimulated HK2 cells, and UBL4A introduction weakened the function of miR-205-5p in LPS-triggered HK2 cell damage. Furthermore, we also manifested circ_0006944 modulated the level of UBL4A by serving as a sponge of miR-205-5p in LPS-mediated HK2 cells.
Overall, our data revealed that circ_0006944 silencing receded LPS-induced damage in HK2 cells through mediating miR-205-5p/UBL4A signalling cascade. Repressing of circ_0006944 might be effective therapeutic strategy of SA-AKI.
Conclusion
In conclusion, our work disclosed that the level of circ_0006944 was higher in serum samples of SA-AKI and LPS-treated HK2 cells, and interference of circ_0006944 facilitated proliferation and inhibited apoptosis, inflammatory injury and oxidative stress injury of LPS-induced HK2 cells. In addition, circ_0006944 could bind to miR-205-5p and downregulation of miR-205-5p effectively rescued the effects of circ_0006944 deficiency on LPS-induced HK2 cell injury. Interestingly, miR-205-5p directly targeted UBL4A, and overexpressed UBL4A eliminated the inhibiting of miR-205-5p on LPS-induced HK2 cell injury. Furthermore, circ_0006944 regulated the level of UBL4A by sponging miR-205-5p. Collectively, the study verified that circ_0006944 depletion restrained HK2 cell injury through regulating UBL4A mediated by miR-205-5p in LPS-induced HK2 cells. This study might provide a novel direction for the therapy of SA-AKI.
Author contributions
Dong Liu designed and performed the research; Junwei Ye and Bingqi Li analysed the data; Fan Zhou wrote the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download MS Word (77 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Gyawali B, Ramakrishna K, Dhamoon AS. Sepsis: the evolution in definition, pathophysiology, and management. SAGE Open Med. 2019;7:1.
- Prescott HC, Angus DC. Enhancing recovery from sepsis: a review. Jama. 2018;319(1):62–9.
- Peerapornratana S, Manrique-Caballero CL, Gómez H, et al. Acute kidney injury from sepsis: current concepts, epidemiology, pathophysiology, prevention and treatment. Kidney Int. 2019;96(5):1083–1099.
- Chancharoenthana W, Leelahavanichkul A, Eiam-Ong S. Sepsis-associated acute kidney injury: sepsis. BMJ. 2017;364:k4891.
- Ma S, Evans RG, Naoya I, et al. Sepsis-induced acute kidney injury: a disease of the microcirculation. Microcirculation. 2019;26(2):e12483.
- Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Brandenburger T, Somoza AS, Devaux Y, et al. Noncoding RNAs in acute kidney injury. Kidney Int. 2018;94(5):870–881.
- Lu H, Chen Y, Wang X, et al. Circular RNA HIPK3 aggravates sepsis-induced acute kidney injury via modulating the microRNA-338/forkhead box A1 axis. Bioengineered. 2022;13(3):4798–4809.
- Wang F, Zhang F, Tian Q, et al. CircVMA21 ameliorates lipopolysaccharide (LPS)-induced HK-2 cell injury depending on the regulation of miR-7-5p/PPARA. Autoimmunity. 2022;55(2):136–146.
- Zhou Y, Qing M, Xu M. Circ-BNIP3L knockdown alleviates LPS-induced renal tubular epithelial cell injury during sepsis-associated acute kidney injury by miR-370-3p/MYD88 axis. J Bioenerg Biomembr. 2021;53(6):665–677.
- Tian C, Liu J, Di X, et al. Exosomal hsa_circRNA_104484 and hsa_circRNA_104670 may serve as potential novel biomarkers and therapeutic targets for sepsis. Sci Rep. 2021;11(1):14141.
- Skube SJ, Katz SA, Chipman JG, et al. Acute kidney injury and sepsis. Surg Infect. 2018;19(2):216–224.
- Verduci L, Tarcitano E, Strano S, et al. CircRNAs: role in human diseases and potential use as biomarkers. Cell Death Dis. 2021;12(5):468.
- Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116.
- Tan M, Bei R. Circ_0091702serves as a sponge ofmiR-545-3pto attenuate sepsis-related acute kidney injury by upregulatingTHBS2. J Mol Histol. 2021;52(4):717–728.
- Dori M, Bicciato S. Integration of bioinformatic predictions and experimental data to identify circRNA-miRNA associations. Genes. 2019;10(9):642.
- Szilágyi B, Fejes Z, Pócsi M, et al. Role of sepsis modulated circulating microRNAs. Ejifcc. 2019;30(2):128–145.
- Wang L, Weng W, Yang S, et al. Circle RNA circ_0007331 promotes colorectal carcinoma by targeting miR-205-5p/high-mobility group A2 axis. Bioengineered. 2022;13(4):9312–9321.
- Huang P, Zhang Y, Wang F, et al. MiRNA-205-5p regulates the ERBB4/AKT signaling pathway to inhibit the proliferation and migration of HAVSMCs induced by ox-LDL. Pathol Res Pract. 2022;233:153858.
- Yang J, Peng W, Zhang M. LncRNA KCNQ1OT1 promotes osteogenic differentiation via miR-205-5p/RICTOR axis. Exp Cell Res. 2022;415(1):113119.
- Zhang Y, Xia F, Wu J, et al. MiR-205 influences renal injury in sepsis rats through HMGB1-PTEN signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23:10950–10956.
- Wang B, Wang Y, Xu K, et al. Resveratrol alleviates sepsis-induced acute kidney injury by deactivating the lncRNA MALAT1/MiR-205 axis. Cent Eur J Immunol. 2021;46(3):295–304.
- Han X, Yuan Z, Jing Y, et al. Knockdown of lncRNA TapSAKI alleviates LPS-induced injury in HK-2 cells through the miR-205/IRF3 pathway. Open Med. 2021;16(1):581–590.
- Chen H, Li L, Hu J, et al. UBL4A inhibits autophagy-mediated proliferation and metastasis of pancreatic ductal adenocarcinoma via targeting LAMP1. J Exp Clin Cancer Res. 2019;38(1):297.
- Zhao Y, Lin Y, Zhang H, et al. Ubl4A is required for insulin-induced Akt plasma membrane translocation through promotion of Arp2/3-dependent actin branching. Proc Natl Acad Sci USA. 2015;112(31):9644–9649.
- He S, Wang G, Pei Y, et al. miR-34b-3p protects against acute kidney injury in sepsis mice via targeting ubiquitin-like protein 4A. Kaohsiung J Med Sci. 2020;36(10):817–824.

