Abstract
MicroRNA (miRNA) plays a regulatory role in periodontitis. This study aimed to explore whether miR-29a could affect lipopolysaccharides (LPSs)-induced injury in human gingival fibroblasts (HGFs) through the competitive endogenous RNAs (ceRNA) mechanism. Periodontal ligament (PDL) tissues and HGFs were derived from patients with periodontitis and healthy volunteers. Periodontitis cell model was established by treating HGFs with LPS. Expression levels of circ_0036490, miR-29a, and DKK1 were evaluated by the reverse transcription quantitative real-time PCR (RT-qPCR) method. Western blotting assay was performed to assess protein expression levels of pyroptosis-related proteins and Wnt signalling related proteins. Cell viability was evaluated by cell counting kit-8 (CCK-8) assay. Concentration of lactate dehydrogenase (LDH), interleukin (IL)-1β, and IL-18 were determined by Enzyme-linked immunosorbent assay (ELISA). Pyroptosis rate were determined by flow cytometry assay to evaluate pyroptosis. The interaction between miR-29a and circ_0036490 or DKK1 was verified by dual-luciferase reporter and RNA pull-down assays. MiR-29a expression was lower in PDL tissues of patients with periodontitis than that in healthy group; likewise, miR-29a was also downregulated in LPS-treated HGFs. Overexpression of miR-29a increased cell viability and decreased pyroptosis of HGFs induced by LPS while inhibition of miR-29a exerted the opposite role. MiR-29a binds to circ_0036490 and elevation of circ_0036490 contributed to dysfuntion of LPS-treated HGFs and reversed the protection function of elevated miR-29a. In addition, miR-29a targets DKK1. Overexpression of DKK1 abrogated the effects of overexpressed miR-29a on cell vaibility, pyroptosis, and protein levels of Wnt signalling pathway of LPS-treated HGFs. Circ_0036490 and DKK1 competitively bind miR-29a to promote LPS-induced HGF injury in vitro. Wnt pathway inactivated by LPS was activated by miR-29a. Thence, miR-29a may be a promising target for periodontitis.
Introduction
Periodontitis is a chronic oral inflammatory disease characterised by the destruction of periodontal tissue, mainly manifested as gingival inflammation, alveolar bone resorption, periodontal pocket formation and tooth loosening, which is one of the main causes of tooth loss [Citation1,Citation2].
As one of the main pathogens of periodontitis, the capsular, pili and cell membrane of P. gingivalis are related to periodontal destruction. In addition, P. gingivalis can accelerate inflammatory destruction and trigger host immune response through secretion of lipopolysaccharide (LPS), outer membrane protein, gingival proteinase and a series of virulence products [Citation3,Citation4]. LPS is reported as an important pathogenic factor of bone loss in periodontitis [Citation5,Citation6]. LPS can stimulate osteoblasts and osteoclasts to secrete a variety of inflammatory factors and then cause immune inflammatory response [Citation7].
Pyroptosis is a kind of inflammatory cell death. The difference between pyroptosis and necrotising apoptosis is that the pore protein of pyroptosis membrane is Gasdermin D (GSDMD), and the inflammatory killing is caused by interleukin 1β (IL-1β) and IL-18 [Citation8]. Periodontal pathogens and their products can significantly upregulate the expression of nucleotide-binding oligomerisation domain like receptor protein (NLRP) in the inflammasome [Citation9]. As a promoter, NLRP plays an important role in the regulation of pyroptosis [Citation10]. Due to the complexity of the peridental microenvironment, the mechanism of pyroptosis in the development of periodontitis has not been clearly determined.
MicroRNA (miRNA) is a class of endogenous, small noncoding RNA (ncRNA), which plays an important role in the regulation of immune response and has become a biological marker of many diseases [Citation11,Citation12]. Studies have reported that increased or decreased expression levels of some miRNAs were detected in inflamed gingival tissues [Citation13,Citation14]. In vivo and in vitro experiments have proved that miRNA-143-3p, miRNA-146a, miRNA-155, miRNA-203 and miRNA-223 were related to periodontal diseases [Citation15–17]. miR-29a is a member of the miR-29 family that regulats immune ability and inflammation [Citation18,Citation19]. miR-29a can exert oncogenic effects on various tumours by binding to key genes that enhance or inhibit cancer progression [Citation20]. Recently, Kapoor et al. [Citation21] found a variety of miRNAs in gingival crevicular fluid of orthodontic patients, among which miR-29a was identified as a potential marker associated with tooth movement. Meanwhile, miR-29a is reported to be involved in the process of periodontal ligament (PDL) stem cells chondrogenesis [Citation22]. Furthermore, miR-29a participates in regulating pyroptosis of diverse inflammatory related diseases [Citation23,Citation24]. Whether miR-29a regulates pyroptosis in periodontitis and the underlying mechanism remains unclear.
Circular RNA (circRNA) is a subset of ncRNA that has recently been emerged as a new regulator of gene expression [Citation25–27]. Meanwhile, circRNA functions as the competitive endogenous RNAs (ceRNA) by spong miRNA in various diseases [Citation28–30]. Dysregulated circRNAs have also been identified in periodontitis, suggesting the potential regulatory effects [Citation31].
In this research, we aimed to investigate the regulating effects of miR-29a on pyroptosis in periodontitis model cells induced by LPS. ceRNA mechanism that may be involved in the process of periodontitis was also investigated.
Materials and methods
PDL tissue
The clinical tissue specimens were provided by the Hospital of Chengdu University of Traditional Chinese Medicine (TCM), approved by the Ethics Committee of Chengdu University of TCM (2022KL-036), and collected in accordance with the principles of the Declaration of Helsinki after the patients signed the informed consent. The PDL tissues (23 cases) were obtained by scraping the root of the tooth during periodontal flap surgery in patients with periodontitis or from the root of the tooth extracted for periodontitis [Citation32]. Meanwhile, healthy PDL tissues from premolars or third molars removed for orthodontic or impacted extraction were obtained from 23 volunteers [Citation33].
The clinical and demographic characteristics of all subjects are listed in supplementary Table 1. The included criteria of periodontitis group: probing depth is 5 mm after periodontal treatment. The exclusion criteria were as follows: 1) patients with other systemic diseases such as diabetes, immune diseases, metabolic diseases, etc.; 2) antibacterial or drug treatment history in the past 3 months and 3) smoking history or bad occlusal habits. After collection, the specimens were stored at −80 °C.
Human gingival fibroblasts (HGFs)
Five healthy gingival tissue pieces (3 males and 2 females, aged 18–27 years) randomly slected from the 23 healthy volunteers mentioned above. The tissue pieces were washed with normal saline and penicillin and streptomycin solution (Solarbio, China) and then digested in 2.5 g/L neutral protease (Solarbio, China) overnight. The connective tissue parts were separated and cut into 1 mm3 fragments and obtained by inverted adhesion method. HGFs cells were cultured in dulbecco’s modified eagle medium (HyClone, Logan, UT, USA) containing 10% FBS (BI) and 1% penicillin and streptomycin solution. When the cell density reached 70%–80%, the medium was changed every other day for 1:2 subculture, and the 3–6 generations of cells in logarithmic growth stage were used for subsequent experiments. Isolated HGFs were then treated with 1 μg/mL P. gingivalis-LPS (Invivogen, San Diego, CA, USA) for 12 h to establish periodontitis model cells [Citation34,Citation35].
Cell transfection
To determine the functional role of miR-29a in periodontitis, miR-29a was upregulated by agomiR-29a (miR-29a agonist) and downregulated by antagomiR-29a (miR-29a inhibitor). agomiR-nc (negative control of miR-29a agonist) and antagomiR-nc (negative control of miR-29a inhibitor) were used as controls. Small interference RNA targeting circ_0036490 (si-circ_0036490), overexpressed circ_0036490 vector (oe-circ_0036490), overexpressed Dickkopf-1 vector (oe-DKK-1), and their negative controls (si-nc and oe-nc). All plasmids and vectors were synthesised by Ribobio Co. (Guangzhou, China). HGFs of each group were transfected with the specified vector and/or sequence using the Lipofectamine 3000 (Invitrogen) regent for 48 h when 60–70% convergence was achieved.
Reverse transcription quantitative real-time PCR (RT-qPCR)
PDL tissues and HGFs samples of each group were selected, and total RNA was extracted in three parallel copies. After determining the RNA concentration, cDNA was synthesised according to the instructions of the reverse transcription kit (18064071; SuperScript ™II, Thermo Fisher). Real-time quantitative RT-PCR was conducted according to the PCR kit instructions (11746500; SuperScript™ III Platinum™ SYBR™ Green; Invitrogen), and GAPDH was selected as the reference gene to detect the mRNA expression of circ_0036490 and DKK1. U6 was used as an internal reference to detect the expression of miR-29a. Primer sequences were shown in . The data were analysed using 2−ΔΔCt relative expression method. All experiments were repeated three times.
Table 1. Primer sequence.
Cell counting kit-8 (CCK-8)
HGFs were seeded into 96 well plates. 10 μL CCK-8 solution (C0038; Beyotime, Shanghai, China) was added to each well and incubated in a cell incubator (37 °C, 5% CO2) for 2 h. Finally, the absorbance value was detected at 450 nm wavelength, and each group of samples was repeated for five times, and the results were averaged.
Lactate dehydrogenase (LDH) measurement
A LDH Cytotoxicity Assay Kit (C0016; Beyotime) was used to measure LDH levels in HGFs under the guidance of manufacture’s instruction. Briefly, cells were inoculated into the 96-well cell culture plates. The LDH release reagents were added to each well and incubated for 1 h. Absorbance values were measured at 495 nm with the microplate reader (BioTek, Vermont, USA).
Enzyme-linked immunosorbent assay (ELISA)
The supernatant of HGFs after centrifugation for 20 min at 900×g were collected. Cytokines (IL-1β and IL-8) were assayed using a IL-1β ELISA kit (PI305; Beyotime) and a IL-18 ELISA kit (PI558; Beyotime) according to the manufacturer’s instructions.
Flow cytometry assay
Cell pyroptosis was detected by a FAM-FLICA® Caspase-1 (YVAD) Assay Kit (Biopike, Beijing, China). HGFs were scraped off and washed with PBS twice, then cells were suspended with caspase-1 staining solution (1:30). After 1 h of incubation in 5% CO2 incubator at 37 °C, PI staining solution (1:100) was then added to suspend the cells.
Western blotting assay
Protein expression of pro-caspase-1, cleaved-caspase-1, and GSDMD-N were detected by western blotting assay. Proteins were extracted from HGFs by RIPA (Sigma-Aldrich, St. Louis, MO, USA) reagent and were detected by BCA method (P0012; Beyotime). 30 μg total protein from each well was separated by 10% SDS-PAGE electrophoresis (P0690; Beyotime) and transfered to PVDF membrane (FFP32; Beyotime). The membranes were blocked with 5% defatted milk for 1 h. Then, primary antibodies, which were listed in were added. The next day, HRP labelled secondary antibody was added after the membrane was washed with PBS, and the membrane was incubated at room temperature for 1 h. ECL developer was developed with GAPDH as control.
Table 2. Antibodies.
Actinomycin D and ribonuclease R (RNase R) treatment
Actinomycin D (2 mg/mL, Sigma-Aldrich) was added into the HGFs to block transcription. As for RNase R treatment, total RNA (2 μg) was added for co-incubation for 1 h at 37 °C with or without RNase R (3 U/μg; Epicentre) [Citation36]. After treatment with actinomycin D and RNase R, qRT-PCR was carried out to determine the expression levels of circ_0036490 mRNA and mortality factor 4-like 1 (MORF4L1) mRNA (circ_0036490 is derived from exons 12 of the MORF4L1 gene).
Bioinformatic analysis
Binding sites between miR-29a and circ_0036490 or DKK1 were predicted through two online datasets, Starbase (http://starbase.sysu.edu.cn/) and TargetScan 7.2 (http://www.targetscan.org/). Kyoto Encyclopaedia of Genes and Genomes (KEGG) analyse was conducted through FunRich [Citation37]. STRING (http://www.string-db.org/) database was applied to construct protein–protein interaction (PPI) network according to the previous research [Citation38].
Dual-luciferase reporter assay
The pGL3 luciferase promoter plasmid (Promega, Madison, Wisconsin, USA) was used to generate constructs containing circ_0036490 or DKK1 3′-UTR [defined as wildtype (wt)]. The circ_0036490 or DKK1 3′-UTR mutation point [defined as mutant-type (mut)] that targeting miR-29a were directly synthesised using the QuickChange Multiple Site-directed Mutagenesis Kit (Stratagene, Santa Clara, CA, USA). Afterwards, all designed plasmids were co-transfected with agomiR-29a and agomiR-nc into HGFs using the Lipofectamine 3000 method (Invitrogen). Fourty-eight hours after transfection, luciferase activities of harvested HGFs were detected by the Dual Luciferase Reporter Assay Kit (Promega). The ratio of firefly to Renilla luciferase activity was subsequently determined.
RNA pull-down assay
The streptavidin magnetic beads (500 μg) combined with 200 pmol biotin-labelled miR-29a were added into total RNA extracted from HGFs of each group. Biotinylated RNAs were prepared using Pierce RNA 3′ End Desthiobiotinylation Kit (Thermo Scientific, # 20163) and T7/T3 RNA in vitro transcription kit (Ambion, # AM1308). The pulled complex was collected after eluting buffer was added and incubation for 30 min. Finally, the circ_0036490 and DKK1 levels were quantitatively analysed by RT-qPCR as previously reported [Citation39].
Nucleus–cytoplasm separation
The nuclear fraction of HGFs was extracted using an Ambion® PARIS™ Kit (AM1921; Austin, TX, USA). HGFs were washed three times with PBS on ice, followed by centrifugation at 300 × g for 5 min. Cells were resuspended in cell fraction buffer, and then centrifuged at 500 × g for 5 min at 4 °C. Nuclear pellets were homogenised with the cell disruption buffer from the PARIS kit. U6, GAPDH, and circ_0036490 levels in the nucleus and cytoplasm sections were evaluated by RT-qPCR assay.
Fluorescence in situ hybridisation (FISH)
HGFs were firstly fixed in 4% paraformaldehyde for 10 min, and then permeabilized with 0.5% Triton X-100 for 15 min at 4 °C. Digoxigenin-labelled circ_0036490 probe was performed to incubate HGFs for 4 h at 37 °C. Afterwards, the signal was detected using HRP-conjugated antidigoxigenin secondary antibodies (Jackson, Ely, Cambs, UK). Images were obtained by the confocal laser scanning microscope (Olympus, Tokyo, Japan). 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) was used for nuclear counterstaining.
Statistical analysis
GraphPad Prism version 8.3 (GraphPad) was applied to analyse all data which were presented as mean ± standard deviation (SD). Student’s t-test (two groups) and one-way analysis of variance (ANOVO) followed by Tukey’s test (multiple groups) were applied for difference analysis. p value less than 0.05 was deemed statistically significant.
Results
MiR-29a protects HGFs from LPS-induced pyroptosis
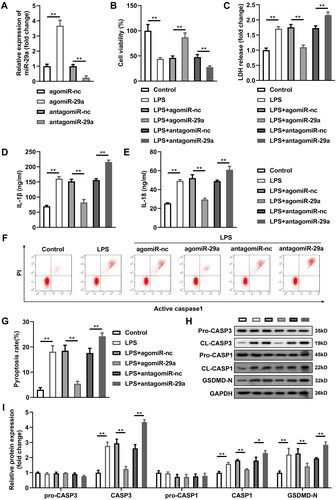
Expression of miR-29a in PDL tissues of two groups was firstly evaluated by RT-qPCR, and the results demonstrated that miR-29a was prominently lowly expressed in PDL samples of periodontitis group compared with that in the healthy control group (p < 0.05) (). Meanwhile, miR-29a was also downregulated in LPS-treated HGFs compared with untreated HGFs (p < 0.05) (). MiR-29a expression significantly increased in HGFs transfected with agomiR-29a and decreased in HGFs transfected with antagomiR-29a (p < 0.05) (). Cell viability of HGFs was prominently inhibited after LPS treatment, whereas secretion of LDH, IL-1β, and IL-18 were markedly increased (p < 0.05) (). Additionally, pyroptosis rate was promoted by LPS, and protein levels of pyroptosis-related proteins including cleaved-caspase-1 and GSDMD-N were also remarkably elevated by LPS exposure in HGFs; meanwhile, apoptosis of HGFs treated with LPS was also promoted according to the protein levels of cleaved-caspase-3 (p < 0.05) (). Upregluation of miR-29a dramatically alleviated LPS-induced cell damage by restoring cell viability, inhibiting LDH, IL-1β, and IL-18 secretion and delaying cell pyroptosis and apoptosis (p < 0.05). Meanwhile, downregulation of miR-29a exerted the opposite role to upregulation of miR-29a, which further aggravated the cell damage caused by LPS (p < 0.05).
Figure 1. MiR-29a is downregulated in periodontitis. MiR-29a expression in (A) PDL tissues from patients with periodontitis and healthy controls, and (B) HGFs before and after LPS treatment evaluated by RT-qPCR. **p < 0.01. PDL: periodontal ligament. HGF: human gingival fibroblast. LPS: lipopolysaccharides. RT-qPCR: reverse transcription quantitative real-time PCR.
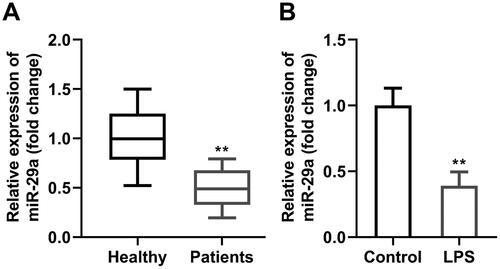
Figure 2. MiR-29a protects HGFs from LPS-induced pyroptosis. (A) miR-29a expression levels were detected by RT-qPCR in HGFs after transfection. (B) Cell viability evaluated by CCK-8 assay. (C-E) Concentration of LDH, IL-1β and IL-18. (F-G) Flow cytometry assay analyses of pyroptosis of HGFs. (H-I) Pyroptosis-related proteins (pro-caspase-1, cleaved-caspase-1, and GSDMD-N) and apoptosis-related proteins (pro-caspase-3, cleaved-caspase-3) analysed by western blotting assay. *p < 0.05, **p < 0.01. RT-qPCR: reverse transcription quantitative real-time PCR. CCK-8: cell counting kit-8. LDH: lactate dehydrogenase. HGF: human gingival fibroblast.
Circ_0036490 sponges miR-29a in HGFs
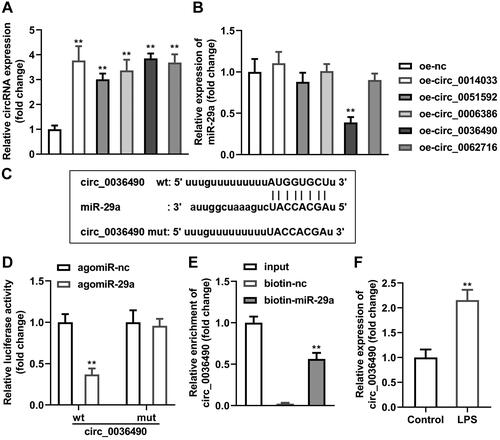
Afterwards, top five circRNAs that bind to miR-29a predicted by StarBase online database were selected as the candidates. Circ_0014033, circ_0051592, circ_0006386, circ_0036490, and circ_0062716 were overexpressed separately in HGFs (p < 0.01) (). miR-29a was significantly downregulated by elevated circ_0036490 (p < 0.01) (). Sequence alignment of miR-29a with the putative binding sites within the wt or mut regions of circ_0036490 were as indicated in . Luciferase activity of HGFs co-transfected with agomiR-29a and wt-circ_0036490 was dramatically decreased (p < 0.05) while there was no significant changes in mut group (p > 0.05) (). The results of RNA pull-down experiment showed that the expression level of circ_0036490 in the biotin-labelled miR-29a group was prominently higher than that in the control group (biotin-nc), which also confirmed the binding relationship between circ_0036490 and miR-29a (p < 0.05) (). Additionally, circ_0036490 level was higher in LPS-treated HGFs compared with control group (p < 0.05) ().
Figure 3. Circ_0036490 sponges miR-29a in HGFs. (A) The top five circRNAs that bind to miR-29a predicted by StarBase online database were upregulated in HGFs separately. (B) MiR-29a levels in HGFs after transfection were evaluated by RT-qPCR. (C) Bioinformatics predicted the binding sites between miR-29a and circ_0036490. Identification of the interaction between miR-29a and circ_0036490 by (D) dual-luciferase reporter assay and (E) RNA pull-down assay. (F) RT-qPCR was performed to determine the circ_0036490 levels in HGFs treated with LPS. **p < 0.01. RT-qPCR: reverse transcription quantitative real-time PCR. HGF: human gingival fibroblast. LPS: lipopolysaccharides.
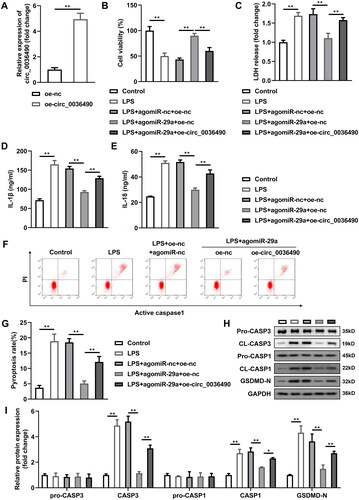
Upregulation of circ_0036490 reverses the effects of miR-29a in HGFs
Subsequently, we found that the expression level of linear type of circ_0036490 was substantially decreased compared with circ_0036490 (p < 0.05) (). Circ_0036490 showed more stable mRNA expression compared with linear group (p < 0.05) (). Furthermore, circ_0036490 was enriched in the cytoplasm (). Images obtained from FISH method indicated that circ_0036490 is enriched in cytoplasm of HGFs (). Then, the circ_0036490 was overexpressed in HGFs to verify the role of circ_0036490 (). After overexpression of circ_0036490, the number of binding miR-29a sites increased, so the number of dissociative miR-29a decreased. As expected, the effect of miR-29a was weakened, which was specifically manifested as the inhibition of cell viability (p < 0.05) () and the increase of LDH, IL-1β, and IL-18 secretion (p < 0.05) (). At the same time, the pyroptosis and apoptosis weakened by miR-29a was promoted by circ_0036490 (p < 0.05) ().
Figure 4. Properties of circ_0036490. (A) mRNA levels of MORF4L1 gene and circ_0138959 in HGFs after RNaseR treatment. (B) RT-qPCR analysis for the expression of circ_0138959 and MORF4L1 gene in HGFs at the indicated time after treatment with Actinomycin D. (C) RNA in the cytoplasm and nucleus was separated, and the expression of circ_0138959 was evaluated by RT-qPCR. (D) Localisation of circ_0036490 was evaluated by FISH assay. **p < 0.01. HGF: human gingival fibroblast. RT-qPCR: reverse transcription quantitative real-time PCR. FISH: fluorescence in situ hybridisation.
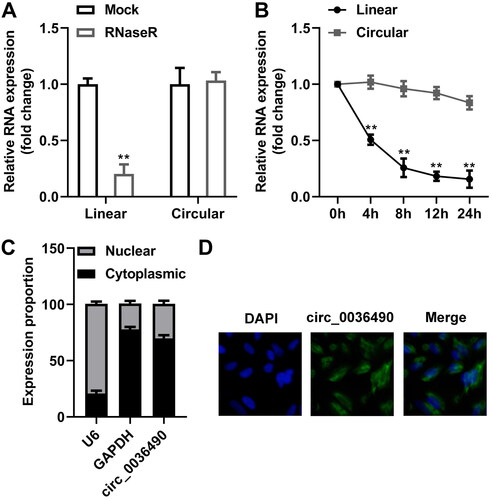
Figure 5. Upregulation of circ_0036490 reverses the effects of miR-29a in HGFs. (A) Circ_0036490 expression levels were detected by RT-qPCR in HGFs before and after transfection. (B) Cell viability evaluated by CCK-8 assay. (C–E) Concentration of LDH, IL-1β, and IL-18. (F–G) Flow cytometry assay analyses of pyroptosis of HGFs. (H–I) Pyroptosis-related proteins (pro-caspase-1, cleaved-caspase-1, and GSDMD-N) and apoptosis-related proteins (pro-caspase-3, cleaved-caspase-3) analysed by western blotting assay. *p < 0.05, **p < 0.01. RT-qPCR: reverse transcription quantitative real-time PCR. HGF: human gingival fibroblast.
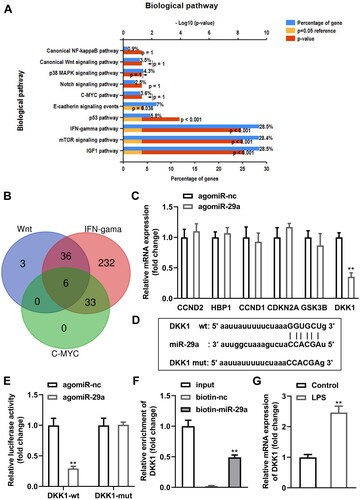
DKK1 is a downstream target gene of miR-29a in HGFs
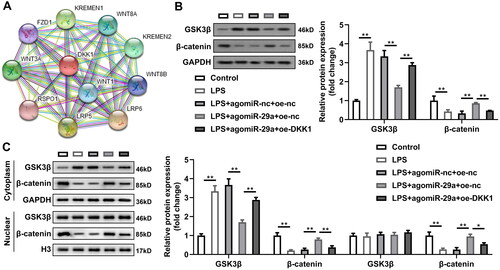
Next, genes that bind with miR-29a predicted by bioinformatics were analysed, and the KEGG results demonstrated that genes were enriched in ten pathways (). Wnt, IFN-gama, and C-MYC pathways shared six genes (), including CCND2, HBP1, CCND1, CDKN2A, GSK3B, and DKK1. Interestingly, DKK1 was prominently downregulated in HGFs transfected with agomiR-29a (p < 0.01) (). The predicted binding sites between DKK1 and miR-29a were as shown in . Dual luciferase reporter and RNA pull-down assays verified the interaction between DKK1 and miR-29a (). Additionally, DKK1 was dramatically upregulated in LPS-treated HGFs (p < 0.05) ().
Figure 6. DKK1 is a downstream target gene of miR-29a in HGFs. (A) KEGG analysis of genes bind with miR-29a (B) Venn diagram of genes shared by Wnt, IFN-gama, and C-MYC pathways. (C) mRNA levels of six shared genes in HGFs transfected with agomiR-29a. (D) Bioinformatics predicted the binding sites between miR-29a and DKK1. Identification of the interaction between miR-29a and DKK1 by (E) dual-luciferase reporter assay and (F) RNA pull-down assay. (G) RT-qPCR was performed to determine the DKK1 levels. **p < 0.01. RT-qPCR: reverse transcription quantitative real-time PCR.
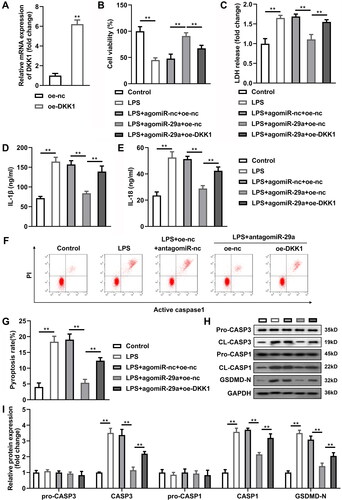
Overexpressed DKK1 alleviates the effects of agomiR-29a
DKK1 expression was prominently increased in HGFs after transfected with oe-DKK1 vectors (p < 0.05) (). After overexpression of DKK1, as DKK1 co-competes with circ_0036490 for miR-29a, the dissociative miR-29a is reduced. As speculated, the effect of miR-29a is weakened, which is specifically manifested as the inhibition of cell viability (p < 0.05) () and the increase of LDH, IL-1β, and IL-18 secretion (p < 0.05) (). At the same time, the pyroptosis and apoptosis weakened by miR-29a is promoted by overexpressed DKK1 (p < 0.05) (). Then, PPI network of DKK1 revealed that multiple Wnt-related proteins interact with DKK1 (). Since DKK1 has been reported to be produced by osteocytes and osteoblasts and to modulate the Wnt signalling [Citation40]; we hypothesised that the interaction between miR-29a and DKK1 affects protein levels of Wnt signalling-related proteins. As indicated in , LPS prominently induced GSK3β expression and decreased β-catenin protein levels in HGFs (p < 0.05). Upregulation of miR-29a reversed the effects of LPS while overexpressed DKK1 significantly reversed the effects of miR-29a to revert to LPS-induced regulatory trends in protein levels of Wnt signalling pathway (p < 0.05) (). Moreover, we isolated the cytoplasm and nucleus of HGFs in each group and detected the expression of GSK3β and β-catenin proteins. The results indicated that the pathway Wnt pathway was significantly activated in the cytoplasm but not in the nucleus (p < 0.05) ().
Figure 7. Overexpressed DKK1 alleviates the effects of agomiR-29a. (A) DKK1 expression levels were detected by RT-qPCR. (B) Cell viability evaluated by CCK-8 assay. (C–E) Concentration of LDH, IL-1β, and IL-18. (F–G) Flow cytometry assay analyses of pyroptosis of HGFs. (H–I) Pyroptosis-related proteins (pro-caspase-1, cleaved-caspase-1, and GSDMD-N) and apoptosis-related proteins (pro-caspase-3, cleaved-caspase-3) analysed by western blotting assay. **p < 0.01. RT-qPCR: reverse transcription quantitative real-time PCR. HGF: human gingival fibroblast.
Discussion
This experiment aimed to explore whether miR-29 regulates pyroptosis of HGFs induced by LPS and the underlying ceRNA mechanism. Currently, we unveiled that miR-29a contributed to the protection of HGFs from LPS. Circ_0036490 and DKK1 competitively binded to miR-29a. Functionally, circ_0036490/miR-29a/DKK1 axis regulated cell viability and pyroptosis of LPS-treated HGFs. Upregulation of miR-29a is a promising method to inhibit pyroptosis thereby to alleviate periodontitis.
Periodontitis is an inflammatory disease characterised by periodontal pocket formation and alveolar bone resorption. It is one of the main causes of tooth loss in adults over 40 years old. The HGFs are the most abundant cells in PDL tissue, and the HGF responses to periodontal pathogens or inflammatory cytokines contribute to the development of periodontitis [Citation41,Citation42]. The expression level of miRNA is related to the development of periodontal inflammation, which can be used as one of the indicators of periodontal diagnosis. MiRNA can reduce the stability of mRNA by binding to target genes, or inhibit the translation of target gene mRNA, so that it can control gene expression after transcription. MiRNAs are involved in the regulation of immune responses, the development of lymphocytes, the regulation of immune factors and the activation of signalling pathways. In periodontitis, miRNA affects the conduction of various signalling pathways such as RANKL and Wnt by regulating the expression of target genes and participates in inflammatory response, osteogenesis and osteoclast. Rencently, miR-29a has been found to be a potential marker associated with tooth movement [Citation21] and is involved in the process of PDL stem cells chondrogenesis [Citation22]. Besides, miR-29a promotes osteoblast proliferation by downregulating DKK1 expression and activating Wnt/β-catenin signalling pathway [Citation43]. DKK1 inhibited miR-29a expression which was induced by the Wnt signalling in osteoblasts [Citation44]. Currently, miR-29a expression was found to be downregulated in periodontitis clinical samples and HGFs treated with LPS. Then whether the aberrant expression of miR-29a accelerated the LPS-induced injury of periodontitis in vitro was studied. Our data suggested that upregulation of miR-29a protects HGFs from LPS induced injury and inhibition of miR-29a further aggravated LPS-induced damage. Therefore, downregulation of miR-29a may be an effective method to alleviate periodontitis in vitro.
Since circRNAs are not easy to be degraded, circRNAs can also be used for disease diagnosis and analysis of biomarkers [Citation25,Citation45]. CircRNA can also act as ceRNA to adsorb miRNA, thus inhibiting the functional role of miRNA in regulating its target genes. Transcripts such as mRNA and circRNA with the same miRNA response element regulate their respective expression by competitive binding to the same miRNA, thus affecting cell function. Among them, RNA with such competitive relationship can be called ceRNA, and miRNA acts as a medium between different transcripts in ceRNA relationship. Zhao et al. [Citation46] declared that ceRNA network in periodontal tissue may be key regulators in periodontitis treatment. Circ_0099630 binds to miR-940 to modulate the pathogenesis of periodontitis [Citation47]. CircLRP6 antagonised the function of miR-145a-5p to promote cementum formation [Citation48]. Likewise, circ_0036490 binded to miR-29a and reversed the effects of miR-29a on cell viability and pyroptosis of LPS-treated HGFs.
Furthermore, DKK1 is a downstream gene of miR-29a and competitively binds to miR-29a with circ_0036490. DKK1 is an antagonist of Wnt3a, and DKK1 could inhibit the Wnt signalling to induce bone loss [Citation49]. The study of Liu Q et al. suggested that DKK1 promotes the osteogenic differentiation of mesenchymal stem cells isolated from PDLs in periodontitis [Citation50]. Thence, DDK1 and Wnt signalling pathway may participate in periodontitis. In this study, overexpression of DKK1 and circ_0036490 exerted the same role of regulating cell viability and pyroptosis of HGFs and alleviates the effects of agomiR-29a.
In the classical Wnt/β-catenin signalling pathway, DKK1 can combine with LRP5/6 in the signalling pathway to form trimers, thereby blocking the Wnt/β-catenin signalling pathway and affecting bone metabolism [Citation51]. Studies have shown that DKK1 can inhibit the osteogenic differentiation of PDL stem cells by inhibiting the Wnt/β-catenin pathway [Citation50]. In current work, the Wnt/β-catenin pathway was inactivated by LPS in HGFs, and miR-29a activated the pathway. Furthermore, DKK1 restored the function of LPS on regulating proteins in the Wnt/β-catenin pathway. Thus, miR-29a/DKK1 axis modulates the activities of Wnt/β-catenin pathway in periodontitis in vitro.
Conclusion
Our research suggested circ_0036490 and DKK1 competitively bind miR-29a to promote LPS-induced HGF injury in vitro. miR-29a may be a potential target for the treatment of periodontitis.
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data and review of the manuscript. Y.W. drafted the work and revised it critically for important intellectual content; B.L., D.D., H.Z., and M.L. were responsible for the acquisition and analysis; H.A., Y.X., and W.H. were responsible for the interpretation of data for the work; L.Z. and L.L. made substantial contributions to the conception or design of the work.
Supplemental Material
Download MS Word (13.2 KB)Disclosure statement of competing interest
No conflict of interest exits in the submission of this manuscript, and manuscript is approved by all authors for publication.
The regulatory mechanism study of Bushen Huoxue Guchi therapy on alveolar bone remodelling of orthodontic tooth with chronic periodontitis based on the miR-29a mediated Dkk-1/Wnt-TNF-α interactive crosstalk (NSFC 81973684).
From the “central-peripheral” holistic perspective to explore the regulatory mechanism of “replenishing qi and opening orifices for reducing phlegm” on Aβ clearance across the blood-brain barrier in Alzheimer’s disease (NSFC82174345).
The study on PDLSCs combined with nanofiber composite scaffold with gradation in cytokines concentration to engineer PDL–bone interface (NSFC 31670992).
The imaging study of syndrome essence and syndrome element evolution in acute ischaemic stroke during over-time window treatment based on DKI (NSFC82074303).
Data availability statement
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Xu W, Zhou W, Wang H, et al. Roles of Porphyromonas gingivalis and its virulence factors in periodontitis. Adv Protein Chem Struct Biol. 2020;120:1–13.
- Sayad A, Ghafouri-Fard S, Shams B, et al. Blood and tissue levels of lncRNAs in periodontitis. J Cell Physiol. 2020;235(12):9568–9576.
- Chiu HC, Fu MM, Yang TS, et al. Effect of high glucose, Porphyromonas gingivalis lipopolysaccharide and advanced glycation end-products on production of interleukin-6/-8 by gingival fibroblasts. J Periodontal Res. 2017;52(2):268–276.
- Lönn J, Ljunggren S, Klarström Engström K, et al. Lipoprotein modifications by gingipains of Porphyromonas gingivalis. J Periodontal Res. 2018;53(3):403–413.
- Zhuang YT, Xu DY, Wang GY, et al. IL-6 induced lncRNA MALAT1 enhances TNF-alpha expression in LPS-induced septic cardiomyocytes via activation of SAA3. Eur Rev Med Pharmacol Sci. 2017;21(2):302–309.
- Du W, Wang L, Liao Z, et al. Circ_0085289 alleviates the progression of periodontitis by regulating let-7f-5p/SOCS6 pathway. Inflammation. 2021;44(4):1607–1619.
- Li X, Luo W, Hu J, et al. Interleukin-27 prevents LPS-induced inflammatory osteolysis by inhibiting osteoclast formation and function. Am J Transl Res. 2019;11:1154–1169.
- Lu F, Lan Z, Xin Z, et al. Emerging insights into molecular mechanisms underlying pyroptosis and functions of inflammasomes in diseases. J Cell Physiol. 2020;235(4):3207–3221.
- Kim YK, Shin JS, Nahm MH. NOD-Like receptors in infection, immunity, and diseases. Yonsei Med J. 2016;57(1):5–14.
- Shi J, Zhao Y, Wang K, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526(7575):660–665.
- Polonio CM, Peron J. ZIKV infection and miRNA network in pathogenesis and immune response. Viruses. 2021;13(10):1992.
- Zhang Z, Huang Q, Yu L, et al. The role of miRNA in tumor immune escape and miRNA-based therapeutic strategies. Front Immunol. 2021;12:807895.
- Kebschull M, Papapanou PN. Mini but mighty: microRNAs in the pathobiology of periodontal disease. Periodontol 2000. 2015;69(1):201–220.
- Saito A, Horie M, Ejiri K, et al. MicroRNA profiling in gingival crevicular fluid of periodontitis-a pilot study. FEBS Open Bio. 2017;7(7):981–994.
- Schmalz G, Li S, Burkhardt R, et al. MicroRNAs as salivary markers for periodontal diseases: a new diagnostic approach? Biomed Res Int. 2016;2016:1027525.
- Wang X, Sun H, Liao H, et al. MicroRNA-155-3p mediates TNF-alpha-Inhibited cementoblast differentiation. J Dent Res. 2017;96(12):1430–1437.
- Venugopal P, Lavu V, RangaRao S, et al. Evaluation of a panel of Single-Nucleotide polymorphisms in miR-146a and miR-196a2 genomic regions in patients with chronic periodontitis. Genet Test Mol Biomarkers. 2017;21(4):228–235.
- Alizadeh M, Safarzadeh A, Beyranvand F, et al. The potential role of miR-29 in health and cancer diagnosis, prognosis, and therapy. J Cell Physiol. 2019;234(11):19280–19297.
- Horita M, Farquharson C, Stephen LA. The role of miR-29 family in disease. J Cell Biochem. 2021;122(7):696–715.
- Wang JY, Zhang Q, Wang DD, et al. MiR-29a: a potential therapeutic target and promising biomarker in tumors. Biosci Rep. 2018;38(1):BSR20171265.
- Kapoor P, Chowdhry A, Bagga DK, et al. MicroRNAs in oral fluids (saliva and gingival crevicular fluid) as biomarkers in orthodontics: systematic review and integrated bioinformatic analysis. Prog Orthod. 2021;22(1):31.
- Wang P, Li Y, Meng T, et al. KDM6A promotes chondrogenic differentiation of periodontal ligament stem cells by demethylation of SOX9. Cell Prolif. 2018;51(3):e12413.
- Chen J, Zhu Z, Xu S, et al. HDAC1 participates in polycystic ovary syndrome through histone modification by regulating H19/miR-29a-3p/NLRP3-mediated granulosa cell pyroptosis. Mol Cell Endocrinol. 2023;573:111950.
- Ning J, He K, Cheng F, et al. Long non-coding RNA MEG3 promotes pyroptosis in testicular ischemia-reperfusion injury by targeting MiR-29a to modulate PTEN expression. Front Cell Dev Biol. 2021;9:671613.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Jin J, Sun H, Shi C, et al. Circular RNA in renal diseases. J Cell Mol Med. 2020;24(12):6523–6533.
- Wang M, Yu F, Li P. Circular RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10(8):258.
- Zuo H, Li X, Zheng X, et al. A novel circRNA-miRNA-mRNA hub regulatory network in lung adenocarcinoma. Front Genet. 2021;12:673501.
- Miao L, Yin RX, Zhang QH, et al. A novel circRNA-miRNA-mRNA network identifies circ-YOD1 as a biomarker for coronary artery disease. Sci Rep. 2019;9(1):18314.
- Zheng X, Huang M, Xing L, et al. The circRNA circSEPT9 mediated by E2F1 and EIF4A3 facilitates the carcinogenesis and development of triple-negative breast cancer. Mol Cancer. 2020;19(1):73.
- Li J, Xie R. Circular RNA expression profile in gingival tissues identifies circ_0062491 and circ_0095812 as potential treatment targets. J Cell Biochem. 2019;120(9):14867–14874.
- Lei F, Li M, Lin T, et al. Treatment of inflammatory bone loss in periodontitis by stem cell-derived exosomes. Acta Biomater. 2022;141:333–343.
- Wei Y, Fu J, Wu W, et al. Quercetin prevents oxidative Stress-Induced injury of periodontal ligament cells and alveolar bone loss in periodontitis. Drug Des Devel Ther. 2021;15:3509–3522.
- Huang X, Shen H, Liu Y, et al. Fisetin attenuates periodontitis through FGFR1/TLR4/NLRP3 inflammasome pathway. Int Immunopharmacol. 2021;95:107505.
- Yang K, Xu S, Zhao H, et al. Hypoxia and Porphyromonas gingivalis-lipopolysaccharide synergistically induce NLRP3 inflammasome activation in human gingival fibroblasts. Int Immunopharmacol. 2021;94:107456.
- Peng QS, Cheng YN, Zhang WB, et al. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit hippo signaling pathway. Cell Death Dis. 2020;11(2):112.
- Deng JL, Xu YH, Wang G. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front Genet. 2019;10:695.
- Feng H, Gu ZY, Li Q, et al. Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis. J Ovarian Res. 2019;12(1):35.
- Li D, Wang W, Liu B, et al. Characterization of circSEC11A as a novel regulator of iodine-125 radioactive seed-induced anticancer effects in hepatocellular carcinoma via targeting ZHX2/GADD34 axis. Cell Death Discov. 2023;9(1):294.
- Bao J, Yang Y, Xia M, et al. Wnt signaling: an attractive target for periodontitis treatment. Biomed Pharmacother. 2021;133:110935.
- Naruishi K, Nagata T. Biological effects of interleukin-6 on gingival fibroblasts: cytokine regulation in periodontitis. J Cell Physiol. 2018;233(9):6393–6400.
- Wielento A, Lagosz-Cwik KB, Potempa J, et al. The role of gingival fibroblasts in the pathogenesis of periodontitis. J Dent Res. 2023;102(5):489–496.
- Zhang F, Cao K, Du G, et al. miR-29a promotes osteoblast proliferation by downregulating DKK-1 expression and activating wnt/beta-catenin signaling pathway. Adv Clin Exp Med. 2019;28(10):1293–1300.
- Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical wnt signaling. J Cell Biochem. 2009;108(1):216–224.
- Chaichian S, Shafabakhsh R, Mirhashemi SM, et al. Circular RNAs: a novel biomarker for cervical cancer. J Cell Physiol. 2020;235(2):718–724.
- Zhao Q, Wen J, Ouyang X, et al. Whole-transcriptome analysis of periodontal tissue and construction of immune-related competitive endogenous RNA network. BMC Oral Health. 2022;22(1):370.
- Zhao XQ, Ao CB, Yan YT. The circular RNA circ_0099630/miR-940/receptor-associated factor 6 regulation Cascade modulates the pathogenesis of periodontitis. J Dent Sci. 2022;17(4):1566–1576.
- Li M, Du M, Wang Y, et al. CircRNA Lrp6 promotes cementoblast differentiation via miR-145a-5p/Zeb2 axis. J Periodontal Res. 2021;56(6):1200–1212.
- Liu J, Ren X, Zhang M, et al. Roles of Wnt3a and Dkk1 in experimental periodontitis. J Dent Sci. 2017;12(3):220–225.
- Liu Q, Hu CH, Zhou CH, et al. DKK1 rescues osteogenic differentiation of mesenchymal stem cells isolated from periodontal ligaments of patients with diabetes mellitus induced periodontitis. Sci Rep. 2015;5(1):13142.
- Park BM, Kim EJ, Nam HJ, et al. Cyclized oligopeptide targeting LRP5/6-DKK1 interaction reduces the growth of tumor burden in a multiple myeloma mouse model. Yonsei Med J. 2017;58(3):505–513.