Abstract
Objective
Gene mutations in tumor cells can lead to several unique metabolic phenotypes, which are crucial for the proliferation of cancer cells. EGFR mutation (EGFR-mt) is the main oncogenic driving mutation in lung adenocarcinoma (LUAD). HIF-1 α and PKM2 are two key metabolic regulatory proteins that can form a feedback loop and promote cancer growth by promoting glycolysis. Here, the linkage between EGFR mutational status and HIF-1α/PKM2 feedback loop in LUAD were evaluated.
Methods
Retrospective study were performed on LUAD patients (n = 89) undergoing first-time therapeutic surgical resection. EGFR mutation was analyzed by real-time PCR. Immunohistochemistry was used to measure the expressions of HIF-1α and PKM2.
Results
We found that the protein expressions of HIF-1α and PKM2 were significantly higher in LUAD than normal lung tissues. In adenocarcinomas, the two protein expressions were both correlated with worse pTNM stage. Moreover, the correlation between the proteins of HIF-1α/PKM2 feedback loop and the EGFR mutational status were also analyzed. We found that EGFR-mt tumors showed higher HIF-1α and PKM2 proteins compared to tumors with EGFR wild-type. Meanwhile, HIF-1α expression was significantly correlated with higher pTNM stage, and PKM2 showed a similar trend, only in EGFR-mutated tumors. The expression of HIF-1α was positively correlated with PKM2 in LUAD, furthermore, this correlation was mainly in patients with EGFR-mt.
Conclusion
Different expression and clinical features of HIF-1α/PKM2 feedback loop was existed between LUAD and normal lung tissues, especially in EGFR mutational tumors, supporting the relationship between EGFR mutation and the key related proteins of aerobic glycolysis (HIF-1α and PKM2) in lung adenocarcinomas.
Introduction
Lung cancer is the main cause of cancer death in the world.Citation1 Lung adenocarcinoma (LUAD) accounts for over 40% of lung cancer and is the most common histological subtype of lung cancers.Citation2 Despite recent progress in lung cancer research, the prognosis of LUAD patients has not significantly improved.Citation3 Thus, it is urgent to understand the potential molecular mechanisms underlying the pathogenesis of LUAD and discover new prognostic biomarkers and therapeutic targets for LUAD.
Glycolysis, also namely the “Warburg effect,” refers to the conversion of glucose into lactate even aerobic environments, which plays a key role in the growth and metastasis of cancer cells by producing large amounts of metabolic intermediates.Citation4 Pyruvate kinase M2 (PKM2) is a key enzyme in the glycolytic pathway, and its upregulation was found in various of cancers, including bladder cancer and hepatocellular carcinoma.Citation5,Citation6 By increasing the glucose consumption and lactate production,Citation7 PKM2 significantly promoting the development and progression of cancer. Guo et al have shown that increased expression of PKM2 contributes to therapeutic resistance of lung adenocarcinoma (LUAD) stem cells, indicating that PKM2 is a potential therapeutic target for LUAD.Citation8 Hypoxia-inducible factor-1α (HIF-1α) is an important transcriptional factor of glycolysis. HIF-1α can activate pkm2 gene transcription,Citation9 while PKM2 participates in a positive feedback loop that promotes HIF-1 transactivation, thereby enhancing the response of cells to hypoxia or oncogene activation.Citation10 Amounts of data indicate that HIF-1α and PKM2 promote tumor growth by facilitating the Warburg effect.Citation11,Citation12 However, there is still limited research on the relationship between PKM2/HIF-1α feedback loop and the progression of lung adenocarcinoma.
Many studies have found that tumor related gene mutations are closely related to the changes in metabolic activity, genes that was involved like isocitrate dehydrogenase (IDH), BRAF and EGFR.Citation13,Citation14 In the study of Babic et al, they found that the EGFR mutation enhanced glycolysis by promoting glycolytic gene expression in brain cancer.Citation15 And Nilsson et al found that in non-small cell lung cancer cells, the mutation status of EGFR is correlated with high HIF-1α level even under hypoxic conditions, implying that cells with EGFR mutation may have a significant regulatory effect on the expression of HIF-1α.Citation16 A recent study showed that the EGFR signaling activation caused nuclear translocation of PKM2, which in turn activates cyclin D1 and Myc.Citation17 While, the association between EGFR-mt and the protein expressions of PKM2/HIF-1α feedback loop in LUAD still need to be clarified.
In our present study, we found that the expression levels as well as the clinical and pathological characteristics of HIF-1α/PKM2 feedback loop protein were different in LUAD and normal lung tissues, and these differences were also existed between EGFR-mt and EGFR-wt LUAD groups. The above results supporting the relationship between oncogenic EGFR mutation and the key related proteins of aerobic glycolysis (HIF-1α and PKM2) in lung adenocarcinomas.
Material and Methods
Patients and Samples
A total of 89 patients who underwent surgical resection of the primary LUAD in the Second Hospital of Hebei Medical University, and had complete clinical data, were reviewed in this retrospective analysis. All patients provided available tissues, including primary tumors and matched normal lung tissues. All patients did not receive any chemotherapy or radiotherapy before the study. TNM system was used for the staging of cancers and the WHO system for the morphologic classification.
Detection of EGFR Gene Mutation
Genomic DNA was extracted from LUAD tissues sampled from surgically resected specimens. EGFR mutation status was measured by the Strategene Mx3000p Real-Time PCR system (Agilent Technologies, Santa Clara, CA, USA) as previously described.Citation18 Two professional analysts were separately responsible for the assessment of the results.
Immunohistochemistry
4 μm serial sections were cut from formalin-fixed, paraffin-embedded LUAD tissues. The sections were conventional dewaxing to water and performed antigen retrieval using EDTA, after incubated in 3% H2O2 for 10 min, sections were blocked with 1% bovine serum albumin (BSA) for 10 min, and incubated with HIF-1α (1:200) and PKM2 (1:400) at 4 °C overnight. The slides were incubated with anti-rabbit/mouse IgG antibody the next day, after incubating with reaction enhancer, the slides were colored with DAB and counterstained with hematoxylin. Negative control was performed by the absence of the primary antibody.
Immunohistochemical Staining Evaluation
We quantitatively scored the sections through the percentage of positive cells and the staining intensity as previously.Citation19,Citation20 The effect that brown-yellow particles were observed in the nucleus or the cytoplasm was considered as HIF-1α and PKM2 positive. Then percentage and intensity of positive cell were both recorded. The percentage scores of the stained area: 0: ≤5%; 1: 6–25%; 2: 26–50%; 3: 51–75%; 4: ≥76%. The staining intensity: 0: no staining; 1: light yellow; 2: brown yellow; 3: brown. Finally, multiply the two scores to get the final score: negative expression: score < 8; positive expression: score ≥ 8.
Statistical Analysis
Statistical analyses were done with SPSS16.0 Software (SPSS Inc., Chicago, IL, USA).
The Chi-squared test and Fisher exact test were used to evaluate the comparison of PKM2 and HIF-1α protein expression between LUAD and corresponding normal tissue, as well as between EGFR-mt and EGFR-wd groups. The correlation among PKM2 and HIF-1α was analyzed by Pearson’s correlation test. p < 0.05 was considered to indicate a statistically significant difference.
Results
EGFR Mutation Status
Histologically confirmed that all 89 (49 females, average age 61 years) included cases were lung adenocarcinoma (LUAD). EGFR mutations were found in 44 (49.4%) out of 89 patients included in this study (). Among patients with EGFR mutations, the proportion of females and never smoking was significantly higher (34 out of 44 patients, p < 0.001) (36 out of 44 patients, p < 0.001). No significant difference was found in TNM staging between patients with different EGFR mutational status (p = 0.964, p = 0.979) ().
Figure 1. EGFR mutational status of lung adenocarcinoma (LUAD). Abbreviation: EGFR-mt, EGFR mutation adenocarcinoma; EGFR-wd, EGFR wild type adenocarcinoma.
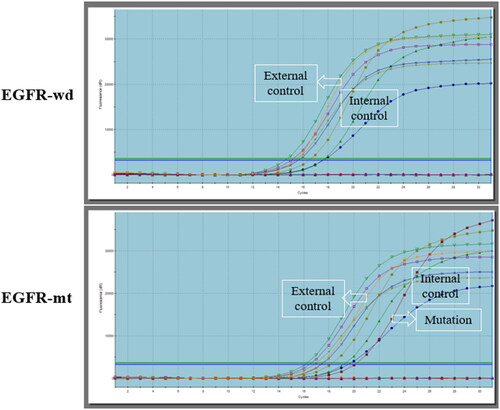
Table 1. The clinicopathological features of 89 LUAD patients.
The Protein Expressions of HIF-1α and PKM2 in Lung Adenocarcinoma Tissues
Immunohistochemical analysis of LUAD tumor tissues showed that HIF-1α and PKM2 were both mainly expressed in the cytoplasm or nucleus of tumor cells. Semiquantitative scoring was utilized to calculate the result. The scores of different groups were compared with Chi-squared test. As shown in and , compared to non-tumor tissues, HIF-1 α And PKM2 protein levels significantly increased in LUAD tissues (p = 0.024, p = 0.001). In addition, spearman correlation analysis showed that the expression of HIF-1α was positively correlated with the expression of PKM2 in LUAD tissues (p = 0.003, r = 0.307) ().
Figure 2. Different expressions of HIF-1α and PKM2 in normal lung tissue (NLT) and lung adenocarcinoma (LUAD).
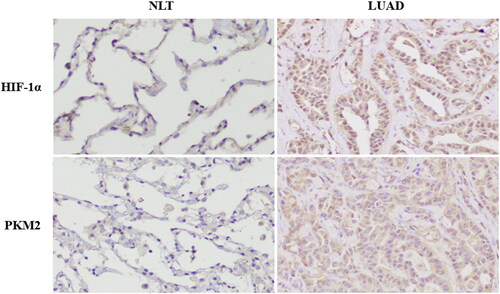
Table 2. Expressions of HIF-1α and PKM2 in normal lung tissue and lung adenocarcinoma.
Table 3. Relationship between the expression of HIF-1α and PKM2 in lung adenocarcinoma.
Relationship between Expressions of PKM2, HIF-1α and Clinical Features of Lung Adenocarcinoma
We explored the relationship between the two proteins of HIF-1α/PKM2 feedback loop and the clinical pathologic features of LUAD in , and we observed that HIF-1α and PKM2 expressions were both correlated with pTNM stage (p = 0.023, p = 0.023). But there was no significant of HIF-1α, PKM2 expression correlation with age, gender and smoking history. Although not statistically significant, HIF-1α and PKM2 had a trend to be highly expressed in lymph node metastases group (p = 0.736, p = 0.117) ().
Table 4. Relationships of HIF-1α/PKM2 feedback loop proteins with clinical pathological characteristics in adenocarcinomas of lung.
HIF-1α Correlated with PKM2 Proteins in EGFR-Mutated LUAD Tissue
EGFR-mutated tumors showed higher HIF-1α protein (p = 0.001) and higher PKM2 protein expression (p = 0.033), compared to tumors with EGFR wild-type ( and ). Spearman correlation test revealed that the expression of HIF-1α were positively correlated with PKM2 in EGFR-mt (r = 0.379, p =0.011) but not in EGFR-wd (r = 0.149, p =0.327) LUAD tissues (). The above data showed that HIF-1α correlated with PKM2 and increases PKM2 expression in EGFR-mt LUAD.
Figure 3. Different expressions of HIF-1α and PKM2 in EGFR wild type (EGFR-wd) and EGFR mutation (EGFR-mt) lung adenocarcinomas.
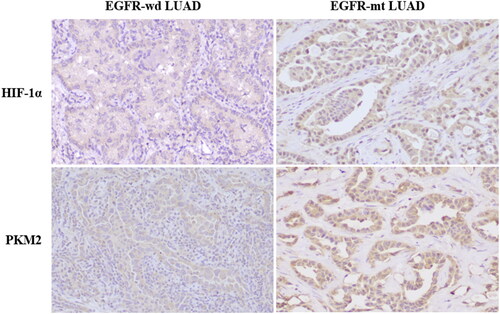
Table 5. HIF-1α and PKM2 protein expressions in EGFR wild-type versus EGFR-mutated lung adenocarcinoma.
Table 6. Relationship between the expression of HIF-1α and PKM2 protein in EGFR wild-type versus EGFR-mutated lung adenocarcinoma.
HIF-1α was Correlated with pTNM Stage Only in EGFR-mt LUAD Tissues
Subsequently, Chi-squared test was used to detect the relationship between PKM2, HIF-1α expressions and clinical characteristics in both EGFR-mt and EGFR-wd groups. We found that high expression of HIF-1α was obviously correlated with pTNM stage only in EGFR-mt group (p = 0.030) but not EGFR-wd group (p = 0.227), and PKM2 showed a similar trend though without statistical significance (p = 0.062) ().
Table 7. Relationships of HIF-1α/PKM2 feedback loop proteins with clinical pathological characteristics in EGFR wild-type versus EGFR-mutated lung adenocarcinoma.
Discussion
HIF-1α and PKM2 are two key metabolic regulatory proteins, they enhance each other’s expression by forming feedback loop, thus playing an important role in the occurrence and development of various human tumors.Citation11,Citation12 Nevertheless, the effect of this feedback loop in lung adenocarcinoma and their relationship with EGFR mutational status is still unclear. This study compared the expression and clinical pathological significance of PKM2, HIF-1α proteins in LUAD and normal lung tissue. We found that the two proteins were both highly expressed in LUAD tissues and related to higher pTNM stage of LUAD. Furthermore, the relationship between HIF-1α, PKM2 expressions and the EGFR mutational status were also analyzed, and our findings indicated higher expressions of HIF-1α and PKM2 in EGFR mutation tumors relative to EGFR wild-type tumors. Meanwhile, the two proteins indicated a higher relationship with higher pTNM stage only in EGFR mutation tumors. A positive association was found between HIF-1α and PKM2 in LUAD, and this correlation was mainly shown in patients with EGFR mutation. Our results firstly revealed that the expressions and clinical pathological significance of HIF-1α, PKM2 were different in LUAD and normal lung tissues, and this difference as more obvious in EGFR mutational tumors, supporting the relationship between EGFR mutation and HIF-1α, PKM2 in lung adenocarcinomas.
Cancer cells reprogram their glucose metabolism from oxidative phosphorylation to glycolysis to accelerate growth and proliferation. This phenomenon caused the “Warburg Effect” and was first described by Otto Warburg in the 1920sCitation21 and has been documented for over 90 years. Glycolysis was modulated by many enzymes of the glycolytic pathway. Pyruvate kinase M2 (PKM2), a glycolytic rate-limiting enzyme, catalyzes the final step of glycolysis, and was revealed to play a crucial role in tumorigenesis by promoting the metabolic reprogramming. It is reported that PKM2 level increases in cancers of hepatocellular, prostate and bladder, and has made an important contribution to the development and metastasis of tumors.Citation9,Citation22,Citation23 Consistent with previous reports, in this study, we found higher expression of PKM2 in LUAD tissue and PKM2 higher expression was correlated with higher pTNM stage in LUAD patients.Citation24 Though with no statistical significance, we found an increasing trend of PKM2 expression in tumors with lymph node metastasis, which may indicate poor clinical outcomes in LUAD patients.
It has been shown that the gene transcription of PKM2 is activated by hypoxia-inducible factor 1α (HIF-1α). While in turn, PKM2 is an essential co-activator that stimulates the transactivation of HIF-1α in tumor cells. Thus, PKM2 and HIF-1α constitute a positive feed-forward loop to reprogram glucose metabolism in cancer cells.Citation25 HIF-1α abnormal expression is increasingly known as a critical step in cancer progression. It has been reported that the deletion of HIF-1α markedly impair metastasis of breast cancer in a mouse model.Citation26 Jacoby et al found that inhibition of HIF-1α with PX-478 suppressed the progression and spread of small cell lung cancer and lung adenocarcinoma in mice.Citation27 And in our present study, we also found an increased expression of HIF-1α in LUAD tissues, the higher expression of HIF-1α was correlated with higher pTNM stage, suggesting a major role of HIF-1α in the pathogenesis of LUAD. Moreover, through Spearman correlation test of IHC results, we found the expression of PKM2 was significantly correlated with HIF-1α, indicating that PKM2 and HIF-1α form a feedback loop in LUAD.
The energy metabolism of cancer cells is mainly relay on aerobic glycolysis rather than mitochondrial oxidative phosphorylation. Many oncogenes, including EGFR, may facilitate the acquisition of the glycolytic phenotype.Citation14,Citation15 The association between glucose metabolism and EGFR mutation in lung cancer has been studied before, with some controversial results. Most of the studies reported that EGFR-mt was associated with low glucose metabolism,Citation28–30 while others considered that EGFR-mt was associated with increased glucose metabolism,Citation31,Citation32 and some studies found that there was no statistical difference of glucose metabolism between EGFR-mt and EGFR-wt groups.Citation33,Citation34 Therefore, more research still needed to elucidate the association. NSCLC may merge with various driving mutations other than EGFR that can affect glucose metabolism, and this may be the reason for the controversial results of glucose metabolism in EGFR-mt patients. In our present study, we found higher expressions of HIF-1α and PKM2 in EGFR mutation tumors relative to EGFR wild-type tumors. These results were supported by Nilsson et al, who recently demonstrated that the expression of EGFR mutations is correlated with higher HIF-1α expressions in NSCLC and NIH-3T3 cells, which means that cells carrying EGFR activated mutations may has a regulatory effect on HIF-1α expression.Citation16 While the relationship of PKM2 and EGFR signaling pathway had already been elucidated, Yang et al revealed that cytosolic PKM2 stabilizes mutant EGFR protein expression through regulating HSP90-EGFR association.Citation17 The positive correlation of HIF-1α and PKM2 expressions in EGFR mutation tumors emphasized the relationship between oncogenic mutated EGFR signaling and HIF-1α, PKM2 in lung adenocarcinomas.
Taken together, our study demonstrated that different expression and clinical features of the proteins of HIF-1α/PKM2 feedback loop was existed between lung adenocarcinoma and normal lung tissues, and this difference was also found in EGFR-mutation and EGFR-wild type tumors, indicating that the EGFR mutation was correlated with the key related proteins of aerobic glycolysis (HIF-1α and PKM2) in lung adenocarcinomas.
Disclosure Statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin North Am. 2019;103(3):1–7. doi:10.1016/j.mcna.2018.12.006.
- Zhang C, Zhang Z, Zhang G, et al. Clinical significance and inflammatory landscapes of a novel recurrence-associated immune signature in early-stage lung adenocarcinoma. Cancer Lett. 2020;479:31–41. doi:10.1016/j.canlet.2020.03.016.
- Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9(2):117. doi:10.1038/s41419-017-0063-y.
- Icard P, Shulman S, Farhat D, Steyaert JM, Alifano M, Lincet H. How the Warburg effect supports aggressiveness and drug resistance of cancer cells? Drug Resist Updat. 2018;38:1–11. doi:10.1016/j.drup.2018.03.001.
- Wang JZ, Zhu W, Han J, et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun (Lond). 2021;41(7):560–575. doi:10.1002/cac2.12158.
- Xu Q, Tu J, Dou C, et al. HSP90 promotes cell glycolysis, proliferation and inhibits apoptosis by regulating PKM2 abundance via Thr-328 phosphorylation in hepatocellular carcinoma. Mol Cancer. 2017;16(1):178. doi:10.1186/s12943-017-0748-y.
- Zahra K, Dey T, Ashish Mishra SP, Pandey U, Pyruvate kinase M2 and cancer: The role of PKM2 in promoting tumorigenesis. Front Oncol. 2020; 10:159. doi:10.3389/fonc.2020.00159.
- Guo C-Y, Yan C, Luo L, et al. Enhanced expression of PKM2 associates with the biological properties of cancer stem cells from A549 human lung cancer cells. Oncol Rep. 2017;37(4):2161–2166. doi:10.3892/or.2017.5438.
- Hasan D, Gamen E, Abu Tarboush N, Ismail Y, Pak O, Azab B. PKM2 and HIF-1α regulation in prostate cancer cell lines. PLoS One. 2018;13(9):e0203745. doi:10.1371/journal.pone.0203745.
- Azoitei N, Becher A, Steinestel K, et al. PKM2 promotes tumor angiogenesis by regulating HIF-1α through NF-κB activation. Mol Cancer. 2016;15(1):3. doi:10.1186/s12943-015-0490-2.
- Hua Q, Mi B, Xu F, et al. Hypoxia-induced lncRNA-AC020978 promotes proliferation and glycolytic metabolism of non-small cell lung cancer by regulating PKM2/HIF-1α axis. Theranostics. 2020;10(11):4762–4778. doi:10.7150/thno.43839.
- Chai XX, Le YF, Wang JC, et al. Carpesium abrotanoides (L.) Root as a potential source of natural anticancer compounds: targeting glucose metabolism and PKM2/HIF-1α axis of breast cancer cells. J Food Sci. 2019;84(12):3825–3832. doi:10.1111/1750-3841.14953.
- Makinoshima H, Takita M, Matsumoto S, et al. Epidermal growth factor receptor (EGFR) signaling regulates global metabolic pathways in EGFR-mutated lung adenocarcinoma. J Biol Chem. 2014;289(30):20813–20823. doi:10.1074/jbc.M114.575464.
- Levine AJ, Puzio-Kuter AM. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science. 2010;330(6009):1340–1344. doi:10.1126/science.1193494.
- Babic I, Anderson ES, Tanaka K, et al. EGFR mutation-induced alternative splicing of Max contributes to growth of glycolytic tumors in brain cancer. Cell Metab. 2013;17(6):1000–1008. doi:10.1016/j.cmet.2013.04.013.
- Nilsson MB, Robichaux J, Herynk MH, et al. Altered regulation of HIF-1alpha in naive- and drug-resistant EGFR-mutant NSCLC: implications for a vascular endothelial growth factor-dependent phenotype. J Thorac Oncol. 2021;16(3):439–451. doi:10.1016/j.jtho.2020.11.022.
- Yang YC, Cheng TY, Huang SM, et al. Cytosolic PKM2 stabilizes mutant EGFR protein expression through regulating HSP90-EGFR association. Oncogene. 2016;35(26):3387–3398. doi:10.1038/onc.2015.397.
- Jin Q, Huang F, Xu X, He H, Zhang Y. High expression of hypoxia inducible factor 1α related with acquired resistant to EGFR tyrosine kinase inhibitors in NSCLC. Sci Rep. 2021;11(1):1199. doi:10.1038/s41598-020-79801-1.
- Lin Y, Zhai H, Ouyang Y, et al. Knockdown of PKM2 enhances radiosensitivity of cervical cancer cells. Cancer Cell Int. 2019;19(1):129. doi:10.1186/s12935-019-0845-7.
- Liu J, Gao L, Zhan N, et al. Hypoxia induced ferritin light chain (FTL) promoted epithelia mesenchymal transition and chemoresistance of glioma. J Exp Clin Cancer Res. 2020;39(1):137. doi:10.1186/s13046-020-01641-8.
- Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270.
- Li TE, Wang S, Shen XT, et al. PKM2 drives hepatocellular carcinoma progression by inducing immunosuppressive microenvironment. Front Immunol. 2020;11:589997. doi:10.3389/fimmu.2020.589997.
- Huang C, Huang Z, Bai P, Luo G, Zhao X, Wang X. Expression of pyruvate kinase M2 in human bladder cancer and its correlation with clinical parameters and prognosis. Onco Targets Ther. 2018;11:2075–2082. doi:10.2147/OTT.S152999.
- Long L, Chen M, Yuan Y, et al. High expression of PKM2 synergizes with PD-L1 in tumor cells and immune cells to predict worse survival in human lung adenocarcinoma. J Cancer. 2020;11(15):4442–4452. doi:10.7150/jca.42610.
- Zhang X, Li Y, Ma Y, et al. Yes-associated protein (YAP) binds to HIF-1alpha and sustains HIF-1alpha protein stability to promote hepatocellular carcinoma cell glycolysis under hypoxic stress. J Exp Clin Cancer Res. 2018;37(1):216. doi:10.1186/s13046-018-0892-2.
- Esteva-Font C, Jin BJ, Verkman AS. Aquaporin-1 gene deletion reduces breast tumor growth and lung metastasis in tumor-producing MMTV-PyVT mice. FASEB J. 2014;28(3):1446–1453. doi:10.1096/fj.13-245621.
- Jacoby JJ, Erez B, Korshunova MV, et al. Treatment with HIF-1alpha antagonist PX-478 inhibits progression and spread of orthotopic human small cell lung cancer and lung adenocarcinoma in mice. J Thorac Oncol. 2010;5(7):940–949. doi:10.1097/JTO.0b013e3181dc211f.
- Cho A, Hur J, Moon YW, et al. Correlation between EGFR gene mutation, cytologic tumor markers, 18F-FDG uptake in non-small cell lung cancer. BMC Cancer. 2016;16(1):224. doi:10.1186/s12885-016-2251-z.
- Na II, Byun BH, Kim KM, et al. 18F-FDG uptake and EGFR mutations in patients with non-small cell lung cancer: a single-institution retrospective analysis. Lung Cancer. 2010;67(1):76–80. doi:10.1016/j.lungcan.2009.03.010.
- Park S, Ha S, Lee SH, et al. Intratumoral heterogeneity characterized by pretreatment PET in non-small cell lung cancer patients predicts progression-free survival on EGFR tyrosine kinase inhibitor. PLoS One. 2018;13(1):e0189766. doi:10.1371/journal.pone.0189766.
- Apostolova I, Ego K, Steffen IG, et al. The asphericity of the metabolic tumour volume in NSCLC: correlation with histopathology and molecular markers. Eur J Nucl Med Mol Imaging. 2016;43(13):2360–2373. doi:10.1007/s00259-016-3452-z.
- Huang CT, Yen RF, Cheng MF, et al. Correlation of F-18 fluorodeoxyglucose-positron emission tomography maximal standardized uptake value and EGFR mutations in advanced lung adenocarcinoma. Med Oncol. 2010;27(1):9–15. doi:10.1007/s12032-008-9160-1.
- Putora PM, Früh M, Müller J. FDG-PET SUV-max values do not correlate with epidermal growth factor receptor mutation status in lung adenocarcinoma. Respirology. 2013;18(4):734–735. doi:10.1111/resp.12083.
- Caicedo C, Garcia-Velloso MJ, Lozano MD, et al. Role of [18 F] FDG PET in prediction of KRAS and EGFR mutation status in patients with advanced non-small-cell lung cancer. Eur J Nucl Med Mol Imaging. 2014;41(11):2058–2065. doi:10.1007/s00259-014-2833-4.

