Abstract
Adipogenesis involves a highly orchestrated series of complex events in which microRNAs (miRNAs) may play an essential role. In this study, we found that the miR-185 expression increased gradually during 3T3-L1 cells differentiation. To explore the role of miR-185 in adipogenesis, miRNA agomirs and antagomirs were used to perform miR-185 overexpression and knockdown, respectively. Overexpression of miR-185 dramatically reduced the mRNA expression of the adipogenic markers, PPARγ, FABP4, FAS, and LPL, and the protein level of PPARγ and FAS. MiR-185 overexpression also led to a notable reduction in lipid accumulation. In contrast, miR-185 inhibition promoted differentiation of 3T3-L1 cells. By target gene prediction and luciferase reporter assay, we demonstrated that sterol regulatory element binding protein 1 (SREBP-1) may be the target of miR-185. These results indicate that miR-185 negatively regulates the differentiation of 3T3-L1 cells by targeting SREBP-1, further highlighting the importance of miRNAs in adipogenesis.
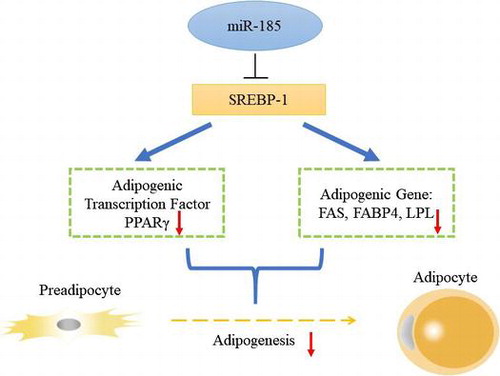
MiR-185 may down-regulate marker genes related to adipocyte differentiation and inhibit 3T3-L1 adipogenesis by targeting SREBP-1.
Key words:
Obesity is a prevalent health challenge in both developed and developing countries.Citation1) Obese individuals are more likely than their lean counterparts to develop a series of pathological disorders, including diabetes, hyperglycemia, hyperlipidemia, atherosclerosis, and chronic inflammation.Citation2−Citation4) The increase in adiposity in these individuals stems from an increase in both adipocyte number and size of individual fat cells. Mature adipocytes arise from preadipocytes and progenitor cells, which is also known as adipogenesis. 3T3-L1 adipocytes have been a popular model for the study of adipogenesis, which is regulated by an elaborate network of transcription factors that coordinate the expression of hundreds of proteins responsible for establishing the mature fat-cell phenotype.Citation5) Therefore, it is important to determine the molecular mechanisms governing adipogenesis.
MicroRNAs (miRNAs) are endogenous noncoding RNAs comprising 19–24 nucleotides that downregulate gene expression at the post-transcriptional level by partially base-pairing with the 3′ untranslated regions (UTRs) of target mRNAs, which leads to mRNA degradation or repression of translation.Citation6,7) It is clear that miRNAs are involved in critical biological processes, including proliferation, differentiation, development, apoptosis, and oncogenesis.Citation8−Citation10) Moreover, an increasing number of studies have indicated that miRNAs may play important roles in adipocyte differentiation by acting on transcription factors and regulating signaling pathways related to adipogenesis.Citation11,12) It has been demonstrated that miR-143 is a facilitator of adipocyte differentiation,Citation13,14) whereas several other studies have found that miR-27 can suppress the terminal differentiation of preadipocytes by targeting peroxisome proliferator-activated receptor γ (PPARγ) and prohibitin.Citation15,16) Other miRNAs, such as miR-21,Citation17) the miR-17-92 cluster,Citation18) miR-378,Citation19) and miR-335,Citation20) have also been discovered to play critical roles in adipogenesis.
MiR-185 is a miRNA shown to be involved in lipid metabolism and insulin sensitivity,Citation21−Citation24) and differential expression of miR-185 has been reported in 3T3-L1 adipocytes and human subcutaneous adipose tissue.Citation25,26) However, the detailed role of miR-185, especially in the development of adipogenesis, remains unknown. In this study, we used miR-185 agomirs to explore its role in adipogenesis and demonstrated that miR-185 overexpression attenuated lipid accumulation during 3T3-L1 preadipocyte differentiation. Knockdown of endogenous miR-185 by antagomirs promoted 3T3-L1 cell differentiation. By target gene prediction and luciferase reporter assay validation, we demonstrated that sterol regulatory element binding protein 1 (SREBP-1) is the target of miR-185. Together, our study suggests that miR-185 is a regulator of adipogenesis, functioning, at least partially, by targeting SREBP-1.
Materials and methods
Cell culture and adipogenic differentiation
3T3-L1 cells and HeLa cells were obtained from Shanghai life science research institute, Chinese academy of sciences. Cells were grown in high-glucose DMEM (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% antibiotic antimycotic solution (Sigma) before differentiation. Cells were maintained at 37 °C in a humidified incubator in 5% CO2, and culture medium was changed every 2 days. For induce differentiation, 3T3-L1 cells were cultured for 2 days in a differentiation medium (DM) containing 10% FBS, 0.5 mM 3-isobutyl-1-methylxanthine, 1 μM dexamethasone, and 1.7 μM insulin after confluence. The medium was then replaced with DMEM containing 10% FBS and 1.7 μM insulin for additional 2 days, after which, normal medium was replaced every other day until day 8.
Transfection of miR-185 agomirs and antagomirs
Agomirs and antagomirs of miR-185 (Ribobio, Guangzhou, China) are synthetic, chemically modified small RNAs, designed to promote or reduce the activity of endogenous miR-185. For transfection followed by adipogenic differentiation, the cell density must reach 70% to ensure that the cells can grow to confluency within two days after transfection. 3T3-L1 adipocytes were subjected to serum starvation for 4 h before transfection. MiR-185 agomirs, antagomirs or the corresponding negative controls (NCs) were transfected into cells using ribo Fect™ CP Transfection Kit (Ribobio, Guangzhou, China) at a concentration of 100 nM per well according to the manufacturer’s instructions.
Luciferase reporter assay
The fragment of the SREBP-1 mRNA 3′ UTR containing the miR-185 target sites was amplified by PCR using primers with the Sac I and Xba I sites. The primers used are listed in Table . The fragment was cloned into the pmirGLO Vector (Promega, Madison, WI, USA) at the multiple cloning sites (MCS), located downstream of the firefly luciferase (luc2) gene. MiR-185 or NC was co-transfected into Hela cells with pmirGLO-SREBP-1-3′ UTR or pmirGLO vectors using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), and the medium was replaced 6 h later. 48 h after transfection, firefly luciferase (luc2) activity was measured and normalized to the Renilla luciferase (hRluc-neo) activity (luc2/hRluc-neo) according to the Dual-Luciferase® Reporter Assay System (Promega).
Table 1. The primer sequences used for RT-PCR and gene cloning.
Oil Red O staining and quantification
For Oil Red O staining, cells were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 30 min. The fixed cells were washed with PBS three times and incubated with 1% filtered Oil Red O solution for 30 min. The cells were washed three times with PBS and then observed under a phase contrast microscope to check for positive cells appearing red. Oil Red O was then also eluted from the stained cells with 100% isopropanol (v/v) and quantified by measuring the optical absorbance at 510 nm.
RNA extraction and real-time quantitative PCR
Total cellular RNA was extracted using TRIzol reagent (TaKaRa, Japan) according to the manufacturer’s instructions. The total RNA concentration was estimated by a spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Approximately 500 ng of total RNA were reverse transcribed to synthesize cDNA using the PrimeScript RT reagent kit (TaKaRa) with random hexomers and oligo dT primers. For miR-185 reverse transcription (RT), specific RT primers and U6 RT primers were synthesized by Ribobio (Guangzhou, China). Real-time quantitative PCR reactions were performed in triplicate using SYBR Premix Ex Taq II (TaKaRa) with a Bio-Rad CFX96 Real-Time PCR Detection system (Hercules, CA, USA). The 2-∆∆Ct method was used to analyze the relative expression of each gene. The internal reference used for mRNA expression was β-actin, and U6 was used to normalize the miRNA expression levels (primers in Table ).
Western blot detection
3T3-L1 adipocytes that transfected with miR-26b agomirs, antagomirs and the corresponding NC were lysed using RIPA buffer (Beyotime, Shanghai, China). The proteins content of different treatment groups were quantified by the BCA method.Citation27) The lysates were boiled in SDS loading buffer (Beyotime) after centrifugation. 40 μg proteins were resolved by 12% SDS-polyacrylamide gels, and then the proteins were transferred onto PVDF membranes (Millipore, Bedford, MA, USA). After blocking in 5% skim milk, the membranes were incubated at 4 °C overnight with primary antibodies. The antidody of Anti- PPARγ (Abcam, Cambridge, UK), Anti-FAS, Anti-β-actin, and Anti-SREBP-1 (Sangon, Shanghai, China) were diluted 1:1000 in bovine serum albumin (Sangon).Then, the PVDF membranes were washed with Tris-buffered saline with Tween 20 (TBST) and hybridized with a secondary antibody at room temperature for 1 h. After three washes with TBST, protein bands were visualized with chemiluminescence reagents (Millipore). The expressed protein bands were normalized and analyzed with Quantity One 4.6.3 software (Bio-Rad Laboratories).
Statistical analysis
All results are presented as mean ± standard error of the mean (SEM) based on at least three separate experiments. GraphPad Prism 6 was used to graph and determine significance. Student’s t- test (parametric test, unpaired test) was used to determine significance. One-way ANOVA followed by Duncan’s test and Bonferroni correction were performed for multiple comparisons. p < 0.05 was considered statistically significant.
Results
MiR-185 is significantly up-regulated during 3T3-L1 cell differentiation
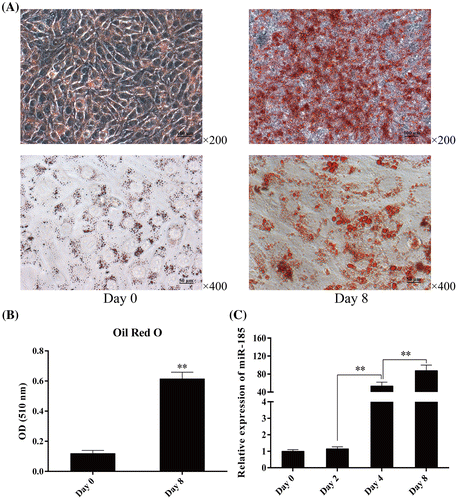
Oil Red O staining and quantitative analysis showed that 3T3-L1 cells differentiated into mature adipocytes after DM stimulation, an adipogenic hormonal cocktail comprising of IBMX, the glucocorticoid, DEX, and insulin (Fig. A and B). To determine the changes in miR-185 expression during 3T3-L1 cell differentiation, we extracted total RNA from adipocytes on day 0, 2, 4, and 8 after differentiation. MiR-185 levels were analyzed using stem-loop qPCR, and U6 was used as a common reference gene for miRNA expression. As shown in Fig. C, miR-185 was expressed at low levels on days 0 and 2 after DM induction. During differentiation, miR-185 expression gradually increased on day 4, and reached the maximum level on day 8. These results indicated that miR-185 may have an important role during 3T3-L1 cell differentiation.
Fig. 1. Expression pattern of miR-185 during 3T3-L1 cell differentiation.
Overexpression of miR-185 using agomirs attenuates lipid accumulation and adipogenic marker genes expression in 3T3-L1 cells
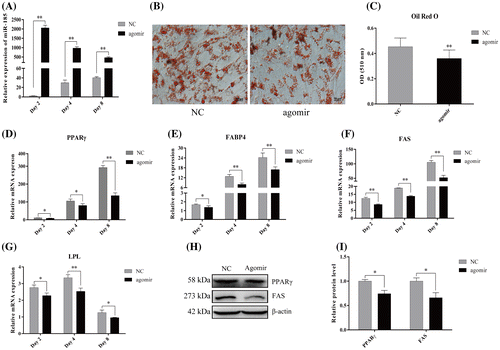
Next, we investigated the role of miR-185 in 3T3-L1 cell differentiation through gain-of-function assays. To this end, we transfected 3T3-L1 cells with synthetic miR-185 agomirs or NC at a cell density of 80–90% and induced differentiation with DM after contact inhibition. MiRNA agomirs are chemically modified miRNA mimics that can improve the stability and transfection. Stem-loop qPCR showed that miR-185 was significantly overexpressed by about 2000-fold following agomir transfection on day 2 after DM induction (96 h post-transfection), and the enhanced miR-185 level was maintained for at least 8 days after adipogenic induction (240 h post-transfection; Fig. A). As shown in Fig. B and C, we found that miR-185 overexpression significantly attenuated fat accumulation by using Oil Red O staining and an extraction assay, suggesting miR-185 has an inhibitory role in adipogenesis. As shown in Fig. D–G, the mRNA levels of the key adipogenesis markers, PPARγ, fatty acid binding protein 4 (FABP4), fatty acid synthase (FAS) and lipoprotein lipase (LPL) (detected on day 2, 4 and 8) were significantly downregulated in cells transfected with miR-185 agomirs compared with NC cells. And this expression alteration was also apparent at the protein level by western blot analysis (Fig. H and I). These results suggest that miR-185 has a negative regulatory role in 3T3-L1 adipocyte differentiation.
Fig. 2. MiR-185 overexpression inhibits 3T3-L1 cell differentiation.
MiR-185 antagomir promotes 3T3-L1 cell differentiation
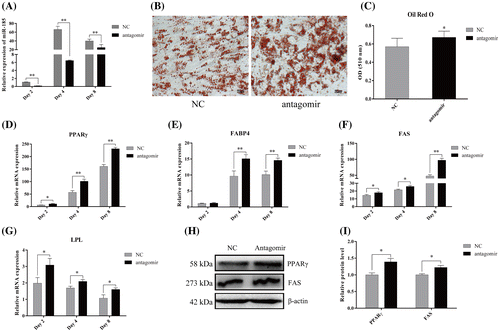
To further determine the role of miR-185 in 3T3-L1 cells during adipocyte differentiation, the anti-sense oligonucleotides (antagomir, chemically modified small RNA) targeting miR-185 or the NC were transfected into 3T3-L1 cells. As shown in Fig. A, miR-185 was effectively inhibited on day 2, 4 and 8 after DM induction. As expected, Oil red O staining on day 8 of differentiation showed an increased neutral lipid accumulation in the antagomir-transfected cells compared with the NC-transfected cells (Fig. B), which was confirmed by quantitative analysis of the intracellular accumulation of neutral lipids (Fig. C). As shown in Fig. D, F and G, the mRNA levels of PPARγ, FAS and LPL were markedly increased on day 2, 4, and 8 after induction of differentiation in cells transfected with miR-185 antagomirs compared with cells transfected with NC. Although there was no significant change in FABP4 mRNA on day 2 compared with NC cells, a significant increase was detected on day 4, 8 after differentiation (Fig. E). In addition, as shown in Fig. H and I, PPARγ and FAS protein levels also showed a significant increase upon miR-185 downregulation on day 8. These results also demonstrated that miR-185 can inhibit the adipogenic differentiation of 3T3-L1 cells.
Fig. 3. MiR-185 antagomism promotes 3T3-L1 cells differentiation.
MiR-185 directly targets the 3′ UTR of SREBP-1 to regulate 3T3-L1 cell differentiation
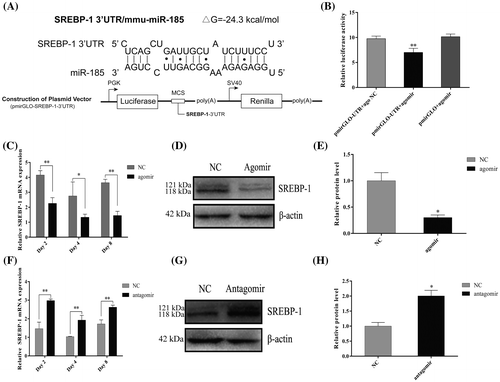
To determine the mechanisms by which miR-185 suppresses 3T3-L1 cell differentiation, bioinformatics analysis was performed to predict direct targets of miR-185. As shown in Fig. A, TargetScan 6.2 (https://www.targetscan.org/) and RNAhybrid software revealed that SREBP-1, a key transcription factor involved in adipocyte differentiation and fatty acid synthesis, contains the target sites for miR-185 in its 3′ UTR. To verify existence of the direct binding between miR-185 and the 3′ UTR of SREBP-1, the fragment of the SREBP-1 mRNA 3′ UTR containing the miR-185 target site was cloned and inserted into the 3′ UTR of the firefly luciferase gene in the pmirGLO reporter plasmid (Fig. A). The luciferase reporter assay showed that co-transfection with miR-185 agomirs significantly reduced the luciferase activity compared with the NC group (Fig. B). We then examined the effect of miR-185 on the endogenous expression of SREBP-1 (including SREBP-1a and SREBP-1c) in 3T3-L1 cells. As shown in Fig. C–E, MiR-185 overexpression caused an obvious decrease in the mRNA and protein level of SREBP-1. Moreover, when the endogenous miR-185 was knocked down with antagomirs, the SREBP-1 mRNA and protein levels were increased compared with the NC group (Fig. G–H). Taken together, these results suggest that miR-185 functions as a negative regulator, which binds to the 3′ UTR of SREBP-1, thereby regulating 3T3-L1 cell differentiation.
Fig. 4. MiR-185 directly targets the 3′ UTR of SREBP-1 to regulate 3T3-L1 cell differentiation.
Discussion
In this study, we found that miR-185 was gradually upregulated during the whole process of 3T3-L1 cell adipogenesis. But in subsequent studies, our results indicated that miR-185 might act as an adipogenesis inhibitory factor. This might be because of compensatory effect and the process of adipocyte differentiation will keep going until all 3T3-L1 cells turn into mature adipocytes once induced differentiation and this irreversible process might in turn induce the expression of miR-185. Overexpression or knockdown of miR-185 caused a significant change the mRNA of the adipogenic marker genes, such as PPARγ, FABP4, FAS, and LPL, as well as the protein expressions of PPARγ and FAS in 3T3-L1 cells. Our results indicate that miR-185 may play a crucial role in 3T3-L1 cell adipogenesis. Recently, miR-185 has been reported to be involved in several cellular processes, such as carcinogenesis, differentiation and apotosis.Citation28−Citation30) For instance, miR-185 suppresses gastric carcinogenesis through regulation the epigenetic effectors DNMT1 and EZH2 and cell cycle.Citation28) MiR-185 overexpression in HEK-293 cells impedes anchorage-independent growth and cell migration by targeting Six1 oncogene.Citation31)MiR-185 is also involved in T cell development through its targeting of several mRNAs.Citation29) Most importantly, Kajimoto et al. have found that miR-185 was significantly upregulated during 3T3-L1 cell differentiationCitation26) and miR-185 was down-regulated in subcutaneous fat samples from obese without Type-2 Diabetes Mellitus women when compared to non-obese subjects.Citation25) It has also been found that miR-185 regulates expression of lipid metabolism genes and improves insulin sensitivity in mice with non-alcoholic fatty liver disease.Citation24) All these findings imply that miR-185 may serve as an important regulator during 3T3-L1 cell differentiation.
Furthermore, we demonstrated that miR-185 suppressed the expression of SREBP-1 through binding to its 3′ UTR using a dual luciferase reporter assay. The results showed that SREBP-1 is a direct target of miR-185 in 3T3-L1 cells. Because of the inhibition of SREBP-1 expression, miR-185 suppressed 3T3-L1 differentiation and exerted a negative role in adipogenesis. Adipogenesis is a complex and highly orchestrated program of gene expression regulated by transcription factors like PPARγ, C/EBPα and SREBP-1, whose function is relatively well established. SREBP-1 is a well-characterized transcription factor, required for adipocyte differentiation and fatty acid synthesis.Citation32)It has been widely recognized that SREBP-1 promotes adipocyte differentiation by directly activating PPARγ transcriptionCitation33) as well as producing an endogenous ligand that activates PPARγ.Citation34) In addition, SREBP-1 may bind both sterol regulatory elements and E-boxes to activate the transcription of lipogenic genes, such as FAS, acetyl CoA carboxylase (ACC) and stearoyl-CoA desaturase 1 (Scd1).Citation35)
Previous study has reported that miR-185 negatively modulated SREBP-1 expression and downregulated its target genes, FAS and 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), thereby suppressing tumorigenicity by blockade of lipogenesis and cholesterogenesis in prostate cancer cells.Citation21) In this study, we mainly focused on the mechanism of miR-185 in adipogenic differentiation of 3T3-L1 cells. And the previous study and our results collectively demonstrated that miR-185 might mediate specific key adipogenic genes such as PPARγ and FAS by negatively regulating the SREBP metabolic pathway, and finally inhibiting 3T3-L1 cell differentiation.
Conclusions
Our study demonstrated the fundamental role and mechanism of miR-185 in the suppression of adipocyte differentiation, which exerts its effects at least in part by targeting SREBP-1. Therefore, miR-185 and its target genes may play a role in the pathological progression of obesity, which may provide a novel research direction for biological therapy of obesity-related diseases.
Author contributions
Chunyou Ning, Guilin Li, Haifeng Liu conceived and designed the expriments. Lu You and Yao Ma performed the experiments. Jideng Ma and Long Jin analyzed the data. Xuewei Li, Mingzhou Li and Haifeng Liu contributed reagents/materials/analysis tools. Guilin Li and Chunyou Ning wrote the paper.
Disclosure statement
No potential conflict of interest was reported by the authors.
Funding
This work was supported by grants from the National High Technology Research and Development Program of China (863 Program) [grant number 2013AA102502]; the National Natural Science Foundation of China [grant number 31522055]; National Program for Support of Top-notch Young Professionals, the Young Scholars of the Yangtze River, and the Program for Innovative Research Team of Sichuan Province [grant number 2015TD0012].
Notes
Abbreviations: miRNAs, microRNAs; UTRs, untranslated regions; SREBP-1, sterol regulatory element binding protein 1; Scd1, stearoyl-CoA desaturase 1; HMGCR, hydroxy-3-methylglutaryl-CoA reductase; PPARγ, peroxisome proliferator-activated receptor gamma; FABP4, fatty acid binding protein 4; FAS, fatty acid synthase; LPL, lipoprotein lipase.
References
- Haslam DW, James WP. Obesity. Lancet. 2005;366(9492):1197–1209.10.1016/S0140-6736(05)67483-1
- Despres JP. Is visceral obesity the cause of the metabolic syndrome? Ann Med. 2006;38(1):52–63.10.1080/07853890500383895
- Trayhurn P. Endocrine and signalling role of adipose tissue: new perspectives on fat. Acta Physiol Scand. 2005;184(4):285–293.10.1111/aps.2005.184.issue-4
- Stefan N, Schick F, Häring HU. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. N Engl J Med. 2014;371(12):1131–1141.
- Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16(4):145–171.10.1146/annurev.cellbio.16.1.145
- Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355.10.1038/nature02871
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.10.1016/S0092-8674(04)00045-5
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006;11(4):441–450.10.1016/j.devcel.2006.09.009
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531.10.1038/nrg1379
- Rottiers V, Näär AM. MicroRNAs in metabolism and metabolic disorders. Nat Rev Mol Cell Biol. 2012;13(4):239–250.10.1038/nrm3313
- Arner P, Kulyté A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat Rev Endocrinol. 2015;11(5):1715–1720.
- Chen Y, Pan R, Pfeifer A. Regulation of brown and beige fat by microRNAs. Pharmacol Ther. 2016;170:1–7.
- Esau C, Kang X, Peralta E, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279(50):52361–52365.10.1074/jbc.C400438200
- Yi C, Xie WD, Li F, et al. MiR-143 enhances adipogenic differentiation of 3T3-L1 cells through targeting the coding region of mouse pleiotrophin. FEBS Lett. 2011;585(20):3303–3309.10.1016/j.febslet.2011.09.015
- Lin Q, Gao Z, Alarcon RM, et al. A role of miR-27 in the regulation of adipogenesis. FEBS J. 2009;276(8):2348–2358.10.1111/ejb.2009.276.issue-8
- Kang T, Lu W, Xu W, et al. MicroRNA-27 (miR-27) targets prohibitin and impairs adipocyte differentiation and mitochondrial function in human adipose-derived stem cells. J Biol Chem. 2013;288(48):34394–34402.10.1074/jbc.M113.514372
- Kim YJ, Hwang SJ, Yong CB, et al. MiR-21 regulates adipogenic differentiation through the modulation of TGF-β signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27(12):3093–3102.
- Wang Q, Li YC, Wang J, et al. miR-17-92 cluster accelerates adipocyte differentiation by negatively regulating tumor-suppressor Rb2/p130. Proc Natl Acad Sci. 2008;105(8):2889–2894.10.1073/pnas.0800178105
- Gerin I, Bommer GT, McCoin CS, et al. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299(2):E198–E206.
- Nakanishi N, Nakagawa Y, et al. The up-regulation of microRNA-335 is associated with lipid metabolism in liver and white adipose tissue of genetically obese mice. Biochem Biophys Res Commun. 2009;385(4):492–496.10.1016/j.bbrc.2009.05.058
- Li B, Sun H. MiR-26a promotes neurite outgrowth by repressing PTEN expression. Mol Med Rep. 2013;8(2):676–680.
- Yang M, Liu W, Pellicane C, et al. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J Lipid Res. 2014;55(2):226–238.10.1194/jlr.M041335
- Jiang H, Zhang J, Du Y, et al. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis. 2015;243(2):523–532.10.1016/j.atherosclerosis.2015.10.026
- Wang XC, Zhan XR, Li XY, et al. MicroRNA-185 regulates expression of lipid metabolism genes and improves insulin sensitivity in mice with non-alcoholic fatty liver disease. World J Gastroenterol. 2014;20(47):17914–17923.
- Ortega FJ, Moreno-Navarrete JM, Pardo G, et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE. 2010;5(2):e9022.10.1371/journal.pone.0009022
- Kajimoto K, Naraba H, Iwai N. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA. 2006;12(9):1626–1632.10.1261/rna.7228806
- Osnes T, Sandstad O, Skar V, et al. Total protein in common duct bile measured by acetonitrile precipitation and a micro bicinchoninic acid (BCA) method. Scand J Clin Lab Invest. 1993;53(7):757–763.10.3109/00365519309092582
- Yoon JH, Choi YJ, Choi WS, et al. GKN1-miR-185-DNMT1 axis suppresses gastric carcinogenesis through regulation of epigenetic alteration and cell cycle. Clin Cancer Res. 2013;19(17):4599–4610.10.1158/1078-0432.CCR-12-3675
- Belkaya S, Murray SE, Eitson JL, et al. transgenic expression of microRNA-185 causes a developmental arrest of T cells by targeting multiple genes including Mzb1. J Biol Chem. 2013;288(42):30752–30762.10.1074/jbc.M113.503532
- Wang J, He J, Su F, et al. Repression of ATR pathway by miR-185 enhances radiation-induced apoptosis and proliferation inhibition. Cell Death Dis. 2013;4(6):e699.10.1038/cddis.2013.227
- Imam JS, Buddavarapu K, Lee-Chang JS, et al. MicroRNA-185 suppresses tumor growth and progression by targeting the Six1 oncogene in human cancers. Oncogene. 2010;29(35):4971–4979.10.1038/onc.2010.233
- Kim JB, Spiegelman BM. ADD1/SREBP1 promotes adipocyte differentiation and gene expression linked to fatty acid metabolism. Genes Dev. 1996;10(9):1096–1107.10.1101/gad.10.9.1096
- Fajas L, Schoonjans K, Gelman L, et al. Regulation of peroxisome proliferator-activated receptor γ expression by adipocyte differentiation and determination factor 1/sterol regulatory element binding protein 1: implications for adipocyte differentiation and metabolism. Mol Cell Biol. 1999;19(8):5495–5503.10.1128/MCB.19.8.5495
- Kim JB, Wright HM, Wright M, et al. ADD1/SREBP1 activates PPARγ through the production of endogenous ligand. Proc Natl Acad Sci. 1998;95(8):4333–4337.10.1073/pnas.95.8.4333
- Eberlé D, Hegarty B, Bossard P, et al. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86(11):839–848.10.1016/j.biochi.2004.09.018

