?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
The COVID-19 pandemic has generated massive data in the healthcare sector in recent years, encouraging researchers and scientists to uncover the underlying facts. Mining interesting patterns in the large COVID-19 corpora is very important and useful for the decision makers. This paper presents a novel approach for uncovering interesting insights in large datasets using ontologies and BERT models. The research proposes a framework for extracting semantically rich facts from data by incorporating domain knowledge into the data mining process through the use of ontologies. An improved Apriori algorithm is employed for mining semantic association rules, while the interestingness of the rules is evaluated using BERT models for semantic richness. The results of the proposed framework are compared with state-of-the-art methods and evaluated using a combination of domain expert evaluation and statistical significance testing. The study offers a promising solution for finding meaningful relationships and facts in large datasets, particularly in the healthcare sector.
1. Introduction
The advancement in technology and increase in data generation has made it imperative for organisations to mine and analyse vast amounts of data to gain valuable insights and hidden relationships. This has become especially crucial in the current scenario where the COVID-19 pandemic has generated massive amounts of data in the healthcare sector, in the work by Abhilash and Mahesh (Citation2021). The sheer volume of data has led researchers and scientists to seek effective methods to uncover the underlying facts and relationships. The importance of uncovering valuable insights and hidden relationships in large datasets cannot be overstated, especially in the healthcare sector where crucial information is required to fight the pandemic. This has prompted the need for innovative approaches and frameworks to analyse and mine large datasets, thereby providing valuable insights and hidden relationships that can assist in making informed decisions (Bringmann et al., Citation2011).
The Knowledge Bases (KBs) created with the domain ontology plays a crucial role in discovering the interesting patterns, which can be mined from logical rules using Association Rule Mining (ARM) with anthologies and Inductive Logic Programming (ILP). Suppose – “If two people test positive for the virus and their cases are classified as community transmission, Then – it is likely that they reside in the same geographical area”. Large KBs have been created due to recent improvements in information extraction. “These knowledge bases typically contain information such as “Beijing is the capital of China”, “Bill Gates was born in Seattle”, and “All doctors are individuals”. YAGO (Suchanek et al., Citation2007), DBpedia (Bizer et al., Citation2009).
The ARM techniques generate rules in the following way: Given a set of items I = , a group of items S =
is created, where S is a subset of I. The association rule A
B is defined over the group G. The interestingness of the rule is measured using metrics such as support, confidence, and Lift. By setting different thresholds for these metrics, the most meaningful rules can be identified from the dataset (Agrawal & Srikant, Citation1994).
To address the problem of uncovering hidden relationships and valuable insights in large datasets, a novel framework is proposed that leverages Resource Description Framework (RDF) data and a semantic approach to extract meaningful facts from COVID-19 data Abhilash and Mahesh (Citation2022b). The framework integrates ontologies to incorporate domain knowledge into data mining, utilising the improved Apriori algorithm for mining semantic association rules. The interestingness of the rules is further evaluated using BERT models for semantic richness. The proposed framework in this research aims to find valuable insights and hidden relationships from the large datasets, particularly in the healthcare sector during the COVID-19 pandemic, which provides a new avenue for data analysis. The approach uses RDF data, ontologies, an improved Apriori algorithm, and BERT models to extract meaningful facts from the data (Gahar et al., Citation2018).
To evaluate the effectiveness of the proposed framework, a combination of domain expert evaluation and statistical significance testing using a Chi-Square test has been employed. The results obtained from the evaluation will provide an insight into the accuracy and reliability of the framework in uncovering interesting facts from large datasets.
The publication “Attention is all you need” by Vaswani et al. (Citation2017) presented the Transformers architecture (2017). The architecture of transformers is encoder-decoder. The Google AI team developed Bidirectional Encoder Representations from Transformers (BERT), a transformer-based pre-trained model (Devlin et al., Citation2018). In this work, the semantic scores are employed as measures of importance, and their distributions, considering the distance measure, are calculated using the BERT models.
More precisely, our contributions are as follows:
An effective data preprocessing technique that introduces semantics at the level of data curation.
An effective Semantic Interestingness Framework using BERT (SIF-B) that incorporates ontology-based methods with ARM techniques to extract meaningful and semantically rich rules from large datasets, particularly in the healthcare sector during the COVID-19 pandemic.
Adoption of healthcare BERT models to introduce semantic interestingness measure to strengthen the framework and makes it novel.
The comparison of the proposed framework with the state-of-the-art results, along with the evaluation through a combination of domain expert assessment and statistical significance testing, demonstrate its effectiveness in uncovering hidden relationships and valuable insights in data.
The structure of this paper is as follows: Section 2 provides a summary of existing methods and techniques. Section 3 outlines the data, preliminary steps, and data pre-processing techniques. Section 4 presents the Semantic Interestingness Framework using BERT (SIF-B). Section 5 presents the results, and compares the semantic-rich rules with the state-of-the-art findings and implementation of BERT models for semantic interestingness. Section 6 includes a significance test and evaluation by domain experts. Finally, Section 7 concludes the paper and suggests future research directions.
2. Related work
Incorporating ontology knowledge into the discovery of interesting patterns in RDF (Resource Description Framework) data enhances the results compared to traditional instance-level data mining approaches by providing a deeper understanding of the relationships and concepts in the data (Abhilash & Mahesh, Citation2022a). The ontology represents the schema-level information and provides a semantic context for the data, which helps in uncovering hidden relationships between entities and making the discovered patterns more meaningful and relevant (Ciotti & Tomasi, Citation2016). The use of ontology information allows for the discovery of interesting patterns that take into account not just the instance-level data, but also the relationships between the concepts and classes in the data. This results in more accurate and relevant insights compared to instance-level data mining approaches, where only the individual instances are analysed without considering the underlying schema information (Su et al., Citation2023; Wikipedia contributors, Citation2021).
In recent years, the field of data mining has seen significant advances in the discovery of interesting patterns in large and complex datasets. One of the most promising developments has been the incorporation of ontology knowledge into these techniques, which allows for a deeper understanding of the semantic relationships between data elements. This has the potential to greatly enhance the results of data mining compared to traditional instance-level approaches, which are limited to only considering the relationships between individual data instances.
One of the key challenges in incorporating ontology knowledge into data mining is the accurate measurement of the semantic significance of the rules generated. This requires not only understanding the relationships between data elements, but also assessing the significance of these relationships in the context of the data.
To address this challenge, researchers have turned to the use of BERT models. These models, based on the transformer architecture, are capable of capturing fine-grained semantic relationships between words and phrases in natural language (Abas et al., Citation2020). By incorporating these models into the rule generation process, researchers can accurately measure the semantic significance of the rules generated from both ontology and instance-level information.
The use of BERT models in this context has been shown to produce promising results. In evaluations, the rules generated using BERT models have been found to be highly correlated with the assessments of domain experts, suggesting that these models are effective at measuring the semantic significance of the rules generated.
Prior research on COVID-19 has concentrated on forecasting case numbers (Arora et al., Citation2020; Qin et al., Citation2020; Tomar & Gupta, Citation2020) and categorising COVID-19 patients from real-world x-ray data sets using sophisticated deep neural network techniques (Apostolopoulos & Mpesiana, Citation2020; Ozturk et al., Citation2020). These techniques, however, focus on examining COVID-19 symptom patterns.
In Bellandi et al. (Citation2007), Bellandi et al. demonstrated how ontologies could improve the rules obtained by ARM systems. The post-processing of ARM results using an ontology for consistency testing is presented by Marinica and Guillet (Citation2010). Filtering the identified rules is proposed by Mangla and Akhare (Citation2015).
Rule-based expert system for self-pre-diagnosis of COVID-19. It analyses symptom data to predict the risk of COVID-19 and provides suggestions for precautionary and supportive actions. The system was tested and found to be effective and efficient in diagnosing and monitoring positive cases during the COVID-19 pandemic (Çelik Ertuğrul & Ulusoy, Citation2022).
Shan et al. (Citation2021) used the transformed-based models for fake news detection. It's observed that the model specific to the healthcare domain yields better results compared to generalised ones.
In Alzubi et al. (Citation2021) COBERT-question answering system design was created to address COVID-19 difficulties and quickly assist researchers and clinical professionals in obtaining legitimate scientific information. For COVID-19-related question answering, cosine similarity measures are applied on the word embedding for categorising the top-K documents (Choi et al., Citation2018; Guo et al., Citation2020; Shen et al., Citation2020).
2.1. Outcome of literature review
The literature review revealed that an ontology-based approach is widely used for generating explicit and implicit facts. However, to the best of our knowledge, the use of ontology-based approaches for rule generation has received relatively little attention. Although previous studies have proposed mining semantic association rules using ARM, it is uncertain whether the remaining rules are semantically meaningful after threshold-based pruning.
2.1.1. Research gaps
With the detailed literature review, the following gaps are identified:
Limited use of ontology-based methods in combination with ARM techniques to extract meaningful and semantically rich rules from large datasets in the healthcare sector.
Limited use of healthcare BERT models to evaluate the interestingness of rules in data mining.
Limited use of a framework that combines ontology-based methods, ARM techniques, and healthcare BERT models to extract meaningful and semantically rich rules from large datasets.
3. Preliminaries, data, and pre-processing technique
This section discusses the dataset with preliminaries and an effective data processing technique designed for this study. Besides the general data analysis and knowledge engineering methods, the ontology-based approach stands aside and has unique importance.
3.1. Preliminaries
Ontology and ARM methods closely work towards the data interestingness (Abhilash & Mahesh, Citation2021). In data mining literature, association rule mining is widely used for rule generation based on frequent patterns.
Definition 3.1
Association Rule: Technique used to mine the frequent patterns in Data. The discovered patterns define the relationship between them.
we call X Y as association rule. To have the strong association rule, we need to compute the support and confidence as indicated in Equations (Equation1
(1)
(1) ) and (Equation2
(2)
(2) ). Rules are defined considering our domain information. AVS refers to the attribute value set.
(1)
(1)
(2)
(2)
Definition 3.2
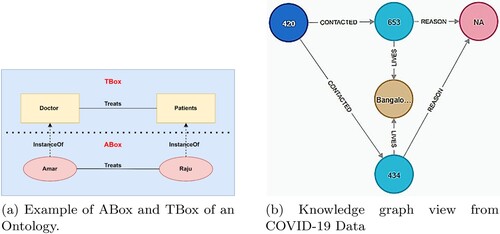
Ontology: An Ontology O is defined as {O = (Tbox + Abox, G)}. Tbox: define the schema or an ontology. Abox refers to RDF triples at the instance level. G is a labelled graph structure produced by connecting the relations with concepts.
Figure represents the ontology and a COVID-19 knowledge graph snippet.
Figure 1. Representation of ontology and knowledge graph.(a) Example of ABox and TBox of an Ontology. (b) Knowledge graph view from COVID-19 Data.
Definition 3.3
Knowledge Graph: A collection of descriptions of concepts, things, relationships, and events that are all linked together.
Definition 3.4
Data Interestingness (DI) Our notion of Data Interestingness is derived by integrating domain ontology (O) with data in RDF (D) and with user interest rules (U) as shown in Equation (Equation3(3)
(3) ).
(3)
(3) Here user interest(U) refers to the unexpectedness and actionability measures. Interestingness framework in Section 4 illustrate Equation (Equation3
(3)
(3) ).
Definition 3.5
Patient Outcomes (PO): This is a measure of the impact that a pattern or relationship has on the health and well-being of patients. You can use patient outcome data such as survival rates, hospital readmission rates, and quality of life scores to evaluate the interestingness of a pattern or relationship
Definition 3.6
Clinical Relevance(CR): This is the extent to which a pattern or relationship is relevant to the treatment and care of patients. You can use clinical knowledge and expert opinion to evaluate the clinical relevance of a pattern or relationship, and use this information to guide your analysis.
Definitions 3.5 and 3.6 are used in domain expert assessment in Section 6.
3.2. Data – COVID-19 corpora and ontology
The proposed framework is suitable for any type of data along with its relevant ontology, therefore, the selection of data sets was based on their availability and relevance to the research objectives. The first data set was selected from a publicly available repository, while the second was from a funded project. The ability to use other data sets depends on their suitability and compatibility with the proposed framework.Footnote1 The data statistics are illustrated in Tables and . We have used two COVID-19 ontology from Bioportal Footnote2 which match the our COVID-19 corpora.
Table 1. BERT model for healthcare domain.
Table 2. COVID-19 corpora description.
Table 3. Dataset and ontology descriptions.
3.3. Data pre-processing technique
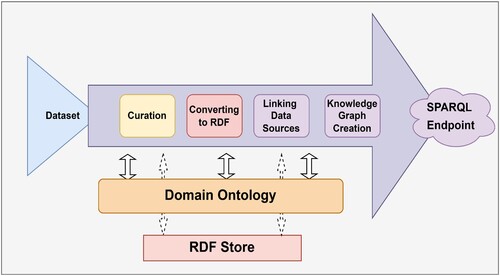
The most crucial step in the interestingness framework is data pre-processing, referred to as RDF data processing. Using ontologies and RDF data modelling to represent the data in a way that makes it more meaningful and easier to understand. This can involve adding additional information such as class and property definitions, and relationships between entities, which can help to clarify the meaning of the data and improve its overall quality. One of the key contributions of this study is the development of a effective data pre-processing technique that enhances the accuracy and effectiveness of the proposed interestingness framework. The structured data is translated to RDF triple form and uses the SPARQL Endpoint for query operations. Implementation of semantic data curation is made available at GitHub .Footnote5 The steps of the proposed data pre-processing technique are as follows:
Data Curating
Converting to RDF
Linking Data Sources
Publishing as Knowledge Graph
3.3.1. Data curation
Introducing semantics during the data curation phase is a unique technique. Semantics serves as a binding agent, defining the data model and relationships before and after the data processing. This process helps to handle data duplication and improves the quality of the data. Additionally, it disambiguates the data items by unifying and classifying multiple labels based on a trained model corpora and identifying semantically equivalent terms.
Example: {Fever, Fiver, FVR, Feever, Fevere} = Fever
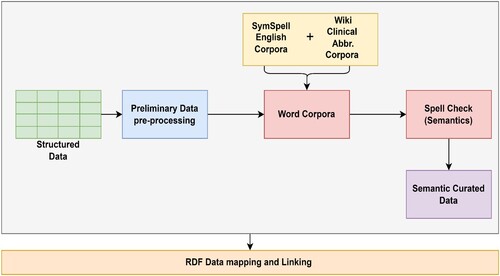
For the data curation, first, we create the corpus of the words from the COVID-19 data illustrated in Table . Now, the data corpus has many misspelled English words and clinical abbreviations.
Revised text: “To correct misspelled English words, we use the SymSpell corpus, which is created from the intersection of Google Ngram datasets and Hunspell dictionary files. Our proposed semantic data curation pipeline is illustrated in Figure . For the clinical abbreviations, we generated a corpus from Wikipedia's page on the list of medical abbreviations. To achieve semantic data curation in the initial stage of RDF data processing, we run a SymSpell check algorithm using the intersection of the generated corpus and the original corpus. This is based on the method described in Garbe's Citation2012 SymSpell paper (Garbe, Citation2012) ”.Footnote6
3.3.2. Converting to RDF
RDF is a structured data representation that provides a standardised way of describing relationships between data entities. Converting datasets to RDF can help in uniform representation, better data organisation, and easier data integration with other systems. It also enables the use of semantic technologies, such as ontologies, to enhance the meaning and context of the data. In this study, datasets are transformed into a uniform representation, RDF, which provides a structured representation of data with its relationships. The data curation pipeline is illustrated in Figure , which converts the curated data into RDF format.
3.3.3. Linking data sources
Linking data is the best practice, and it is done using the web links information using RDF and IRIs. The data linking is done using domain ontology. The ontology concepts with the prefix and URI are used to link the data sources. In our study, we have used Footnote7, Footnote8 , Footnote9 , Footnote10, Footnote11 data sources.
3.3.4. Publishing as knowledge graph
The COVID-19 knowledge base, also known as a COVID-19 triple store, is created by installing the GraphDB tool and a SPARQL endpoint on a Windows PC. SPARQL, a query language similar to SQL, is used to query the knowledge graph. This setup constitutes the COVID-19 knowledge base.
4. Interestingness framework using BERT
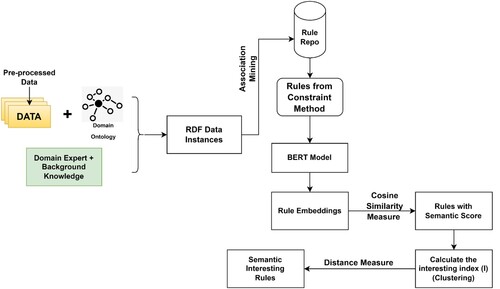
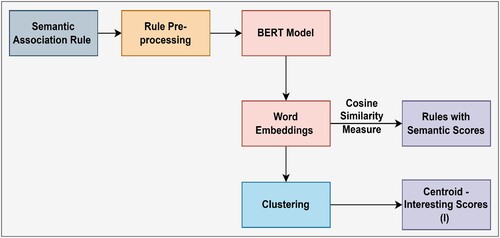
The Figure illustrate the SIF-B framework, the data is converted to RDF format using ontology and domain knowledge. Then, association mining algorithm, such as the improved Apriori algorithm, is applied to generate a rule repository from the RDF data and an OCA Mining algorithm is applied to have the relevant and useful rules required by the decision makers. Its where the interesting rules are selected based on predefined constraints. Further, BERT model to generate semantic embeddings. By applying cosine similarity measure, it measures the similarity between two non-zero vectors of an inner product space and henceforth identify the semantic rich rules. This methodology diagram provides a visual representation of the proposed framework for uncovering interesting insights in large COVID-19 datasets. The use of ontologies, the improved Apriori algorithm, and the BERT model for evaluating the interestingness of the rules makes the framework unique and promising for finding meaningful relationships and facts in large datasets.

The SIF-B algorithm illustrated in Algorithm 1, input the RDF Data (RD) and the ConstApriori Algorithm and Domain Ontology (DO). The algorithm first generates instance-level rules using the ConstApriori Algorithm on RD and the rules generated in the previous step are enriched with schema-level relationships from the domain ontology (DO). Further, semantic scoring is generated for teh interesting rules using cosine similarity measure to enriched the rules using a transformer-based method, such as BERT.

The algorithm procedure for RDF Data Generation (RDG) is a indicated in Procedure 1. The procedure is for converting data instances into RDF data. The algorithm takes in two inputs, the data instances (DI) and the domain ontology (DO) and outputs RDF data (RD). It Extract entities (E) and relationships (R) from the data instances (DI). Annotate the entities (E) and relationships (R) using the domain ontology (DO). For each entity (e) in the entities (E), map the entity to the equivalent resource in the domain ontology (DO). For each relationship (r) in the relationships (R), it maps the relationship to the equivalent property in the domain ontology (DO). Finally it generate triples (subject, predicate, object) from the entities (E) and relationships (R).

The Procedure 2 is a Ontology-based Constraint Apriori (OCA) Algorithm is a method to discover interesting patterns in RDF data while taking into account domain knowledge represented by an ontology. The algorithm starts by generating frequent itemset from the RDF data using Apriori algorithm with a minimum support value. The next step is to generate association rules from the frequent itemset with a minimum confidence and minimum lift value. After that, each association rule is then checked against the ontology constraint. If the rule meets the constraint, it is added to the set of interesting rules. Finally, the set of interesting rules is returned as the output.
Example 4.1
Suppose you have a COVID-19 patient information and want to find the frequently occurred symptoms. Normally, the Apriori algorithm would find the frequent item sets without any constraints. But in this example, we will add a constraint. Let's say, we want to find the pattern sets that are only with diabetic and high blood pressure symptoms. This makes the mining method filter based on the constraints defined. The constraint are derived from ontology and verified by the domain experts to avoid the user bias in generating the rules.
Complexity and Overhead Analysis: The complexity and overhead analysis can be represented in LaTeX as follows:
Let's assume the number of items in a transaction be represented by n and the number of transactions in the database be represented by m. The time complexity of Apriori algorithm is given as . The use of constraints from ontology can reduce the search space and thus improve the efficiency of the Apriori algorithm. Let's assume the reduction in search space to be represented by k. Therefore, the improved time complexity of the Apriori algorithm with constraints from ontology can be represented as
. The use of BERT for generating semantic scores also brings additional computational overhead. Let's assume the overhead to be represented by o. Therefore, the total computational overhead of the proposed approach can be represented as
. The above representation is just an approximation and the exact overhead and complexity of the proposed approach will depend on the specific implementation and the size of the database being analysed. Overall, the proposed approach may have higher computational overhead compared to traditional association rule mining techniques. However, the added value of incorporating constraints from ontology and semantic scores can offset the overhead and result in improved results.
Motivating Example: The Motivation behind this work is identifying interesting inferences
John lives in New York.
John suffers from Diabetic and Hypertension.
Jack had a cardiac problem for the past year.
John and Jack are treated at Apollo Healthcare centre.
Jack was treated in New York.
Apollo Healthcare centre is located in New York.
Diabetic, Hypertension, and Cardiac-related treatments are provided at Apollo Healthcare centre.
5. Result and discussions
The experiment results were obtained on an Intel Core i7-8565U machine with 16GB RAM. Protégé was used for mapping the data to an ontology and generating RDF data. The rule generation and interestingness mining algorithm and procedures discussed in Section 4 were coded in Python on Google Collaboratory. Footnote12Footnote13.
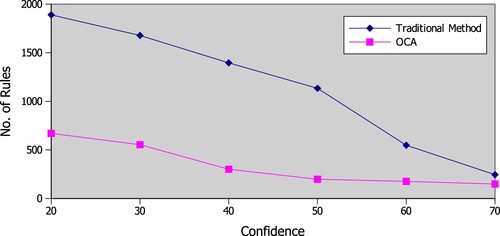
Unlike traditional methods of association rule mining, ontology-based constraint apriori incorporates domain knowledge in the form of ontologies to filter and refine the set of association rules generated. This not only ensures the high quality of the rules but also significantly reduces the number of rules generated, making it easier to understand and interpret the results. Figure illustrate visually.
In a traditional method of association rule mining, the main focus is on generating a large number of association rules, which often results in a large number of irrelevant or redundant rules. These rules not only make it difficult to interpret the results but also increase the computational overhead required to mine the rules. In contrast, OCA employs a set of constraints defined in the ontology to restrict the rule generation process, reducing the number of rules generated and improving the quality of the rules.
5.1. Rules from ontology
In this study, we utilised the ontology that was developed in a prior study. This ontology provides the necessary structure and relationships for our analysis. We have reused the ontology and integrated the BERT model to enhance its functionality.
Generating rules from an ontology can be achieved through the use of a rule language, such as Semantic Web Rule Language (we), which is built on top of OWL (Web Ontology Language). These rules can be used to make inferences, or deductions, based on the relationships and concepts represented in the ontology. This can lead to a more automated and efficient process of reasoning and knowledge discovery, as well as increased consistency and accuracy in the resulting inferences.
SWRL rule that operate on the COKPME ontology are listed below:
A patient diagnosed with COVID-19 suffering from a particular condition, can be represented as:

If a patient is diagnosed with COVID-19 and resides in an area with high infection rates, then they are more likely to have severe symptoms.

If a patient is over the age of 60 and has a pre-existing medical condition, then they are at higher risk for severe COVID-19 outcomes.

If a patient receives a specific treatment for COVID-19, then their symptoms are likely to improve.

If a patient has travelled from a location with high COVID-19 transmission and they are suffering from a respiratory illness, they are at high risk of having COVID-19.

If a patient has a primary contact who has tested positive for COVID-19, the patient is at risk of having COVID-19.

If a patient is diagnosed with COVID-19, they should receive treatment according to the COVID-19 treatment protocol.

If a patient is suffering from a respiratory illness and has travelled from a location with high COVID-19 transmission, they should be tested for COVID-19.

The SWRL rules listed above illustrate the complex relationships in COVID-19 dataset. Example: “If a patient has a property of “suffersFrom” COVID-19 and also has a property of “residesAt” a place with high cases of COVID-19 and “hasAge” in a certain range, then they are at high risk of contracting the infection”.
Table indicates the sample key entities used in Procedure 2 for the OCA algorithm. To begin, ConstApriori mines frequent patterns from RDF data and then uses the constraint file to generate interesting rules. The entity items from the constraint file are cross-checked with domain experts to avoid bias in the analysis.
Table 4. Items from the constraint file.
5.2. Semantic interestingness in COVID-19 corpora
The interestingness framework aims to generate interesting rules from the data and the domain ontology. The semantic association rules are extracted using the OCA algorithm, which is based on the interestingness framework.
Table indicates the rules from the OCA algorithm. The generated rules were as shown below:

Table 5. Few semantic association rules from COKPME and KATrace dataset.
To make it interpretable, we have truncated the Universal Resource Identifier (URI) of the rule from both antecedent and consequent. Table shows the interesting entities of the rules that were identified from the data using different confidence values. The symptoms listed at Center for Disease Control and Prevention (CDC) has high correlation with our OCA generated symptoms Footnote14.
Table 6. Interesting entities of the rules with different confidence values.
5.3. Comparison to the state-of-the-art
There have been several recent works in the literature that apply association rule mining on COVID-19 data, however, most of these works focus on analysing X-ray or CT-scan data and are specific to patients with co-existing conditions such as diabetes. As a result, limited comparables are available in the literature for comparison with our methodology. Despite this, we have summarised the comparisons we found in the available literature in a Tables and .
Table 7. Comparative analysis of rules I.
Table 8. Comparative analysis of rules II.
In Çelik Ertuğrul and Ulusoy (Citation2022), the authors generate inferences from COVID-19 patient's self-reported symptoms using a domain ontology. Table compares the results of the proposed OCA algorithm with those from Çelik Ertuğrul and Ulusoy (Citation2022), as this work is closely related to our study. Our results show a high degree of correlation between the two sets of rules, both in terms of how they are framed. However, it is important to note that the state-of-the-art study in this domain focuses solely on patient-identified symptoms, whereas our study considers additional factors for symptom identification.
The traditional method for deriving rules from the COVID-19 dataset employs association rule mining, as presented in Tandan et al. (Citation2021). The resulting rules are noteworthy due to the diverse attributes considered, such as age, gender, and symptoms, to uncover patterns. However, the proposed method takes a more innovative approach by utilising constraint-based and SWRL query-based techniques to mine interesting patterns.
5.4. Semantic interestingness using transformers
To choose the appropriate BERT model for the semantic interestingness technique, we did a thorough literature study as indicated in Table . We decided to leverage the pre-trained transfer learning models as they have significant relevance to the domain and the trained corpora. According to the study, we conclude that Transformer-based methods are much less explored to their full potential to detect interestingness in the rules. Two such models, BioClinicalBERT and CovidBERT, are selected for this research work to find the most interesting rules from the input rule set. We must design a practical and reasonable model that justifies our claim for interestingness. With GPU-enabled computing resources, these pre-trained models are fine-tuned using the Simple Transformers library on Google Collaborator. The detailed workflow of the transformer-based interestingness method in our study is shown in Figure . The rules obtained from semantic association rule mining are processed using BioClinicalBERT and CovidBERT models to generate rule embeddings. Next, using cosine similarity measures for a total input of 1242 unique rules, we generated 752151 rules with a similarity index. This huge corpus is obtained after mapping each rule with a different rule. Further, we identify the interesting centroids by applying clustering on the average rule embeddings and cosine similarity index of all the rules. The centroid is computed in two aspects of interest, first on the average value of the rule embeddings score and second on the cosine similarity index of all the rules. Both aspects are compared for interestingness in data.
5.4.1. Tokenization and embeddings
All of the pre-trained models require a specific format for the input text. In our case, for the semantic association rules, tokenizers break the input text into smaller tokens. Before fine-tuning the model, these tokens are then transformed into embeddings. Each transfer learning model has undergone pre-training on a particular corpus with a predetermined vocabulary. The model's input text may include terms not part of its set vocabulary. The normal BERT model employs a WordPiece tokenizer to handle these terms that are out of the vocabulary (OOV) while retaining information from the input data (Schuster & Nakajima, Citation2012). It is trained on lower-cased English text with a vocabulary size of about 30,000 tokens. Each transformer block has 768 hidden layers and 12 self-attention heads, and 110M parameters for training.
5.4.2. Interesting centroids
Interesting cluster centroids are identified by applying the k-nearest neighbour (KNN) algorithm on the average word embedding. Tables and illustrate the interesting rules derived using the healthcare BERT models. The interesting centroids from CovidBERT are found to represent the following concise demonstrations of the rules:
Table 9. Rules from cluster centroid-CovidBERT.
Table 10. Rules from cluster centroid – BioClinicalBERT.
Symptoms = {Fever}
Treatment = {Medicine prescribed with home quarantine advice}
Age = { 2, 3, 10}
Category = {ILI}
Symptoms = {Fever}
Treatment = {Medicine prescribed with home quarantine advice, Admit to another Hospital}
Age = { 20, 35 }
Category = {ILI, SARI }
By comparing the rules from both models, the BioClinicalBERT has detailed rules that have more interesting facts. Whereas CovidBERT indicates the basic and frequent level of attributes only.
5.4.3. Rules with semantic score
The cosine similarity index computes the degree of similarity between two vectors in an inner product space. It determines if two vectors point in the same general direction by computing the cosine of the angle between them. Referring to Tables and , the semantic rules represent the most interesting ones to the decision makers compared with the centroid method. The semantic rules illustrate the broad level of information like:
Symptom = {Acute Febrile illness, Diabetic, Hypertension, Breathlessness, SARI }
Treatment = {Admitted to own hospital, Home quarantine advice, medicines prescription for the symptoms }
Age = {55, 70, 43}
Category = {ILI, SARI, COVID-19}
Table 11. Rules from CovidBERT with semantic scores.
Table 12. Rules from BioClinicalBERT with semantic scores.
The summary of the semantic interesting rules covers the larger scope for the decision-makers. Also, by comparing the BERT models' rules, it's observed that BioClinicalBERT has semantic-rich information compared to the CovidBERT model. The similarity index is high with the rules about the SARI and breathlessness. Also, diabetes has high relevance with ILI patients.
5.4.4. Rules using distance measure
In the literature, the use of distance-based measures for identifying interesting rules is widely acknowledged. In this study, we have incorporated a distance-based measure, cosine similarity, for evaluating the semantic rules generated. The results presented in Tables and from the CovidBERT model and Tables and from the BioClinicalBERT model represent the most interesting rules from the analysed COVID-19 corpora. These tables compare two rules from the input rule set and demonstrate the distance between them, emphasising the significance and relevance of the generated rules. The distance-based measure is applied to the five clusters created through K-means clustering, as discussed in Section 5.4.2. These findings assist in identifying semantically similar rules for decision-makers. For example, referring to Table , rule 1 highlights the relevance of treatment provided, while rule 2 highlights the relevance between patients.
Table 13. Semantic rules with absolute distance measure using CovidBERT for Cluster 0, 1 and 2.
Table 14. Semantic rules with absolute distance measure using CovidBERT for cluster 3 and 4.
Table 15. Semantic rules with absolute distance measure using BioClinicalBERT for cluster 0, 1 and 2.
Table 16. Semantic rules with absolute distance measure using BioClinicalBERT for cluster 3 and 4.
5.4.5. Industrial significance
An ontology-based approach for semantic data interestingness has been applied in a variety of real-world domains, including:
Healthcare: To support clinical decision making and improve patient outcomes.
Social media: To support the analysis of public opinion and sentiment.
Business intelligence: To support the analysis of market trends and customer behaviour.
Bioinformatics: To support the analysis of genetic and proteomic data.
Environmental monitoring: To support the analysis of climate change, air and water quality, and biodiversity.
6. Rule evaluation
An effective rule evaluation framework is proposed to indicate the level of rule's interestingness. The framework has two dimensions: Statistical analysis and domain expert's evaluation.
6.1. Statistical significance
The statistical method for evaluating these rules involves analysing them using the Chi-square measure, and deciding on the defined hypothesis based on the cross-tabulation of the inferred rules. Next, domain experts are consulted to assess the relevance, significance, and usefulness of the top rules. Finally, the inferred rules are ranked based on the input from the domain experts, to indicate their level of interest.
Referring to Table , chi-square statistic is 51.457. The p-value is .000013. The result is significant at p<.05.
Table 17. Chi-square test for rule significance on sample rules.
6.1.1. Domain expert evaluation
For the evaluation of the proposed framework, domain experts were selected from authorised healthcare facilities in the medical field. These domain experts were chosen for their expertise and experience in the relevant area of medicine. The experiment was conducted by presenting the extracted rules to the domain experts, who then rated the level of interestingness based on the usefulness and relevance of the rules. The evaluation was performed by conducting a survey where the domain experts were asked to rate each rule on a scale of 1–5 as illustrated in Table . This approach was chosen as it provides a subjective measure of the interestingness of the extracted rules, which is considered to be an important aspect of evaluating the effectiveness of the proposed framework. The experiment was carried out in a controlled environment to ensure the accuracy of the results.
Table 18. Evaluation scale range.
To have a potential method to use domain experts for rule evaluation, measuring the level of interestingness is proposed on a scale of one to five. Two domain experts carried out the evaluation. The more important rules are thought to be more interesting rules. We ask the domain expert to rate a hypothesis based on its importance, relevance, and usefulness. The following Hypothesis cloud is considered:
Rules that are with sufficient confidence than experts knowledge.
Rules and experts' knowledge have the same phase or confidence.
Rules with a lower level of confidence than expert's knowledge.
Table 19. Domain expert evaluation summary.
6.2. Implication and limitations of our framework
The propagation of interestingness through ontology-based methods has been a new area of research in recent years. As in the healthcare domain, it mainly focuses on symptom analysis, diagnosis, and analysing the consequences of the spread, symptom patterns, etc. As a result, it is critical to find interesting facts that could help the decision-makers. The interesting patterns lie as implicit facts in the data. With domain ontology, it can be uncovered with semantic knowledge. However, this is only the first step toward detecting interestingness in data using domain ontology, and there is always an opportunity for improvement. Our Semantic Interesting Framework using BERT (SIF-B) has some shortcomings that will be addressed in future work. To highlight few implications:
Improved discovery of meaningful relationships and facts in large RDF datasets.
Enhanced quality of data insights by incorporating domain knowledge through ontologies.
Some of the limitations are as follows.
Potential limitations in performance for extremely large datasets.
Dependence on the quality of the ontology and the domain knowledge encoded.
Limitations in the interpretation of the results may arise due to the complex nature of the transformer-based methods.
7. Conclusion
This study presents a novel approach for uncovering interesting insights in RDF data using instance and schema-level information. The current research in this field primarily focuses on mining instance-level data to discover interesting relationships. However, our proposed approach, called SIF-B, addresses this issue by incorporating knowledge encoded at the schema level through an ontology's relationships and using the Ontology ConstApriori algorithm (OCA) for instance-level rules. The semantically enriched rules are generated using the schema level relationships such as “rdf:type” and “rdf:subClassOf”. The rules and their semantic scores are generated using a transformer-based method.
Additionally, COVID-BERT, Clinical-BERT, and Bio-BERT models are employed to identify the most interesting rules through the equivalence of variance method. The results showed that the variance was equally distributed among all the interesting clusters. To evaluate the generated rules, we used the Chi-square test for significance and a domain expert evaluation mechanism. It was found that the semantic rules from SIF-B were significant at a p-value of 0.5 and the transformer-model identified and domain expert ranked rules showed a high level of correlation.
Acknowledgments
We also extend our special thanks to the e-Health section of HFWS, Government of Karnataka, for providing all the necessary support and encouragement. We extended our gratitude to JSS Mahavidyapeeta, Mysore and my parent institute, JSSATE Bangalore, for allowing me to pursue my doctoral degree. Also, we would like to thank five anonymous reviewers for commenting on earlier versions of this paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Notes
References
- Abas, A. R., El-Henawy, I., Mohamed, H., & Abdellatif, A. (2020). Deep learning model for fine-grained aspect-based opinion mining. IEEE Access, 8, 128845–128855. https://doi.org/10.1109/Access.6287639
- Abhilash, C., & Mahesh, K. (2021). Graph analytics applied to COVID19 karnataka state dataset. In 2021 The 4th International Conference on Information Science and Systems (pp. 74–80). Association for Computing Machinery.doi:10.1145/3459955.3460603
- Abhilash, C. B., & Mahesh, K. (2022a). Ontology-based interestingness in covid-19 data. In Metadata and Semantic Research: 15th International Conference, MTSR 2021, Virtual Event, November 29–December 3, 2021, Revised Selected Papers (pp. 322–335). Springer.
- Abhilash, C. B., & Mahesh, K. (2022b). Ontology-based method for semantic association rules. In IEEE 19th India Council International Conference (pp. 1–7). IEEE.
- Agrawal, R., & Srikant, R. (1994). Fast algorithms for mining association rules. In Proceedings 20th International Conference Very Large Data Bases, VLDB (Vol. 1215, pp. 487–499). Citeseer.
- Alzubi, J. A., Jain, R., Singh, A., Parwekar, P., & Gupta, M. (2021). COBERT: COVID-19 question answering system using BERT. Arabian Journal for Science and Engineering, 1–11.
- Apostolopoulos, I. D., & Mpesiana, T. A. (2020). Covid-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine, 43(2), 635–640. https://doi.org/10.1007/s13246-020-00865-4
- Arora, P., Kumar, H., & Panigrahi, B. K. (2020). Prediction and analysis of COVID-19 positive cases using deep learning models: A descriptive case study of India. Chaos, Solitons & Fractals, 139, 110017. https://doi.org/10.1016/j.chaos.2020.110017
- Bellandi, A., Furletti, B., Grossi, V., & Romei, A. (2007). Ontology-driven association rule extraction: A case study. Contexts and Ontologies Representation and Reasoning, 10.
- Bizer, C., Lehmann, J., Kobilarov, G., Auer, S., Becker, C., Cyganiak, R., & Hellmann, S. (2009). Dbpedia-a crystallization point for the web of data. Journal of Web Semantics, 7(3), 154–165. https://doi.org/10.1016/j.websem.2009.07.002
- Bringmann, B., Nijssen, S., & Zimmermann, A. (2011). Pattern-based classification: A unifying perspective. arXiv preprint arXiv:1111.6191.
- Çelik Ertuğrul, D., & Ulusoy, D. C. (2022). A knowledge-based self-pre-diagnosis system to predict covid-19 in smartphone users using personal data and observed symptoms. Expert Systems, 39(3), e12716.
- Choi, E., He, H., Iyyer, M., Yatskar, M., Yih, W.t., Choi, Y., Liang, P., & Zettlemoyer, L. (2018). QuAC: Question answering in context. arXiv preprint arXiv:1808.07036.
- Ciotti, F., & Tomasi, F. (2016). Formal ontologies, linked data, and TEI semantics. Journal of the Text Encoding Initiative, 9.
- Devlin, J., Chang, M.-W., Lee, K., & Toutanova, K. (2018). Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805.
- Gahar, R. M., Arfaoui, O., Hidri, M. S., & Hadj-Alouane, N. B. (2018). An ontology-driven mapreduce framework for association rules mining in massive data. Procedia Computer Science, 126, 224–233. https://doi.org/10.1016/j.procs.2018.07.236
- Garbe, W. (2012). SymSpell (Vol. 6). https://github.com/wolfgarbe/SymSpell
- Guo, X., Mirzaalian, H., Sabir, E., Jaiswal, A., & Abd-Almageed, W. (2020). Cord19sts: Covid-19 semantic textual similarity dataset. arXiv preprint arXiv:2007.02461.
- Mangla, M., & Akhare, R. (2015). Association rules filtration using dynamic methods. International Research Journal of Engineering and Technology, 2(3), 1103–1106.
- Marinica, C., & Guillet, F. (2010). Knowledge-based interactive postmining of association rules using ontologies. IEEE Transactions on Knowledge and Data Engineering, 22(6), 784–797. https://doi.org/10.1109/TKDE.2010.29
- Ozturk, T., Talo, M., Yildirim, E. A., Baloglu, U. B., Yildirim, O., & Acharya, U. R. (2020). Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine, 121, 103792. https://doi.org/10.1016/j.compbiomed.2020.103792
- Qin, L., Sun, Q., Wang, Y., Wu, K.-F., Chen, M., Shia, B.-C., & Wu, S.-Y. (2020). Prediction of number of cases of 2019 novel coronavirus (COVID-19) using social media search index. International Journal of Environmental Research and Public Health, 17(7), 2365. https://doi.org/10.3390/ijerph17072365
- Schuster, M., & Nakajima, K. (2012). Japanese and korean voice search. In 2012 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (pp. 5149–5152). IEEE.
- Shan, G., Zhou, L., & Zhang, D. (2021). From conflicts and confusion to doubts: Examining review inconsistency for fake review detection. Decision Support Systems, 144, 113513. https://doi.org/10.1016/j.dss.2021.113513
- Shen, I., Zhang, L., Lian, J., Wu, C.-H., Fierro, M. G., Argyriou, A., & Wu, T. (2020). In search for a cure: Recommendation with knowledge graph on CORD-19. In Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining (pp. 3519–3520).
- Su, H., Li, H., & Li, D. (2023). Knowledge reasoning with multiple relational paths. Connection Science, 1–21. https://doi.org/10.1080/09540091.2022.2161480
- Suchanek, F. M., Kasneci, G., & Weikum, G. (2007). Yago: A core of semantic knowledge. In Proceedings of the 16th International Conference on World Wide Web (pp. 697–706).
- Tandan, M., Acharya, Y., Pokharel, S., & Timilsina, M. (2021). Discovering symptom patterns of COVID-19 patients using association rule mining. Computers in Biology and Medicine, 131, 104249. https://doi.org/10.1016/j.compbiomed.2021.104249
- Tomar, A., & Gupta, N. (2020). Prediction for the spread of COVID-19 in India and effectiveness of preventive measures. Science of The Total Environment, 728, 138762. https://doi.org/10.1016/j.scitotenv.2020.138762
- Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., Kaiser, Ł., & Polosukhin, I. (2017). Attention is all you need. Advances in Neural Information Processing Systems, 30.
- Wikipedia contributors. (2021). FAIR data – Wikipedia, The Free Encyclopedia. Online: Retrieved August 24, 2021, from https://en.wikipedia.org/w/index.php?title=FAIR_data&oldid=1038845392