Abstract
Objective
This study aims to investigate the association between miR-203 expression and the prognostic value in patients with esophageal cancer by the method of systematic review and meta-analysis.
Methods
We searched PubMed, Web of Science, Embase, and Cochrane Library to collect studies on the relationship between miR-203 expression and the prognostic value of esophageal cancer up to July 2023. Stata 15.0 statistical software was used for data analysis. Hazard ratio (HR) and 95% confidence interval (CI) were used as effect sizes.
Results
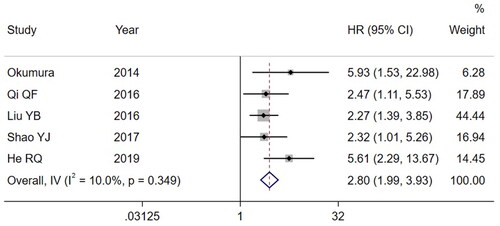
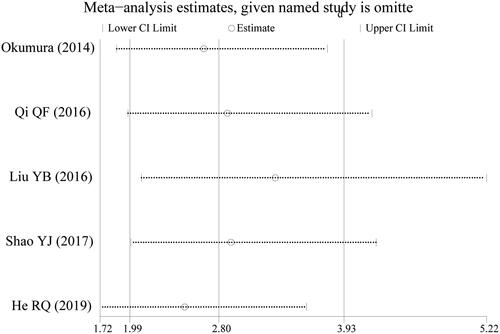
A total of 6 studies were included in this review, including 476 patients with esophageal cancer. The results showed that miR-203 low expression was associated with worse overall survival (OS) in patients with esophageal cancer compared with miR-203 high expression (HR = 2.80, 95%CI: 1.99 ∼ 3.93, p < 0.001). The results of Egger’s (p = 0.154) and Begg’s Tests (p = 0.221) indicated no obvious publication bias. Sensitivity analysis verified the robustness of the results obtained in this study.
Conclusion
The expression of miR-203 is significantly correlated with the prognostic value in patients with esophageal cancer. Esophageal cancer patients with high expression of miR-203 had better prognosis than those with low expression of miR-203. Due to the limited studies included in this meta-analysis, more trials are needed to confirm the conclusions of this study in the future.
Keywords:
1 Introduction
Esophageal cancer, as one of the most aggressive gastrointestinal malignancies, is also the sixth leading cause of current cancer-related death [Citation1], whose incidence rate is still on the rise. According to Global Cancer Statistics 2020, there were 604,100 cases of esophageal cancer diagnosed worldwide in one year, of which 544,076 patients died from esophageal cancer [Citation2]. The histological types of esophageal cancer mainly include squamous cell carcinoma and adenocarcinoma. The main treatment methods for esophageal cancer involve surgery, radiotherapy, chemotherapy, etc. At present, surgical treatment is still the first choice for esophageal cancer [Citation3]. However, due to the biological characteristics of the esophagus, the symptoms of patients with early esophageal cancer are often not obvious, so patients with esophageal cancer are often diagnosed in a relatively advanced stage of the disease [Citation4]. With the improvement of medical technology, the survival time and quality of life of patients with esophageal cancer have been significantly improved, but the five-year survival rate is still about 15%-20% [Citation5]. There are many factors affecting the prognosis of patients with esophageal cancer. To further improve the survival of patients, and reduce local recurrence and metastasis, it is still essential to investigate the factors affecting the prognosis of patients with esophageal cancer.
MicroRNAs (miRNAs), a class of non-protein-coding, endogenous, are evolutionarily conserved small RNAs with a length of approximately 20 to 24 nucleotides, playing a key role in numerous biological processes. Mature miRNAs binding RNA-induced silencing complex (RISC) are transported to the corresponding target mRNAs, regulating more than one-third of human protein-coding genes. MiRNAs targeting tumor suppressor genes or oncogenes can affect tumor formation once they are abnormal, so these miRNAs play a crucial role in regulating tumor suppressor genes or oncogenes in vivo [Citation6,Citation7]. In 2018, a meta-analysis by Gao et al. [Citation8] displayed that high expression of miR-203 in tissues was significantly correlated with better overall survival (OS) of patients with esophageal cancer. However, in the meta-analysis by Gao et al. [Citation8], there were only two studies on miR-203 expression related to the prognosis of esophageal cancer. Later, more studies on the correlation between miR-203 expression and prognosis of esophageal cancer were published.
Therefore, we used the method of systematic review and meta-analysis to explore the correlation between miR-203 expression and prognostic value in patients with esophageal cancer.
2 Methods
This systematic review and meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [Citation9].
2.1. Literature retrieval
We searched PubMed, Web of Science, Embase, and Cochrane Library databases to collect studies on the correlation between miR-203 expression and prognostic value of esophageal cancer. The search deadline is July 2023. The main search strategies are as follows: (“microRNA-203” OR “miR-203” OR “miRNA-203”) AND (“Esophageal cancer” OR “Esophageal tumor” OR “Esophageal neoplasm” OR “Esophageal carcinoma” OR “Esophageal malignant” OR “Esophageal oncology” OR “Oesophageal cancer” OR “Oesophageal tumor” OR “Oesophageal neoplasm” OR “Oesophageal carcinoma” OR “Oesophageal malignant” OR “Oesophageal oncology” OR “Esophagus cancer” OR “Esophagus tumor” OR “Esophagus neoplasm” OR “Esophagus carcinoma” OR “Esophagus malignant” OR "Esophagus oncology" OR "Cancer of the esophageal" OR "Cancer of the esophageal"). Moreover, the literature is traced through the review, supplemented by a manual search. The language was restricted to English. The two researchers conducted the search independently, checked the differences encountered in the process, and reached a consensus through discussion.
2.2. Inclusion and exclusion criteria
2.2.1. Inclusion criteria:
(1) Study type: retrospective cohort study; (2) Population: patients diagnosed with esophageal cancer; (3) Exposure factors: The study group was defined as low expression of miR-203 in tissues, while the control group was defined as high expression of miR-203 in tissues; (4) Outcome indicators: Overall survival (OS), and Disease-free survival (DFS).
2.2.2. Exclusion criteria
(1) Conference abstract, animal experiment, cell experiment; (2) Insufficient data; (3) Research not published in English.
2.3. Literature quality evaluation and data extraction
Two researchers conducted literature quality evaluation and data extraction independently, and finally cross-checked the results. If there was a disagreement, it’s discussed to reach an agreement. According to the Newcastle-Ottawa Scale (NOS) [Citation10] cohort analysis scale, the quality of the included studies was assessed from three aspects: selection, comparability and outcome measurement. The total NOS score is 9. High-quality research is defined when NOS is not less than 6 scores.
Information extracted from the literature included: first author, year of publication, region, assay method, tumor TNM staging, sample source, tissue type, threshold definition, sample size, hazard ratio (HR) of OS or DFS and 95% confidence interval (CI).
2.4. Statistical method
Stata 15.0 statistical software was used for data analysis. The pooled HR and 95%CI were used to evaluate the relationship between miR-203 expression and prognosis of esophageal cancer. If HR was not directly provided in the literature, but survival curves were, then Engauge Digitizer 11.3 software was adopted to extract the data and calculate its HR and 95%CI. Heterogeneity analysis was performed by Chi-square test and I2 statistics. If I2 <50% and p > 0.05, there was good homogeneity among studies, and then a fixed-effects model (FEM) was chosen for combined analysis. On the other hand, if there was heterogeneity (I2 ≥50%, or p ≤ 0.05), a random-effects model (REM) was used for analysis. The funnel plot was not employed to assess publication bias, for less than 10 studies were included in this review [Citation11]. Therefore, Egger’s and Begg’s Tests were used to determine potential publication bias. Finally, sensitivity analysis was performed to verify the robustness of the results obtained.
3 Results
3.1. Literature screening results
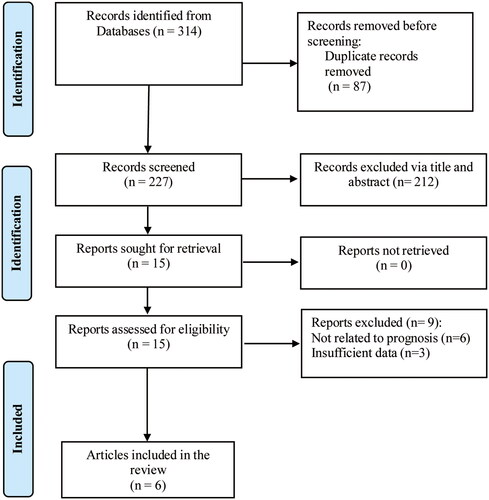
After strict screening, a total of six published papers [Citation12–17] were entered into this review (), including 476 patients with esophageal cancer. Five of them reported OS, and one reported DFS. Besides, one was from Japan [Citation12], one from Czech [Citation13], and the other four from China [Citation14–17]. The basic characteristics of the included publications are shown in . The included articles were published from 2014 to 2019. All the samples were derived from esophageal cancer tissue. The tumor stages covered I to IV. The histopathologic type in the three articles was esophageal squamous cell carcinoma [Citation12,Citation14,Citation17], one esophageal adenocarcinoma [Citation13], and the other two esophageal cancer without definite histopathological classification [Citation15,Citation16].
Table 1. Basic features included in the meta-analysis.
3.2. Quality assessment results of the included literature
The results of the quality assessment of the included literature are shown in Supplementary Table S1. The NOS scores of the included studies were all above 5, indicating high-quality publications included. All studies scored four for case selection, which was rated as “low risk of bias”. Four studies scored 2 points for comparability between groups [Citation12–14, Citation16], and the other two scored 1 point [Citation15, Citation17]. Five articles scored 1 point in exposure factor measurement [Citation12–14, Citation16,Citation17], and one scored 2 points [Citation15].
3.3. Main results of meta-analysis
Five published papers reported OS [Citation12, Citation14–17]. There was no significant heterogeneity across the studies (I2= 10.0%, p = 0.349), so a FEM was used for pooled analysis. The pooled results showed () that compared with the high expression of miR-203, the low expression of miR-203 could reduce the OS of patients with esophageal cancer (HR = 2.8, 95%CI: 1.99 ∼ 3.93, p < 0.001).
3.4. Effect of miR-203 expression on DFS in patients with esophageal cancer
Only one paper reported the correlation between miR-203 expression and DFS in patients with esophageal cancer [Citation17]. The study by Qi et al. [Citation17] showed that patients with esophageal cancer with low expression of miR-203 had worse DFS compared with high expression of miR-203 (HR= 2.47, 95%CI: 1.11 ∼ 5.53, p = 0.026).
3.5. Publication bias detection
Publication bias of included studies for OS was determined by Egger’s and Begg’s tests. The P-value of Egger’s test was 0.154, and the P-value of Begg’s test was 0.221, indicating no obvious publication bias in this meta-analysis.
4 Discussion
As a member of the miRNAs family, miR-203 is specifically expressed in epithelial tissues [Citation18], and its expression level is significantly different in malignant tumors from different tissues [Citation15, Citation19]. MiR-203 can act as an oncogene or a tumor suppressor gene, interacting with multiple mRNA target genes and participating in the occurrence and development of tumors at different stages. MiR-203 is located in the 14q32.33 region, which is an unstable region on chromosomes, and the loss of heterozygosity in this region may be a key factor leading to malignant tumors [Citation20,Citation21]. Two members of this family are miR-203a and miR-203b. In previous studies, the abnormal expression of miR-203 has been shown to be closely related to the progression of various tumors, such as reduced expression in nasopharyngeal carcinoma [Citation22], melanoma [Citation23], cervical cancer [Citation24], and lung cancer [Citation25], etc. When studying the expression of miR-203 in esophageal cancer, Feber et al. [Citation26] found that compared with normal esophageal tissues, the expression of miR-203 in esophageal squamous cell carcinoma and esophageal adenocarcinoma decreased by 1/10-1/2 times, suggesting that miR-203 may play an essential role as a tumor suppressor gene in esophageal cancer. Zhang et al. [Citation27] reported that circ-PRMT5 could stimulate migration in esophageal cancer by binding miR-203. The study by Wang et al. [Citation28] showed that, through regulating miR-203/Slug, circRNA-0008717 could promote cell proliferation, migration, and invasion in esophageal cancer cells.
The results of this meta-analysis showed that miR-203 expression in tissues was significantly correlated with the prognosis of patients with esophageal cancer. High expression of miR-203 was associated with better OS prognosis in patients with esophageal cancer. Although only 5 records were included, the homogeneity of these 5 publications was good, and the conclusion of the combined analysis was robust. This was confirmed by sensitivity analysis. There was also no significant publication bias in this meta-analysis based on Egger’s and Begg’s Tests. There was only one study on the relationship between miR-203 expression and DFS in patients with esophageal cancer. The results of DFS were also consistent with the conclusions of OS. Hezova et al.'s study showed that high expression of miR-203 in patients with esophageal cancer was associated with better DFS prognosis [Citation13]. The conclusions of this meta-analysis are consistent with those of Gao et al. [Citation8]. However, compared with the study of Gao et al. [Citation8], there were 5 studies on OS in our meta-analysis, which reached a more robust conclusion. Therefore, miR-203 in cancer tissues has the potential to be one of the prognostic markers of esophageal cancer. In actual clinical practice, miR-203 can be combined with other markers, such as serum markers or other miRNAs, to comprehensively judge the prognosis of patients with esophageal cancer. Thus, the accuracy of judgment can be provided. Then, corresponding treatment measures can be taken to improve the prognosis of patients with esophageal cancer.
Although we have searched four English databases comprehensively and conducted systematic analysis, there were still several limitations. First, there were few publications included in this study, only 5 related to OS There was only one paper involving DFS, and no more studies can be combined for analysis at present, which needs to be verified by more studies in the future. Limitations in the number of references may impact the robustness of the results obtained. Second, this study included only published studies in English, and excluded potential high-quality studies in other languages, which might lead to certain selection bias. Third, the sample size of this study was limited. Fourth, the research population included in this study was limited, namely China, Japan and Czech. Therefore, whether the conclusions of this study could be extrapolated to populations in other countries needs more trials to confirm.
In conclusion, the expression of miR-203 is significantly correlated with the prognostic value of esophageal cancer. Esophageal cancer patients with high expression of miR-203 had better prognosis than those with low expression of miR-203. miR-203 is expected to be a prognostic marker for patients with esophageal cancer. Therefore, this study has important clinical significance. Considering the limitations of this meta-analysis, such as the small sample size and number of included studies, more trials with more rigorous designs are needed to confirm the conclusions of this review in the future.
Authors’ contribution
QRC, LPN conceived of the study and participated in its design. QRC, LPC, LPN conducted the systematic literature review. QRC, LPC performed data analyses. QRC, LPC drafted the article. QRC, LPC, LPN critically revised the manuscript. All authors have approved the final version of the manuscript.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval
Ethical approval was not needed because this study was based the published literature.
Supplemental Material
Download MS Word (38.5 KB)Acknowledgements
None.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Watanabe M, Suehara N, Koga K, et al. Outcomes of endoscopic submucosal dissection and esophagectomy for early and superficial carcinoma of the esophagus. ESOPHAGUS-TOKYO. 2010;7(4):1–6. doi:10.1007/s10388-010-0249-1.
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660.
- Watanabe M, Otake R, Kozuki R, et al. Recent progress in multidisciplinary treatment for patients with esophageal cancer. Surg Today. 2020;50(1):12–20. doi:10.1007/s00595-019-01878-7.
- Xu QL, Li H, Zhu YJ, Xu G. The treatments and postoperative complications of esophageal cancer: a review. J Cardiothorac Surg. 2020;15(1):163. doi:10.1186/s13019-020-01202-2.
- Jemal A, Bray F, Center M, Ferlay J, Forman D. Global cancerstatistics. CA A Cancer Journal for Clinicians. 2011;6(2):169–190.
- Galasso M, Sana ME, Volinia S. Non-coding RNAs: a key to future personalized molecular therapy? Genome Med. 2010;2(2):12. doi:10.1186/gm133.
- Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci. 2020;21(5):1723.
- Gao S, Zhao ZY, Zhang ZY, Zhang Y, Wu R. Prognostic value of MicroRNAs in esophageal carcinoma: a meta-analysis. Clin Transl Gastroen. 2018;9(11):203.
- Moher D, Liberati A, Tetzlaff J, Altman DG, PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int J Surg. 2010;8(5):336–341. doi:10.1016/j.ijsu.2010.02.007.
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. doi:10.1007/s10654-010-9491-z.
- Sterne JAC, Sutton AJ, Ioannidis JPA, et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ. 2011;343(jul22 1):d4002–d4002. doi:10.1136/bmj.d4002.
- Okumura T, Shimada Y, Moriyama M, et al. MicroRNA-203 inhibits the progression of esophageal squamous cell carcinoma with restored epithelial tissue architecture in vivo. Int J Oncol. 2014;44(6):1923–1932. doi:10.3892/ijo.2014.2365.
- Hezova R, Kovarikova A, Srovnal J, et al. Diagnostic and prognostic potential of miR-21, miR-29c, miR-148 and miR-203 in adenocarcinoma and squamous cell carcinoma of esophagus. Diagn Pathol. 2015;10(1):42. doi:10.1186/s13000-015-0280-6.
- Liu Y, Dong Z, Liang J, et al. Methylation-mediated repression of potential tumor suppressor miR-203a and miR-203b contributes to esophageal squamous cell carcinoma development. Tumour Biol. 2016;37(4):5621–5632. doi:10.1007/s13277-015-4432-9.
- Shao Y, Gu W, Ning Z, et al. Evaluating the prognostic value of microRNA-203 in solid tumors based on a meta-analysis and the cancer genome atlas (tcga) datasets. Cell Physiol Biochem. 2017;41(4):1468–1480. doi:10.1159/000470649.
- He R, Wang J, Ye K, et al. Reduced miR-203 predicts metastasis and poor survival in esophageal carcinoma. Aging (Albany NY). 2019;11(24):12114–12130. doi:10.18632/aging.102543.
- Qi Q, Ling Y, Zhu M, Zhang Y, Liu Y. Hypermethylation and low expression of miR-203 in patients with esophageal cancer in Chinese population. Int J Clin Exp Pathol. 2016;9(6):6245–6251.
- You A, Fu L, Li Y, Li X, You B. MicroRNA-203 restrains epithelial-mesenchymal transition, invasion and migration of papillary thyroid cancer by downregulating AKT3. Cell Cycle. 2020;19(10):1105–1121. doi:10.1080/15384101.2020.1746490.
- Zhao F, Wei C, Cui MY, et al. Prognostic value of microRNAs in pancreatic cancer: a meta-analysis. Aging (Albany NY). 2020;12(10):9380–9404. doi:10.18632/aging.103214.
- Ahir BK, Lakka SS. Elucidating the microRNA-203 specific biological processes in glioblastoma cells from comprehensive RNA-sequencing transcriptome profiling. Cell Signal. 2019;53:22–38. doi:10.1016/j.cellsig.2018.09.014.
- Bueno MJ, Pérez De Castro I, Gómez De Cedrón M, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13(6):496–506. doi:10.1016/j.ccr.2008.04.018.
- Jiang N, Jiang X, Chen Z, et al. MiR-203a-3p suppresses cell proliferation and metastasis through inhibiting LASP1 in nasopharyngeal carcinoma. J Exp Clin Canc Res. 2017;36(1):138.
- Conde-Perez A, Gros G, Longvert C, et al. A caveolin-dependent and PI3K/AKT-independent role of PTEN in β-catenin transcriptional activity. Nat Commun. 2015;6(1):8093. doi:10.1038/ncomms9093.
- Mao L, Zhang Y, Mo W, Yu Y, Lu H. BANF1 is downregulated by IRF1-regulated microRNA-203 in cervical cancer. PLoS One. 2015;10(2):e117035. doi:10.1371/journal.pone.0117035.
- Wang C, Wang X, Liang H, et al. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PLoS One. 2013;8(9):e73985. doi:10.1371/journal.pone.0073985.
- Feber A, Xi L, Luketich JD, et al. MicroRNA expression profiles of esophageal cancer. J Thorac Cardiovasc Surg. 2008;135(2):255–260; discussion 260. doi:10.1016/j.jtcvs.2007.08.055.
- Zhang LW, Wang B, Yang JX, Yang H. Circ-PRMT5 stimulates migration in esophageal cancer by binding miR-203. Eur Rev Med Pharmaco. 2020;24(19):9965–9972.
- Wang T, Wang J, Ren W, et al. CircRNA-0008717 promotes cell proliferation, migration, and invasion by regulating miR-203/Slug in esophageal cancer cells. Ann Transl Med. 2020;8(16):999–999. doi:10.21037/atm-20-5205.