Abstract
Our previous study found that Cullin 4B (CUL4B) inhibited rheumatoid arthritis (RA) pathology through glycogen synthase kinase-3beta (GSK3β)/canonical Wnt signalling pathway. In this work, pre-experiment and bioinformatics analysis suggested that circ_0011058 may lead to the up-regulation of CUL4B expression by inhibiting miR-335-5p. Therefore, we studied whether circ_0011058 can promote the expression of CUL4B through sponging the miR-335-5p and further promote the pathological development of RA. Bioinformatics prediction, real-time quantitative PCR (RT-qPCR), western blot (WB), double luciferase reporter gene and other relevant methods were used to study the inhibition of circ_0011058 on RA pathology and its molecular mechanism. Results showed that the expression of circ_0011058 was significantly increased in adjuvant arthritis (AA) rats and RA fibroblast-like synoviocytes (FLS). The knockout of circ_0011058 inhibited the proliferation of AA FLS and RA FLS, decreased the levels of interleukin-1 beta (IL-1β), interleukin 6 (IL-6), interleukin 8 (IL-8), and inhibited the expression of matrix metalloproteinase 3 (MMP3), fibronectin, which showed that circ_0011058 had a strong role in promoting RA pathology. Furthermore, miR-335-5p expression was reduced in AA rats and RA FLS. The highly expressed circ_0011058 directly sponged the miR-335-5p, which led to the increase of CUL4B expression and promoted the activation of the GSK3β/canonical signalling pathway. Finally, we confirmed that miR-335-5p mediated the roles of circ_0011058 in promoting RA pathological development, which showed that the circ_0011058/miR-335-5p/CUL4B signal axis was involved in RA pathology. This work was of great significance for clarifying the roles of circ_0011058 in RA pathology, and further work was needed to establish whether circ_0011058 was a potential therapeutic target or diagnostic marker for RA.
1. Introduction
Rheumatoid arthritis (RA) is an autoimmune disease caused by complex causes. Its main symptoms are symmetrical and aggressive arthritis of small joints of hands and feet, accompanied by damage of extraarticular organs and positive rheumatoid factor in peripheral blood [Citation1]. Inflammatory cell infiltration, as well as persistent joint synovial hyperplasia and inflammation, increases the release of inflammatory factors such as interleukin, further destroys the balance of joint synovial microenvironment, aggravates the proliferation of fibroblasts-like synoviocytes (FLS) and angiogenesis, leading to pannus formation, cartilage erosion and bone damage [Citation2]. Extraarticular manifestations of RA include fever, rheumatoid nodules, rheumatoid vasculitis and lymph node enlargement. Prolonged RA can also cause heart and kidney damage such as chronic endocarditis, primary glomerulonephritis, and tubulointerstitial nephritis [Citation3].
RA eventually leads to joint deformity and loss of function, affects the quality of life of patients, and also brings a great economic burden to patients’ families and society. The aetiology of RA is very complex, which is related to heredity, environment and hormones. The current research attempts to explain the aetiology of RA from different perspectives, but the pathogenesis of RA is still not clarified [Citation4].
Circular RNAs (circRNAs) are a special class of non-coding RNA molecules, which is a new research hotspot in the field of epigenetics. CircRNAs are not affected by RNA exonuclease and have a closed ring structure, characterised by stable expression. Studies have shown that circRNAs are rich in microRNAs (miRNAs) binding sites and play the roles of sponging miRNAs in physiological mechanism, relieving the inhibition of miRNAs on target genes. CircRNAs play an important role in regulating the pathological mechanism of human diseases by sponging disease-related miRNAs, which determines that circRNAs are potential targets for treating human diseases [Citation5,Citation6].
The wnt signalling pathway can be divided into canonical Wnt signalling pathway and non canonical Wnt signalling pathway, which is a complex intracellular signal transduction pathway. After Wnt proteins bind frizzled (FZD), the activated disevelled (Dvl) dissociates from the macromolecular complex of β-catenin, Dvl, casein kinase 1 (CK1), axin, adenomatous polyposis coli (APC), glycogen synthase kinase-3beta (GSK3β), leading to the inactivation of GSK3β, the disintegration of the complex, the transfer of free β-catenin into the nucleus, and the initiation of gene transcription [Citation7,Citation8]. In addition to intervening in normal physiological processes such as embryonic development, the Wnt signalling pathway is common in the pathological mechanism of tumours and plays an important regulatory role in a variety of other human diseases. Studies have shown that the canonical Wnt signalling pathway is involved in the pathogenesis of RA and plays an important role in promoting the disease [Citation9].
In our previous study, we find that the expression of CUL4B is increased in RA pathology. CUL4B directly targets GSK3β to activate the canonical Wnt signalling pathway (Wnt/β-catenin) and promotes joint synovial hyperplasia and inflammation. We confirm that CUL4B is a promoter of RA pathological development [Citation10].
In this work, the analysis of literature and pre-experimental results suggested that CUL4B may be the direct target of miR-335-5p, and miR-335-5p may be directly regulated by circ_0011058 [Citation11,Citation12], that was, there may be circ_0011058/miR-335-5p/CUL4B signal axis in RA pathology.
Therefore, we used real-time quantitative PCR (RT-qPCR), western blot (WB), bioinformatics and other methods to study the roles of circ_0011058 in RA pathology and whether circ_0011058 inhibited RA through the miR-335-5p/CUL4B signal axis. This work was of great significance for clarifying the roles of circ_0011058 in RA pathology and evaluating whether circ_0011058 was a potential therapeutic target or diagnostic marker for RA.
2. Methods
2.1. Materials and reagents
β-catenin antibody (ab32572) was purchased from Abcam Company (Waltham, MA, USA), and the gene primer used in this work was purchased from Sangon Biotech (Shanghai) Co., Ltd (Shanghai, China). Short hairpin RNA (shRNA) sh-circ0011058, miR-335-5p mimics and miR-335-5p inhibitors and corresponding negative control (NC) sequences were purchased from Shanghai GenePharma Co., Ltd (Shanghai, China), and dual-Luciferase® Reporter Assay System (E1910) and RIP detection kit (Bes5101) were purchased from Promega (Beijing) Biotech Co., Ltd (Beijing, China) and BersinBio Biotechnology Co., Ltd (Guangzhou, China) respectively. Antibody and primer information were provided in Supplementary material 1.
2.2. Adjuvant arthritis rats and fibroblast-like synoviocytes culture
All experimental animals were approved by the Animal Ethics Committee of Anhui University of Chinese Medicine. The experimental animals used in this work were sprague dawley (SD) rats, male, weighing 160-180 g, provided by the Liaoning Changsheng Biotechnology Co., Ltd [Citation13,Citation14]. Complete Freund’s adjuvant (CFA) was purchased from Sigma-Aldrich (St. Louis, MO, USA). The AA rat model was prepared by injecting CFA 0.1 mL into the plantar of rats [Citation15]. Twenty SD rats were randomly divided into normal control group and AA model group, with 10 rats in each group. AA rats were selected as RA models on the basis of their similar pathological characteristics. RA clinical samples were provided by the Department of Orthopaedics of the First Affiliated Hospital of Anhui Medical University. The synovium of AA rat joints and human synovium were isolated, and AA FLS and RA FLS were cultured by tissue block method. The RA clinical sample study protocol has been approved by the Ethics Committee of the First Affiliated Hospital (North District) of Anhui Medical University (No. PJ-YX2021-026) and followed the principle of informed consent of the donor (Supplementary Material 2).
2.3. Cell transfection
In the six well plate, 500 μL of the fourth generation FLS was inoculated into each well, and 1 mL of the medium was added for culture. After the cell density reached 80%, it was used for transfection. The transfection reagent Lipofectamine 3000 was diluted (Invitrogen, Carlsbad, CA, USA). We diluted each well by adding 1 μL Lipofectamine 3000 and 50 μL optimal minimal essential medium (opti-MEM), mixed gently after dilution and incubated at room temperature for 5 min. Diluted siRNA, took an appropriate amount of siRNA, and diluted it with 50 μL opti-MEM, that was, mixed 12.5 μL siRNA with 200 μL opti-MEM. The diluted Lipofectamine 3000 and siRNA were gently mixed and incubated at room temperature for 20 min. The mixture was added to the cell culture medium, and the orifice plate was gently shaken to mix. The model group was added with 400 μL opti-MEM as the control. The orifice plate was placed in a 37 °C CO2 incubator for 6 h, and then the complete medium was replaced. After 6 h of transfection, the transfection efficiency was detected by fluorescence microscope [Citation10,Citation16].
2.4. Cell counting kit-8 (CCK8)
CCK8 was used for simple and accurate cell proliferation and toxicity analysis. The amount of formazan generated was in direct proportion to the number of living cells. We uniformly inoculated FLS into 96 pore plates. After the cells fully adhered to the cell wall, the knockdown circ_0011058 was added and treated for 24 h [Citation11]. According to the instructions of CCK8 kit (Beyotime, China), we added 10 μL CCK8 reagent into each well, incubated it in the incubator for 1 h, and measured the absorbance at 450 nm with the microplate reader [Citation17].
2.5. Real-time qPCR (RT-qPCR)
Total RNA was extracted using Trizol reagent (Invitrogen, USA). After the FLS reached 95%, added 1 mL Trizol reagent to each bottle of FLS to make the lysate evenly distributed on the cell surface, and then blew the cell with a rubber tip dropper for 5 min to make the cell fully lyse and fall off. Reverse transcription of total RNA was performed according to the instruction of the reverse transcription kit (Biosharp, China), and gene amplification was performed with reference to RT-qPCR amplification kit (Biosharp, China). Finally, calculated the amplification results with 2−ΔΔCt method [Citation18,Citation19]. Amplification parameters were provided by Supplementary Material 3.
2.6. Western blot
The total FLS protein was extracted according to the kit method (Beyotime, China). After the phosphate-buffered solution in the cell culture bottle was removed by the pipette, 0.4 mL of the lysate (including 4 μL phenylmethanesulfonyl fluoride 100 mM) was added and blew repeatedly for 5 min. After cracking, used a pipette to transfer the cracking solution into a 1.5 mL centrifuge tube. 100 μL of sample buffer was added, blew and mixed well. Boiled for 10 min and stored at −80 °C. Prepared separation gel and concentrated gel and added samples. 80 V electrophoresis for 30 min, then adjusted the voltage to 120 V until bromophenol blue reached the bottom of the separation gel [Citation20]. PolyVinylidene Fluoride (PVDF) was sealed in 5% skim milk solution for 2 h and then washed three times. PVDF was incubated with anti-β-actin (Abcam, ab226, 1:10000) and anti-β-catenin (Abcam, ab32572, 1:1000) at 4 °C overnight. After the primary antibody was incubated, PVDF was incubated with the secondary antibody (ZSBg-Bio, 1:1000) at room temperature for 1 h. Finally, took out the PVDF film, used a pipette to add colour developing solution (Enhanced Chemiluminescence, Thermo Scientific to the PVDF film, used the Western blotting imaging system to take photos and analysed the gray value of the target strip [Citation21].
2.7. Enzyme-linked immunosorbent assay (ELISA)
The expression levels of inflammatory cytokines IL-1β, IL-6, and IL-8 were detected by ELISA kit. Added 50 μL sample analysis buffer (ColorfulGene, Wuhan, China) on 96 well plate, and then added 50 μL sample to the corresponding hole and incubated at room temperature for 120 min. Washed the board for five times. Added 100 μL/well of antibody and incubated at room temperature for 60 min. Washed the board for five times. Added horseradish peroxidase labelled Streptavidin 100 μL/well and incubated at room temperature in dark for 20 min. Washed the board for 5 times and patted it on the thick absorbent paper for the last time. Added 100 μL/hole of developer tetramethylbenzidine solution and incubated at room temperature and away from light for 20 min. Added 50 μL/well of termination solution and measured A450 value with microplate reader (Shanghai Peiou 318 C+, Shanghai, China) immediately after mixing. Drew the standard curve with the standard concentration as the abscissa and the A450 value as the ordinate. Calculated the concentration of the factor to be measured according to the standard curve [Citation22].
2.8. RNA immunoprecipitation (RIP)
RIP kit was provided by Boxin Biotechnology Co., Ltd (Guangzhou, China). Collected the FLS sample, added 0.9 mL polysome lysis buffer, 9 μL Protease inhibitor, and 4.5 μL RNase inhibitor into the cell precipitation, and mixed well. The FLS were lysed after 10 min on ice. 4.5 μL DNase salt stock and 10 μL DNase (10 U) were added to the cell lysate and incubated at 37 °C for 10 min to remove DNA. 20 μL protein A/G beads were prepared for each group of RIP samples, and 0.5 mL polysome lysis buffer was added to the magnetic beads. After 10 times washing, the magnetic beads were collected by the magnetic rack and the supernatant was removed. We divided the cell lysis sample into two parts according to 0.8 mL (IP) and 0.1 mL (Input) and kept the input sample at − 80 °C for standby. IP samples were added with experimental antibodies (anti-IgG, anti-Ago2), incubated at 4 °C in a vertical mixer for 16 h (overnight), and then immunoprecipitated. Added 200 μL phenol chloroform isoamyl alcohol mixture to IP and Input samples to extract RNA. According to the manufacturer’s instructions, the reverse transcription of DNA enzyme RNA processing [Citation23] is used.
2.9. Dual-Luciferase reporter gene System
Cloned circ_0011058 or CUL4B 3’UTR containing miR-335-5p binding site into the pmirGLO vectors (Lianmai Biotechnology Co., Ltd, Shanghai, China) to form wild-type luciferase reporter genes (circ_0011058 wild type or CUL4B 3’UTR wild type). Mutant luciferase reporter genes (circ_0011058 mutant type or CUL4B 3’UTR mutant type) were formed through mutation binding sites. Co-transfected the corresponding reporter genes with miR-NC or miR-335-5p into FLS. Firefy and Renilla luciferase activities were detected by Dual Luciferase® Reporter Assay System (No. E1910, Promega, Madison, WI, USA).
The Luciferase Assay Recipient II (LAR II) was prepared by resuspending the Luciferase Assay Substrate (provided by the kit) with 10 mL Luciferase Assay Buffer II provided by the kit (Promega, Madison, WI, USA). Prepared Stop & Glo® reagent. Added 1 volume of 50X Stop & Glo® substrate to the glass tube containing 50 volume of Stop & Glo® buffer solution. Allocated 100 μL LAR II to a suitable number of light-emitting detection tubes for the required number of DLR™ tests. The program of the luminescence detector was set, and carefully transferred the 20 μL cell lysate to the luminous detector tube with LAR II. Blew it for three times to make it mix evenly without vortex oscillation. Placed the tube in the luminous detector and started reading the data. Took out the sample tube in the luminous detector, then add 100 μL Stop & Glo® reagent and mixed it for a short time. Then placed the tube in the luminous detector again and started reading the data. The activity of firefly luciferase was standardised through the activity of Renila luciferase [Citation24].
2.10. Data statistics
All the experimental data in this work were calculated by SPSS 26.0 software, and the data were presented as mean ± standard deviation. Two independent sample t-test was used to compare the two groups of data, and analysis of variance was used to compare multiple groups. According to the statistical principle, the number of experimental animals in each group was determined as 10. p < 0.05 indicated that there was a difference in the mean value between the two groups.
3. Results
3.1. Circ_0011058 expression significantly increased in AA rats and RA FLSs
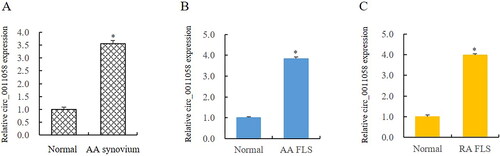
Through literature analysis, the expression of circ_0011058 in tissues and cells of papillary thyroid cancer was abnormally elevated [Citation11]. In view of the uncontrolled proliferation of RA FLS, we detected the expression of circ_0011058 in RA tissues and cells. The results showed that the expression of circ_0011058 in synovium was upregulated by 3.56 times (). Circ_0011058 was upregulated 3.83 times in FLS of AA rats (). Furthermore, the expression of circ_0011058 in FLS of RA patients was 3.98 times that of the control group (). This suggested that circ_0011058 was related to RA pathology.
Figure 1. Circ_0011058 expression significantly increased in AA rats and RA FLS.
(A and B) RT-qPCR showed that circ_0011058 was upregulated in synovium and FLS of AA rats. (C) Circ_0011058 was significantly upregulated in RA FLS, further confirming the expression change of circ_0011058 in RA. *AA/RA group vs normal, n = 3 (3 different samples). AA, adjuvant arthritis; RA, rheumatoid arthritis; RT-qPCR, real time qPCR; FLS, fibroblast-like synoviocytes
3.2. Circ_0011058 knockdown inhibited RA pathology
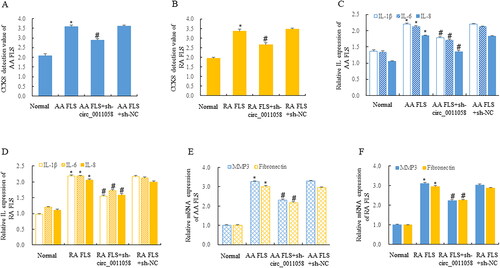
We used CCK8 to detect the effect of circ_0011058 knockdown on the proliferation of AA FLS and RA FLS. The results showed that AA FLS proliferation was significantly inhibited after circ_0011058 knockdown (). After circ_0011058 was knocked down, RA FLS was also inhibited (). ELISA detection showed that circ_0011058 knockdown inhibited the expression of IL-1β, IL-6, and IL-8 in supernatant of AA FLS () and RA FLS (), and RT-qPCR detection showed that circ_0011058 knockdown also inhibited the expression of matrix metalloproteinase 3 (MMP3) and fibronectin in these two types of cells (). This suggested that circ_0011058 has an obvious anti-RA effect.
Figure 2. Circ_0011058 knockdown inhibited RA pathology.
CCK8 detection showed that sh-circ0011058 inhibited the proliferation of AA FLS (A) and RA FLS (B). ELISA showed that sh-circ0011058 inhibited the expression of IL-1β, IL-6, and IL-8 in AA FLS (C) and RA FLS (D). RT-qPCR showed that sh-circ0011058 inhibited the expression of MMP3 and fibronectin in AA FLS (E), and also inhibited these two RA genes in RA FLS (F). *AA FLS/RA FLS vs normal, # group 3 mean vs group 2 mean (from left to right), n = 3.
3.3. Circ_0011058 directly targeted miR-335-5p
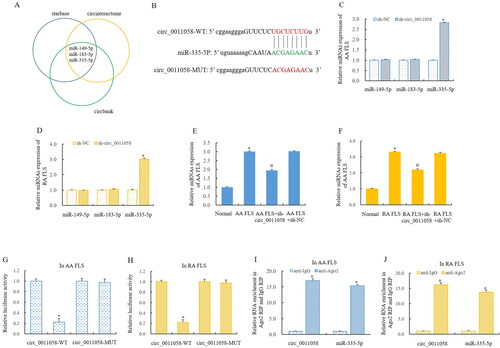
Bioinformatics predicted that miR-335-5p was the direct target of circ_0011058 (). To verify this regulatory relationship, we examined the effect of knockdown circ_0011058 on the expression of miR-149-5p, miR-183-5p and miR-335-5p in AA rats and RA FLS. The results showed that the expression of miR-149-5p and miR-183-5p was not significantly affected. Importantly, the expression of miR-335-5p was significantly upregulated in both AA FLS and RA FLS (). This suggested that circ_0011058 may play an important regulatory roles in RA pathology through miR-335-5p. Furthermore, RT-qPCR showed that circ_0011058 knockdown up-regulated miR-335-5p expression in AA FLS () and RA FLS (). We performed double luciferase reporter gene detection in these two types of cells, and the results also indicated that miR-335-5p was the direct target of circ_0011058 (). Furthermore, the RIP experiment confirmed this discovery ().
Figure 3. Circ_0011058 directly targeted miR-335-5p.
(A, B) Bioinformatics predicted that miR-335-5p was the direct target of circ_0011058.
(C, D) The expression of miR-149-5p and miR-183-5p was not significantly affected, and the expression of miR-335-5p was significantly upregulated. (E, F) Circ_0011058 knockdown upregulated miR-335-5p expression. Double luciferase reporter gene detection showed that miR-335-5p was the direct target of circ_0011058 (G and H), and RIP experiment confirmed this discovery (I, J). *The mean of group 2 vs the mean of group 1, # group 3 mean vs group 2 mean (from left to right), n = 3. RIP, RNA Immunoprecipitation
3.4. CUL4B was a target of miR-335-5p
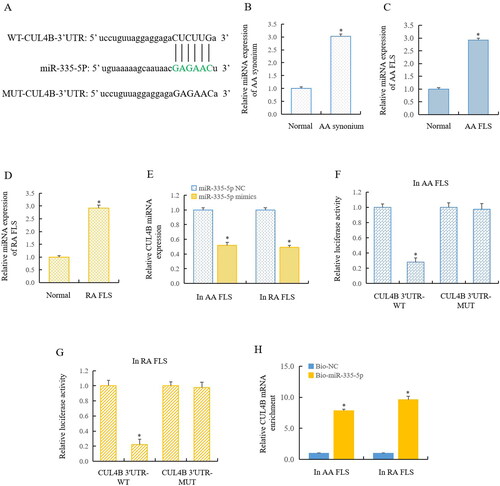
Our previous research has confirmed that the expression of CUL4B was significantly increased in RA pathology. A high level of CUL4B inhibited GSK3β, making GSK3β unable to phosphorylate and degraded the β-catenin, leading to the increase of free β-catenin and entering the nucleus to start the canonical Wnt signal pathway [Citation10]. In this work, we used bioinformatics to predict that CUL4B may be the target of miR-335-5p (). We detected that the expression of circ_0011058 was upregulated 3.03-fold in the synovium of AA rats () and 2.92-fold in the FLS of AA rats (). Our detection showed that CUL4B expression was significantly up-regulated in AA rats () and RA FLS (). RT-qPCR showed that the transfer of miR-335-5p mimics significantly inhibited CUL4B expression (). Furthermore, double luciferase reporter gene detection showed that miR-335-5p directly regulated the CUL4B in AA FLS () and RA FLS (). RIP detection further confirmed that CUL4B was a target of miR-335-5p (). This suggested that circ_0011058 increased the expression of CUL4B and further activated the canonical Wnt signalling pathway by inhibiting the miR-335-5p.
Figure 4. CUL4B was a target of miR-335-5p.
(A) Bioinformatics predicted that CUL4B may be the target of miR-335-5p. RT-qPCR showed that CUL4B expression was significantly upregulated in AA rats (B and C) and RA FLS (D). (E) MiR-335-5p mimics significantly inhibited CUL4B expression. Double luciferase reporter gene detection showed that miR-335-5p directly regulated the CUL4B in AA FLS (F) and RA FLS (G), and RIP detection confirmed the findings (H). *The mean of group 2 vs the mean of group 1, n = 3.
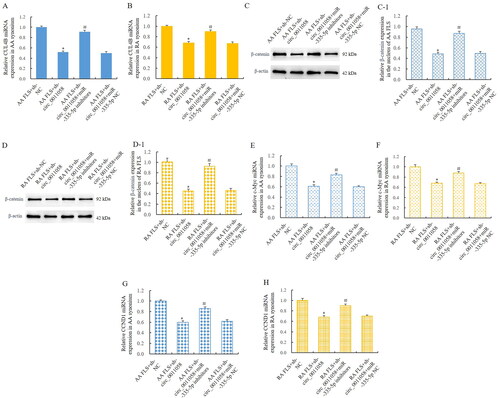
3.5. MiR-335-5p mediated the effect of circ_0011058 on CUL4B and canonical Wnt signalling pathway
To verify that miR-335-5p mediated the effect of circ_0011058 on CUL4B and the canonical Wnt signal pathway, we knocked down the circ_0011058 in AA FLS and RA FLS, respectively, and detected the interference of miR-335-5p inhibitors on the effect of circ_0011058 knockdown. RT-qPCP detection showed that circ_0011058 knockdown inhibited the expression of CUL4B. After miR-335-5p inhibitors were added, the above effect was reversed (). WB test showed that the knockdown of circ_0011058 resulted in the decrease of β-catenin protein level in the nucleus of AA and RA FLS. After miR-335-5p inhibitors were added, the above effect was reversed (). Furthermore, circ_0011058 knockdown inhibited the expression of c-Myc. After miR-335-5p inhibitors were added, the above phenomenon was reversed (). Similarly, CCND1 was similarly regulated (). β-catenin, c-Myc and CCND1 were key members of the classical Wnt signalling pathway [Citation25]. Therefore, we confirmed that miR-335-5p mediated the effect of circ_0011058 on CUL4B and canonical Wnt signalling pathway.
Figure 5. MiR-335-5p mediated the effect of circ_0011058 on CUL4B and canonical Wnt signalling pathway.
RT-qPCR showed that sh-circ_0011058 inhibited CUL4B expression in AA FLS (A) and RA FLS (B), and miR-335-5p inhibitors reversed these effects. WB showed that sh-circ_0011058 inhibited the expression of β-catenin in the nuclei of AA FLS (C) and RA FLS (D), and miR-335-5p inhibitors reversed. (C-1) Quantitative results of β-catenin in AA FLS. (D-1) Quantitative results of β-catenin in RA FLS. (E and F) Sh-circ_0011058 inhibited c-Myc expression, and miR-335-5p inhibitors could reverse them. (G and H) Sh-circ_0011058 inhibited CCND1 expression and miR-335-5p inhibitors reversed. *The mean of group 2 vs the mean of group 1, # group 3 mean vs group 2 mean (from left to right), n = 3.
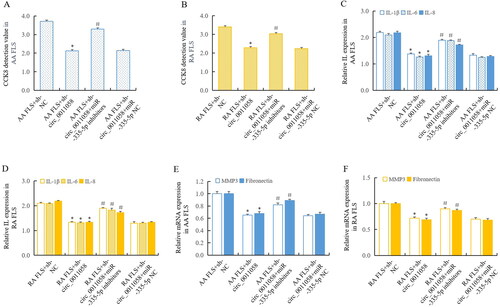
3.6. MiR-335-5p mediated the influence of circ_0011058 on RA pathology
To verify that circ_0011058 affected RA pathology through miR-335-5p, we knocked down circ_0011058 again in AA FLS and RA FLS, respectively, and detected the interference of miR-335-5p inhibitors on the effect of circ_0011058 knockdown. CCK8 detection showed that circ_0011058 knockdown inhibited the proliferation of AA FLS and RA FLS. After miR-335-5p inhibitors were added, the above inhibition was reversed (). ELISA detection showed that circ_0011058 knockdown led to the decrease of IL-1β, IL-6, and IL-8 in culture supernatant, and miR-335-5p inhibitors reversed these effects (). Further, RT-qPCR detection showed that circ_0011058 knockdown inhibited the expression of MMP3 and fibronectin. After miR-335-5p inhibitors were added, the above effects were reversed (). These results confirmed that miR-335-5p mediated the influence of circ_0011058 on RA pathology. Combined with the previous research findings, it was certain that there was the circ_0011058/miR-335-5p/CUL4B/GSK3β signal axis in RA pathology, which played important roles in promoting RA.
Figure 6. MiR-335-5p mediated the influence of circ_0011058 on RA pathology.
CCK8 showed that sh-circ_0011058 inhibited FLS proliferation in AA FLS (A) and RA FLS (B), and miR-335-5p inhibitors reversed. ELISA showed that sh-circ_0011058 inhibited the expression of IL-1β, IL-6, and IL-8 in AA FLS (C) and RA FLS (D), and miR-335-5p inhibitors reversed. E. F, sh-circ_0011058 inhibited the expression of MMP3 and fibronectin, and miR-335-5p inhibitors could reverse them. *The mean of group 2 vs the mean of group 1, # group 3 mean vs group 2 mean (from left to right), n = 3.
4. Discussion
RA is a chronic, invasive autoimmune disease characterised by joint synovitis. RA mainly occurs in the small joints of the hands and feet, showing the characteristics of multijoints, symmetry and aggressiveness, and is often accompanied by extraarticular organ damage and positive rheumatoid factor in the peripheral blood. Persistent synovitis causes invasive proliferation of synovial cells, resulting in cartilage erosion and bone loss [Citation26]. If RA cannot be controlled, it will lead to joint deformity, which will easily lead to disability, reduce the quality of life of patients, and bring a serious burden to patients’ families and society [Citation27].
RA first affects the synovium of the joint, then affects articular cartilage, bone tissue, and even affects joint ligaments and muscle bonds. In addition to joints, RA can also induce extensive inflammatory lesions in the connective tissue of heart, lungs, eyes, and other organs. The general manifestations of RA include fever, fatigue, pleuritis, pericarditis, arteritis, subcutaneous nodules, and peripheral neuropathy. These complications increase the pain and mortality of patients [Citation28].
The aetiology of RA is very complex, and the pathogenesis has not been fully clarified, but the authoritative pathogenesis of RA is the damage caused by autoimmune disorder. For example, the formation of extravascular immune complexes stimulates inflammatory reaction, and cellular immunity causes the release of inflammatory factors such as interleukin, which will cause the imbalance of the joint synovial microenvironment and lead to synovial proliferation and cartilage erosion [Citation29]. The study finds that those with human leukocyte antigen (HLA) RD4 and DW4 antigens are highly sensitive to external environmental factors, as well as internal infection, neuropsychiatric, and endocrine factors. By inducing changes in HLA epitopes, synovial tissue components of joints become targets of abnormal immune response. Macrophages recognise external stimulators and also stimulate T cells to increase the release of a series of inflammatory factors, thus producing abnormal immune responses [Citation30].
The synovium of RA patients not only has a large number of lymphocyte infiltration, but also is accompanied by the elevation of various cytokines, especially IL-1β, IL-6, IL-8, and tumour necrosis factor [Citation31,Citation32]. Interleukin can stimulate lymphocyte activation, FLS proliferation and osteoclast activation in joint synovium, which is an important factor to promote the occurrence and development of RA [Citation33]. Our research shows that the levels of IL-1β, IL-6, and IL-8 are significantly increased after RA. If RA is effectively treated and the symptoms improve, the levels of the above-mentioned interleukins will be significantly reduced [Citation34]. Similarly, the expression of MMP3 and fibronectin in RA cases is also directly related to RA pathology [Citation35,Citation36]. In this work, after the expression of circ_0011058 is knocked down, the levels of IL-1β, IL-6, and IL-8 are significantly reduced in the culture supernatant, and MMP3 and fibronectin are also reduced, suggesting that circ_0011058 is a significant RA pathological promoter.
CircRNAs have a closed ring structure, are not affected by RNA exonuclease, are stable in expression, and are not easy to degrade [Citation37]. CircRNA is rich in miRNA binding sites, which can inhibit its expression by sponging miRNA, thereby affecting the expression of target genes [Citation38]. CircRNAs play important roles in physiological and pathological mechanisms through the interaction of disease-related miRNAs [Citation39,Citation40]. CircRNA CDR1as plays a role as a competitive endogenous RNA to promote the progression of hepatocellular carcinoma [Citation41]. CircRNA_0016624 can regulate the expression of BMP2 in postmenopausal osteoporosis by sponge miR-98 [Citation42]. CircRNA_0092516 regulates the pathology of osteoarthritis through miR-337-3p [Citation43].
In our study, circ_0011058 expression is significantly increased in RA model rats and RA FLS. Through bioinformatics analysis, it is suggested that circ_0011058 may directly regulate miR-335-5p, that is, miR-335-5p is the direct target of circ0011058. Our experiment confirmed the direct interaction between circ_0011058 and miR-335-5p. Bioinformatics analysis and our research also confirm that miR-335-5p and CUL4B also have a direct relationship, so there is circ_0011058/miR-335-5p/CUL4B signal axis in RA pathological mechanism, which plays an important role in pathological promotion of RA.
CUL4B protein participates in the formation of the Cullin-Ring ubiquitin ligase complex, is an important skeleton protein of this complex, and plays an important regulatory role in the pathogenesis of tumours [Citation44]. For example, CUL4B promotes the occurrence of breast cancer and is a possible diagnostic marker of the tumour [Citation45]. CUL4B is also associated with tumour drug resistance. For example, CUL4B affects the expression of ER-α36 by sponging miR-32-5p and mediates the resistance of breast cancer cells to tamoxifen [Citation46]. We find for the first time that CUL4B participates in the pathogenesis of RA and is an important RA promoter. The abnormally expressed CUL4B targets GSK3β and promotes the increase of free β-catenin, which leads to the activation of canonical Wnt signalling pathway. Interestingly, CUL4B is not only capable of regulating downstream genes but also directly regulated by upstream miRNAs. Our research reveals that miR-101-3p is the upstream regulator of CUL4B [Citation10]. Furthermore, circ_0015756 increases CUL4B by sponging miR-942-5p, thereby activating the Wnt signalling pathway in RA pathology [Citation17]. Therefore, it can be assumed that if the expression of miR-101-3p and miR-942-5p is up-regulated, the expression of CUL4B will be inhibited, further inhibiting the pathological development of various tumours and RA.
There is no doubt that the canonical Wnt signalling pathway not only plays an important role in human development, physiology and tumour pathology but also is involved in the pathological mechanism of RA. It has been clear that Wnt proteins binding to FZD outside the cell membrane trigger the Wnt signalling pathway, including canonical and non-canonical Wnt signalling pathways [Citation47]. For the canonical, Wnt proteins bind FZD and activate the Dvl, leading to the disintegration of the complex composed of Dvl, β-catenin, CK1, axin, APC, GSK3β. β-catenin cannot be degraded by phosphorylation and ubiquitination, and the free β-catenin is transferred into the nucleus and starts gene transcription [Citation48]. The expressions of β-catenin and c-Myc in RA-FLS were significantly upregulated. High expression of β-catenin leads to the proliferation of FLS activated by Wnt/β-catenin signalling [Citation49]. Our research also fully proved the promotion of canonical Wnt signalling pathway in RA. For example, miR-148b-3p affects the canonical Wnt signal pathway by directly targeting DNA methyltransferase 1 (DNMT1), thereby interfering with the pathological mechanism of RA [Citation50], and miR-375 regulates this pathway by silencing FZD8 to affect RA [Citation51]. The DNA methylation modification of SFRP2 is also involved in RA pathology through this pathway [Citation52].
In this work, we found that the expression of circ_0011058 was significantly increased in AA rats and RA FLS. When circ_0011058 was knocked down, the proliferation of AA FLS and RA FLS was inhibited, the levels of IL-1β, IL-6, and IL-8 in culture supernatant were inhibited, and the expression of MMP3, fibronectin, the pathological genes of RA, were decreased, showing a strong role in promoting RA pathology. Bioinformatics analysis also suggested that circ_0011058 may play a role through miR-335-5p, and miR-335-5p inhibited the canonical Wnt signalling pathway through CUL4B/GSK3β. Our experiments confirmed the above conjecture. In order to further verify the above mechanism, we transfected miR-335-5p inhibitors to AA FLS and RA FLS, respectively, to verify whether the knockdown of miR-335-5p would affect the knockdown effect of circ_0011058. It was found that the addition of miR-335-5p inhibitors reversed the knockdown effect of circ_0011058.
Through this study, we confirmed that CUL4B mediated the roles of circ_0011058/miR-335-5p in promoting RA pathological development and confirmed that circ_0011058/miR-335-5p/CUL4B/GSK3β signal axis exists in RA pathology. This is of great significance for clarifying the roles of circ_0011058 in RA pathology and evaluating whether circ_0011058 is a potential therapeutic target or diagnostic marker for RA, and further work is needed to establish whether circ_0011058 is a potential therapeutic target or diagnostic marker for RA.
Supplemental Material
Download ()Acknowledgements
We thank the National Natural Science Foundation of China, Anhui Provincial Department of Science and Technology, Anhui Provincial Department of Education and Anhui University of traditional Chinese medicine for their support to this work.
Disclosure statement
Ethics approval and consent to participate
The clinical sample study was approved by the Fourth Affiliated Hospital of Anhui Medical University. All participants signed the informed consent, and the ethics review approval document No.: PJ-YX2021-026. The animal experiment was approved by the animal ethics committee of Anhui University of Chinese medicine (AHUCM-rats-2022115).
Consent for publication
This manuscript was published with the approval of all authors, and there was no conflict of interest in the submission.
Availability of data and material
Study raw data were provided by the corresponding authors upon request.
Competing interests
We confirm that there are no conflicts of interest in this publication and no financial support that has an impact on this work.
Additional information
Funding
References
- Lin YJ, Anzaghe M, Schülke S. Update on the pathomechanism, diagnosis, and treatment options for rheumatoid arthritis. Cells. 2020;9(4):1.
- Scherer HU, Häupl T, Burmester GR. The etiology of rheumatoid arthritis. J Autoimmun. 2020;110:102400.
- Dai Y, Wang W, Yu Y, et al. Rheumatoid arthritis-associated interstitial lung disease: an overview of epidemiology, pathogenesis and management. Clin Rheumatol. 2021;40(4):1211–11.
- Deane KD, Holers VM. Rheumatoid arthritis pathogenesis, prediction, and prevention: an emerging paradigm shift. Arthritis Rheumatol. 2021;73(2):181–193.
- Zhang M, Bai X, Zeng X, et al. circRNA-miRNA-mRNA in breast cancer. Clin Chim Acta. 2021;523:120–130.
- Chen L, Shan G. CircRNA in cancer: fundamental mechanism and clinical potential. Cancer Lett. 2021;505:49–57.
- Zhang Y, Wang X. Targeting the wnt/β-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13(1):165.
- Hu HH, Cao G, Wu XQ, et al. Wnt signaling pathway in aging-related tissue fibrosis and therapies. Ageing Res Rev. 2020;60:101063.
- Miao CG, Yang YY, He X, et al. Wnt signaling pathway in rheumatoid arthritis, with special emphasis on the different roles in synovial inflammation and bone remodeling. Cell Signal. 2013;25(10):2069–2078.
- Miao C, Chang J, Zhang G, et al. CUL4B promotes the pathology of adjuvant-induced arthritis in rats through the canonical wnt signaling. J Mol Med (Berl). 2018;96(6):495–511.
- Zhang Z, Wang W, Su Z, et al. Circ_0011058 facilitates proliferation, angiogenesis and radioresistance in papillary thyroid cancer cells by positively regulating YAP1 via acting as miR-335-5p sponge. Cell Signal. 2021;88:110155.
- Yu C, Ying J, Yu K, et al. Circ_0074027 contributes to non-small cell lung cancer progression by upregulating CUL4B expression through miR-335-5p. Cancer Biother Radiopharm. 2022;37(2):73–83.
- Qin Y, Cai ML, Jin HZ, et al. Age-associated B cells contribute to the pathogenesis of rheumatoid arthritis by inducing activation of fibroblast-like synoviocytes via TNF-α-mediated ERK1/2 and JAK-STAT1 pathways. Ann Rheum Dis. 2022;81(11):1504–1514.
- Liu W, Zhang Y, Zhu W, et al. Sinomenine inhibits the progression of rheumatoid arthritis by regulating the secretion of inflammatory cytokines and monocyte/macrophage subsets. Front Immunol. 2018;9:2228.
- Chen H, Wen Y, Pan T, et al. Total glucosides of paeony improve complete freund’s adjuvant-induced rheumatoid arthritis in rats by inhibiting toll-like receptor 2-mediated tumor necrosis factor receptor-associated factor 6/nuclear factor-kappa B pathway activation. J Tradit Chin Med. 2019;39(4):566–574.
- Duckert B, Vinkx S, Braeken D, et al. Single-cell transfection technologies for cell therapies and gene editing. J Control Release. 2021;330:963–975.
- Wang X, Chang J, Zhou G, et al. The traditional chinese medicine compound huangqin qingre chubi capsule inhibits the pathogenesis of rheumatoid arthritis through the CUL4B/wnt pathway. Front Pharmacol. 2021;12:750233.
- Hawkins SFC, Guest PC. Multiplex analyses using Real-Time quantitative PCR. Methods Mol Biol. 2017;1546:125–133.
- Forero DA, González-Giraldo Y, Castro-Vega LJ, et al. qPCR-based methods for expression analysis of miRNAs. BioTechniques. 2019;67(4):192–199.
- Mishra M, Tiwari S, Gomes AV. Protein purification and analysis: next generation Western blotting techniques. Expert Rev Proteomics. 2017;14(11):1037–1053.
- Kim B. Western blot techniques. Methods Mol Biol. 2017;1606:133–139.
- Tabatabaei MS, Ahmed M. Enzyme-Linked immunosorbent assay (ELISA). Methods Mol Biol. 2022;2508:115–134.
- Gagliardi M, Matarazzo MR. RIP: RNA immunoprecipitation. Methods Mol Biol. 2016;1480:73–86.
- Sun Y, Zhao J, Patil SB, et al. Improved dual luciferase reporter (DLR) assay to determine the protein stability. Anal Biochem. 2021;612:114021.
- Liu J, Xiao Q, Xiao J, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7(1):3.
- Zaiss MM, Joyce Wu HJ, Mauro D, et al. The gut-joint axis in rheumatoid arthritis. Nat Rev Rheumatol. 2021;17(4):224–237.
- Kolarz B, Podgorska D, Podgorski R. Insights of rheumatoid arthritis biomarkers. Biomarkers. 2021;26(3):185–195.
- Buch MH, Eyre S, McGonagle D. Persistent inflammatory and non-inflammatory mechanisms in refractory rheumatoid arthritis. Nat Rev Rheumatol. 2021;17(1):17–33.
- Akiyama M, Kaneko Y. Pathogenesis, clinical features, and treatment strategy for rheumatoid arthritis-associated interstitial lung disease. Autoimmun Rev. 2022;21(5):103056.
- Wu F, Gao J, Kang J, et al. B cells in rheumatoid arthritis: pathogenic mechanisms and treatment prospects. Front Immunol. 2021;12:750753.
- Chikanza IC. Mechanisms of corticosteroid resistance in rheumatoid arthritis: a putative role for the corticosteroid receptor beta isoform. Ann N Y Acad Sci. 2002;966(1):39–48.
- Arunsi UO, Chioma OE, Etusim PE, et al. Indigenous Nigeria medicinal herbal remedies: a potential source for therapeutic against rheumatoid arthritis. Exp Biol Med (Maywood). 2022;247(13):1148–1178.
- Miao C, Bai L, Yang Y, et al. Dysregulation of lncRNAs in rheumatoid arthritis: biomarkers, pathogenesis and potential therapeutic targets. Front Pharmacol. 2021;12:652751.
- Jung YK, Kang YM, Han S. Osteoclasts in the inflammatory arthritis: implications for pathologic osteolysis. Immune Netw. 2019;19(1):e2.
- Miao CG, Shi WJ, Xiong YY, et al. MicroRNA-663 activates the canonical wnt signaling through the adenomatous polyposis coli suppression. Immunol Lett. 2015;166(1):45–54.
- Lerner A, Neidhöfer S, Reuter S, et al. MMP3 is a reliable marker for disease activity, radiological monitoring, disease outcome predictability, and therapeutic response in rheumatoid arthritis. Best Pract Res Clin Rheumatol. 2018;32(4):550–562.
- Patop IL, Wüst S, Kadener S. Past, present, and future of circRNAs. Embo J. 2019;38(16):e100836.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Li Y, Chen J, Xie M, et al. Identification of a circRNA-miRNA-mRNA network to explore the effects of circRNAs on renal injury in systemic lupus erythematosus. Autoimmunity. 2023;56(1):2193361.
- Lin D, Wang Y, Lei L, et al. Circ_0003645 serves as miR-335-5p sponge to promote the biological process of diffuse large B-cell lymphoma by upregulating NFIB. Autoimmunity. 2022;55(2):127–135.
- Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11(19):8183–8203.
- Yu L, Liu Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem Biophys Res Commun. 2019;516(2):546–550.
- Huang Z, Ma W, Xiao J, et al. CircRNA_0092516 regulates chondrocyte proliferation and apoptosis in osteoarthritis through the miR-337-3p/PTEN axis. J Biochem. 2021;169(4):467–475.
- Li Y, Wang X. The role of cullin4B in human cancers. Exp Hematol Oncol. 2017;6(1):17.
- Huang W, Zhang J, Huo M, et al. CUL4B promotes breast carcinogenesis by coordinating with transcriptional repressor complexes in response to hypoxia signaling pathway. Adv Sci (Weinh). 2021;8(10):2001515.
- Wang Y, Pan X, Li Y, et al. CUL4B renders breast cancer cells tamoxifen-resistant via miR-32-5p/ER-α36 axis. J Pathol. 2021;254(2):185–198.
- Zhou Y, Xu J, Luo H, et al. Wnt signaling pathway in cancer immunotherapy. Cancer Lett. 2022;525:84–96.
- Chatterjee A, Paul S, Bisht B, et al. Advances in targeting the WNT/β-catenin signaling pathway in cancer. Drug Discov Today. 2022;27(1):82–101.
- Xiao CY, Pan YF, Guo XH, et al. Expression of β-catenin in rheumatoid arthritis fibroblast-like synoviocytes. Scand J Rheumatol. 2011;40(1):26–33.
- Miao C, Yu H, Chang J, et al. miR-148b-3p affects the pathogenesis of adjuvant-induced arthritis rats through the direct target DNMT1. Autoimmunity. 2018;51(2):43–52.
- Miao CG, Shi WJ, Xiong YY, et al. miR-375 regulates the canonical wnt pathway through FZD8 silencing in arthritis synovial fibroblasts. Immunol Lett. 2015;164(1):1–10.
- Miao C, Chang J, Dou J, et al. DNA hypermethylation of SFRP2 influences the pathology of rheumatoid arthritis through the canonical wnt signaling in model rats. Autoimmunity. 2018;51(7):1–14.


