Abstract
Clopidogrel is one of the most frequently used drugs in patients to reduce cardiovascular events. Since patients with different genetic variations respond quite differently to clopidogrel therapy, the related genetic testing plays a vital role in its dosage and genetic testing related to clopidogrel therapy is currently considered as routine test worldwide. In this study, we aim to use two different methods MALDI-TOF mass spectrometry and pyrosequencing to detect gene variant of CYP2C19 and ABCB1. Six single nucleotides polymorphisms (SNP) within CYP2C19 (*2, *3, *4, *5, *17) and ABCB1 C3435T in 458 Chinese Han patients were determined using both MassARRAY and Pyrosequencing. Sanger sequencing was used for verification. Results of both methods were analyzed and compared. Allele frequencies of each SNP and distribution of different genotypes were calculated based on the MassARRAY and Sanger sequencing results. Both methods provided 100% call rates for gene variants, while results of six samples were different with two methods. With Sanger sequencing as the reference results, MassARRAY generated all the same results. The minor allele frequencies of the above six SNPs were 27.1% (CYP2C19*), 5.9% (CYP2C19*3), 0% (CYP2C19*4), 0% (CYP2C19*5), 1.1% (CYP2C19*17), 40.9% (ABCB1), respectively. MassARRAY provides accurate clopidogrel related genotyping with relatively high cost-efficiency, throughput and short time when compared with pyrosequencing.
1. Introduction
Clopidogrel is considered as standard antiplatelet drug in medical care for patients with acute coronary syndromes (ACS) and percutaneous coronary intervention (PCI) undergoing stent implantation (Antman et al., Citation2008). Over 80% of ischemic events and post-PCI cardiovascular events could be prevented by clopidogrel therapy (Cayla et al., Citation2011). Through inhibiting the purinergic ADP receptor P2Y12, clopidogrel reduces adenosine diphosphate-induced platelet aggregation and decreases the risk of cardiovascular events (Rytkin et al., Citation2017). However, a large number of patients continue to suffer recurrent ischemic events, and this clinical phenomenon has been correlated with lesser degrees of platelet inhibition (Udell et al., Citation2016). This failure of the antiplatelet drug to inhibit its target of action is called clopidogrel non-responsiveness or clopidogrel resistance (Nguyen et al., Citation2005). Although novel drugs including prasugrel and ticagrelor have been shouwn to be more rapid and effective P2Y12 receptor inhibitor, they tend to induce more reverse complications such as bleeding events (Kubica et al., Citation2014; Wiviott et al., Citation2007). Thus, clopidogrel remains to be one of the most extensively used antiplatelet drugs and research focused on how to diagnose the inter-individual variability in clopidogrel disposition and efficacy is of vital significance.
Clopidogrel is a prodrug that requires a two-step hepatic metabolism by several cytochrome P450 isoforms (Kazui et al., Citation2010). This procedure is firstly induced by CYP2C19 enzyme followed by further oxidation where the intermediate metabolite turns into an active metabolite by CYP3A4, CYP2C19, CYP2B6, and CYP2C9 isoenzymes. Accordingly, polymorphisms of CYP2C19 gene can partially explain variability in response to clopidogrel (Mega et al., Citation2009). In addition, clopidogrel is an oral drug, whose absorption in the duodenum requires P-glycoprotein involvement in the intestinal transport. The ATP-binding cassette subfamily B member 1 (ABCB1) gene encodes the enteric intestinal efflux transporter pump P-glycoprotein, which limits the oral bioavailability of clopidogrel and modulates its absorption in the intestine (Beitelshees et al., Citation2015). Currently, more than 50 SNPs residing in the coding region of this gene have been described (Fung & Gottesman, Citation2009). Among these, ABCB1 c.3435C > T (rs1045642) polymorphism has been extensively studied and some investigations have shown that this polymorphism influences the function of P-glycoprotein and, consequently, alter the absorption of clopidogrel (Simon et al., Citation2009). Results of systematic review also suggested that ABCB1 C3435T polymorphism may increase the risk of bleeding in Asian patients treated with clopidogrel (Zhai et al., Citation2017).
Pharmacogenetics (PGx)-study of variations in DNA sequences affecting drug response has been gradually applied in personalized patient care (Jani et al., Citation2015; Yip et al., Citation2015). CYP2C19 genotyping detection for clopidogrel therapy has been recommended by Clinical Pharmacogenetics Implementation Consortium (Scott et al., Citation2013). Various techniques involving fast and accurate detection of gene detection have been used in hospitals, such as TaqMan PCR, Sanger sequencing, gene-chip, high-melting curve and MALDI-TOF mass spectrometry. Since some of the patients may suffer from complications, clinical doctors hope that the genotype result is available at the time of prescription. In addition, they also require that the therapy be decided in a very short time (Kubica et al., Citation2014). The ability to identify more SNPs in a single test is also highly desirable in clinical practice.
Taking these factors into consideration, we aim to compare the most commonly used two methods: MassARRAY and pyrosequencing. MassARRAY technology is reported to shorten the turn-around-time to 12 h when compared with Sanger sequencing, the later of which uses about 24 h to provide clinicians with results. In addition, MassARRAY could obtain results of several SNPs per test. In our current study, we tested the loss of function (LOF) allele of CYP2C19 (*2, *3, *4, *5, *17) and ABCB1 (C3435T) with both MassARRAY and Pyrosequencing and compared the accuracy of these two techniques.
2. Methods and materials
2.1. Study volunteers and DNA sample preparation
This study included 458 unrelated Chinese Han volunteers who underwent routine examination of clopidogrel related genotyping test in our department from February 2016 to October 2017. 2 mL peripheral blood was collected using EDTA anticoagulant tube without coagulation or hemolysis. All volunteers have signed the informed consent and the study was approved by Ethics Committee of our hospital.
Genome DNA was extracted based on protocol of DNA isolation kit (Hipure Blood DNA Mini Kit, Magen). DNA extraction must be completed within 4 hours of blood collection. Then DNA was quantified by NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, USA) and diluted to the concentration between 2.5–5.0 ng/μL and stored at −80 °C.
2.2. Genotyping of CYP2C19 and ABCB1 by MassARRAY and pyrosequencing
Genotyping was performed using MassARRAY platform integrating iPLEX and Mass ARRAY technology (Agena Bioscience, San Diego, CA, USA)- based MALDI-TOF MS assay and with the kit designed for gene loci (Bioyong Technology Ltd, Beijing, China). We performed MassARRAY on the DNA extraction from 458 samples and focused on the following SNPs: CYP2C19*1-*5, CYP2C19*17 and ABCB1 C3435T. Our MassARRAY procedure was based on the previously described study of Stacey G (Gabriel et al., Citation2009). Primer pairs for amplification and sequencing were designed and provided by Bioyong Technology Ltd (Beijing). This technique contained five major procedures: Firstly, initial locus-specific PCR amplification was performed. Then, shrimp alkaline phosphatase was used to neutralize the uncombined deoxynucleotides (dNTPs). Thirdly, single base extension (SBE) uses mass-modified dideoxynucleotide (ddNTP) terminators of an oligonucleotide primer that anneals immediately upstream of the target polymorphic site. Then we used MALDI-TOF mass spectrometry to analyze the distinct mass of extended primer, which could trace the alternative alleles. Positive and negative template control samples were included in each assay plate. Any assay found as positive in negative template control was removed from the study.
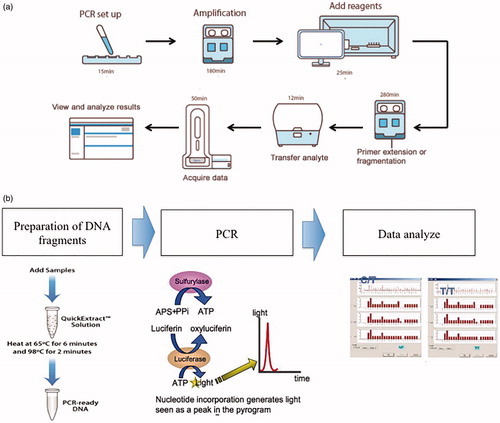
Pyrosequencing was performed strictly according to the protocol of Real time quantitative Pyrosequencing analyzer (PyroMark Q24 MDx, Germany, Qiagen) and the supporting kit (Qiagen, Germany). further illustrates the key steps of these two methods.
2.3. Sanger sequencing for verification
After comparing the genotyping results of MassARRAY and Pyrosequencing, inconsistent samples were selected for Sanger sequencing verification (Shanghai Biological Engineering Inc, Shanghai).
2.4. Statistical analysis
SPSS 22.0 was used for statistical analysis. Samples that produce results in the first run were regarded as determinate while others as indeterminate. The call rates of MassARRAY and Pyrosequencing were assessed by the percentage of determinate samples (Call rate = the number of determinate samples/(the number of determinate + the number of indeterminate samples)). The Chi-square test was applied to assess the consistency of MassARRAY and Pyrosequencing. Hardy-Weinberg equilibrium (HWE) was also calculated. p < 0.05 (two-tailed) was considered as statistical significance. The genotyping results of each SNP were calculated.
3. Results
3.1. Call rate evaluation
We tested all 458 samples with both MassARRAY and Pyrosequencing. For all the SNPs in our study: CYP2C19*2 (rs4244285), CYP2C19*3 (rs4986893), CYP2C19*4 (rs28399504), CYP2C19*5 (rs56337013), CYP2C19*17 (rs12248560) and ABCB1 C3435T (rs1045642), the call rates of both these two methods were 100%. Thus, there was no significant difference in call rates between MassARRAY and Pyrosequencing (p > 0.05).
3.2. Distribution of genotyping in Chinese Han population
Among all the included samples, the distribution of genotypes and alleles of the CYP2C19 and ABCB1 variants in our sample is summarized in . The minor allele frequencies of the above six SNPs were 27.1% (CYP2C19*2, rs4244285), 5.9% (CYP2C19*3, rs4986893), 0% (CYP2C19*4, rs28399504), 0% (CYP2C19*5, rs56337013), 1.1% (CYP2C19*17, rs12248560), 40.9% (ABCB1, rs1045642), respectively. The genotype distributions of all SNPs (except rs28399504 and rs56337013) were in Hardy-Weinberg equilibrium (p > 0.05). Due to the absence of the variant for rs28399504 and rs56337013, X2 value could not be defined. According to the results in our study, mutation rates of ABCB1 (rs1045642) is the highest in current population, followed by CYP2C19*2, *3 and *17. The observed CYP2C19 and ABCB1 variant frequencies in our study were also compared with those reported for both Chinese and European populations (Borecki et al., Citation2017; Santos et al., Citation2011; Zhuo et al., Citation2018).
Table 1. Genotype and allele frequencies of CYP2C19 and ABCB1 variants in Chinese Han population and comparison with European and Asian countries in previous studies.
Based on the MassARRAY results and Sanger sequencing verification, seven genotypes were found in our study: *1/*1(636GG, 681GG), *1/*2(636GG, 681GA), *2/*2(636GG, 681AA), *1/*3(636GA, 681GG), *2/*3(636GA, 681GA), *3/*3 (681GG, 636AA) and *1/*17 (-806CT). The frequencies of each genotype were: 43.01%, 35.15%, 8.07%, 8.51%, 2.83%, 0.21% and 2.18%, respectively (). Among the included 458 samples, we also divided genotypes according to their relevance for clopidogrel metabolism, which could help to predict risk of cardiovascular complications. The highest metabolic type is intermediate metabolism (*1/*2, *1/*3), followed by extensive metabolism (*1/*1), poor metabolism (*2/*2, *2/*3, *3/*3) and ultra metabolism (*1/*17, *17/*17). The frequencies of each type were 43.67%, 43.01%, 11.13% and 2.18%, respectively.
Table 2. Distribution of CYP2C19 genotypes according to their relevance for clopidogrel metabolism phenotypes in Chinese Han population.
3.3. Performance assessment of MassARRAY and pyrosequencing
Then we evaluated performance of MassARRAY and Pyrosequencing, considering Sanger sequencing as the reference test. We found that concerning CYP2C19*2 and CYP2C19*3, 6 samples have different results (). Then we performed Sanger sequencing on these 6 samples, all results were the same as MassARRAY, which indicated that the accuracy of MassARRAY is higher than that of Pyrosequencing. Then we analyzed the difference between MassARRAY and Pyrosequencing. Kappa values of CYP2C19*2 and CYP2C19*3 were 0.948 and 0.931, respectively, indicating a concordance between these two methods.
Table 3. Comparison of MassARRAY iPLEX and PyroMark Q24 on CYP2C19 genotypes.
3.4. Operational characteristics
We compared the operational characteristics of these two methods (). Turnaround time of each sample depends on the throughput and working time per run of the equipment. For both MassARRAY iPLEX and PyroMark Q24, the turnaround times of genotyping of clopidogrel were similar (average 7–8 h). The highest throughput of MassARRAY iPLEX is 384, which means 384 samples are allowed per plate. For each plate, multiplex PCR is available, thus allowing for multiple SNPs detection. As for PyroMark Q24, 24 samples are allowed each run and for each plate only one locus can be tested. Thus, compared with PyroMark Q24, MassARRAY iPLEX can be used for larger number of samples with more genotypes identified and less turnaround time. For both methods, the sensitivity is the same and both require 2ul DNA extraction. In addition, MassARRAY iPLEX can directly analyze the molecular weight of amplification product, which is rarely influenced by outside factors, thus generating more precise results. However, PyroMark Q24 analyze through capturing fluorescent signal, which might be affected by efficiency of fluorescence combination. Reagent cost per plate of MassARRAY iPLEX is much lower than PyroMark Q24 since the former method does not require fluorescent reagent and has higher throughput. According to recent studies (Svidnicki et al., Citation2015), cost of each sample with MassARRAY iPLEX for six SNPs (CYP2C19*2, *3, *4, *5, *17 and ABCB1-3435) is $5, while cost of each sample with PyroMark Q24 for 2 SNPs (CYP2C19*2 and *3) is about $12.3.
Table 4. Characteristics of the two methods and the gold standard (Sanger sequencing) Available for Clopidogrel Efficacy Genotyping.
4. Discussion
Aspirin and clopidogrel are the standard care for patients experiencing acute coronary syndrome (ACS) and/or percutaneous coronary intervention (PCI). Despite the unambiguous clinical benefit of aspirin plus clopidogrel, a number of patients continue to experience major adverse cardiac events (MACE). In order to promote the prediction and accuracy of clopidogrel efficiency in clinical practice, several rapid and precise gene-testing methods have been applied in clinics. However, each method has its defect: Sanger sequencing requires complex operation and longer time to generate results (Zhang et al., Citation2013); TaqMan requires expensive probes and are vulnerable to lab contaminant (Carlquist et al., Citation2013; Cervinski et al., Citation2013); gene chip, with its low flexibility, needs specific equipment, which is difficult to realize in common clinical labs (Buchan et al., Citation2011; Chae et al., Citation2013; Choi et al., Citation2014). Since doctors require the detection time to be as short as possible, a technique with a high call rate and shorter turnaround time is required.
After the discovery of SNPs associated with a particular phenotype in a genome-wide association studies (GWAS) study, replication of findings will usually be required in a second population. Often only tens of SNPs require genotyping for this phase. MassARRAY is one such platform that allows for tens to hundreds of user-defined SNPs to be genotyped in hundreds to thousands of DNA samples in a high-throughput and cost-effective manner (Ellis & Ong, Citation2017). MassARRAY is based on MALDI-TOF mass spectrometry, with the soft-ionization technology that allows the analysis of biomolecules. The whole workflow includes mainly the following procedures. Firstly, PCR is employed to amplify the regions of the genome containing each SNP. An ‘extension’ PCR reaction is then performed, in which an extension primer anneals just proximal to the polymorphic base, and a single ‘terminator’ nucleotide base extends the DNA fragment by one additional base that is specifically complementary to the polymorphic base. The terminator bases are ‘mass-modified’, whose masses are different between fragments, which is distinguished by a single base are detectable by mass spectrometry(Oeth et al., Citation2009). Its advantages include the wide range of molecular weight, the fast speed of scanning, the simple steps of operation, and particularly higher sequencing flux when compared with Sanger sequencing.
In the last several years, the range of applications for this platform was significantly expanded. In a multicenter study concerning patients with lung adenocarcinoma suspected of EGFR mutation, when comparing MassARRAY with pyrosequencing and PNAc, it was indicated that MassARRAY has a higher accuracy for the detection of EGFR mutations in tissue specimens (Min et al., Citation2016). In another meta-analysis, Tania et, al. (Fleitas et al., Citation2016) found that when used in combination with the OncoCarta™ panels and other customized panels, MassARRAY is cost-effective in terms of screening for somatic mutations in solid tumors. It would allow a rapid decision making, which would improve patient clinical trial inclusion. In addition, the initial cost of MassARRAY (Agena Bioscience, San Diego, CA, USA) ranges between $300,000 and $800,000 but it rarely requires routine maintenance. The cost of Pyrosequencing analyzer (PyroMark Q24 MDx, Germany, Qiagen) ranges between $70,000 and $100,000 and the routine maintenance costs about $100 per month. Thus, mass spectrometry has the potential to become an applicable approach in clinical diagnosis and can be used in genetic individualized therapy. Based on MassARRAY platform and iPLEX GOLD technique, it can conveniently analyze as many as 40 genotypes, with flexible and cheap design.
In our study, Chinese Han population was chosen as subjects. Among them, 27.1% carried the loss-of-function allele CYP2C19*2 A, who were poor clopidogrel metabolizers. CYP2C19*2 is the first allele discovered and it contains a single base pair mutation on exon 5 (681G > A), leading to an aberrant splice site. Since the A allele is associated with reduced conversion of clopidogrel to active metabolite. Patients with this allele are more vulnerable for cardiovascular complications including stent thrombosis (Mega et al., Citation2010). CYP2C19 *3 A was found to account for 5.9% of our sample, which is reported to be present only in Asians with frequency from 1 to 6% (Genomes Project et al., Citation2015). CYP2C19*4 and *5 were not observed in our current research. CYP2C19*17 (TT homozygote) accounted for 1%; these “ultra-rapid metabolizers” may suffer from increased risk of bleeding events (Li et al., Citation2012). Almost one-third of our patients possessed CYP2C19 variants phenotypically expressed by alteration of clopidogrel metabolism. In addition, an unexpected 41.5% of patients presented with ABCB1 mutation, which is the gene encoding P-glycoprotein multidrug resistance spontaneous transporters. Therefore, identification of these variants at early stage would allow consideration of alternative drugs for treatment.
To date, at least 28 alleles of CYP2C19 have been identified, and there are ethnic and geographical differences in the distribution of CYP2C19 variants(Kurose et al., Citation2012). When compared with Caucasian and African populations, CYP2C19*1 allele (wild-type) frequency was significantly lower, while the frequency of CYP2C19*3 allele was higher in the Chinese Han subjects (Ganoci et al., Citation2017; Masimirembwa et al., Citation1995). Zhou et al. (Citation2009) compared allele frequencies of Han samples (Beijing) with five other geographical subpopulations of Han Chinese (Hong Kong, Shanghai, Shantou, Shenyang and Xi’an) and observed that the frequencies of CYP2C19*1, *2 and *3 were significantly different from that of Shengyang and Xi’an subpopulations. However, Hu et al. (Hu et al., Citation2012) reported that the frequency of the CYP2C19*2 allele and the CYP2C19*3 allele found between southern and northern Chinese Han populations are not significantly different. When compared with European countries, the allele frequencies of CYP2C19*4, CYP2C19*5, and CYP2C19*17 in our cohort were much lower (Ferguson et al., Citation1998; Ghasemi et al., Citation2016). Three common genotypes that cause the poor metabolizer (PM) phenotype include CYP2C19*2/*2, CYP2C19*3/*3 and one heterozygous genotype CYP2C19*2/*3. The frequencies of CYP2C19*1/*1, *1/*2, *1/*3, *2/*2, *3/*3 and CYP2C19*2/*3 were not significantly different between our current study and previous studies (Dai et al., Citation2014; Zhou et al., Citation2009). In addition to the common allele, CYP2C19*6, *18, *23, *24, *25, *27, *29, *33, *34 although quite rare, have also been identified in Chinese Han population (Dai et al., Citation2014; Zhou et al., Citation2009).
5. Conclusion
In this study, we investigated two commonly used methods for CYP2C19 and ABCB1 polymorphisms in clinical laboratories among clopidogrel-treated Chinese patients. We found that with MALDI-TOF MS as the detection method, there are opportunities for providing genotyping results in a reasonable time frame that can facilitate their application. Thus, MassARRAY based on mass spectrometry is an excellent tool for the study of SNPs in patients who are potential candidates for targeted therapies, which should facilitate in the near future its approval for clinical testing. In the future, we aim to collect the clinical data to investigate the association between CYP2C19 and ABCB1 polymorphisms and the antiplatelet effect of clopidogrel.
Authors’ contribution
guarantor of integrity of the entire study:Juan Liu
study concepts:Zesheng Xu
study design:Ya Li
definition of intellectual content:Shipeng Dai
literature research:Junying Liu
clinical studies:Junjun Pan
experimental studies:Yang Jiang
data acquisition: Yang Jiang
data analysis:Junjun Pan
statistical analysis:Junying Liu
manuscript preparation:Juan Liu
manuscript editing:Ya Li
manuscript review:Shipeng Dai
| List of abbreviation | ||
| SNP | = | single nucleotides polymorphisms |
| ACS | = | acute coronary syndromes |
| PCI | = | percutaneous coronary intervention |
| ABCB1 | = | ATP-binding cassette subfamily B member 1 |
| PGx | = | Pharmacogenetics; LOF: loss of function |
| dNTPs | = | deoxynucleotides |
| SBE | = | single base extension |
| ddNTP | = | dideoxynucleotide |
| HWE | = | Hardy-Weinberg equilibrium |
| MACE | = | major adverse cardiac events |
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
References
- Antman EM, Hand M, Armstrong PW, Bates ER, Green LA, Halasyamani LK, et al. 2008. 2007 Focused Update of the ACC/AHA 2004 Guidelines for the Management of Patients With ST-Elevation Myocardial Infarction: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines: developed in collaboration With the Canadian Cardiovascular Society endorsed by the American Academy of Family Physicians: 2007 Writing Group to Review New Evidence and Update the ACC/AHA 2004 Guidelines for the Management of Patients With ST-Elevation Myocardial Infarction, Writing on Behalf of the 2004 Writing Committee. Circulation 117:296–329.
- Beitelshees AL, Voora D, Lewis JP. 2015. Personalized antiplatelet and anticoagulation therapy: applications and significance of pharmacogenomics. Pharmgenomics Pers Med 8:43–61.
- Borecki K, Zawada I, Pawinska-Matecka A, Salkic NN, Karakiewicz B, Adler G. 2017. ABCB1 3435C > T and 2677G > T/A polymorphisms in Polish and Bosnian patients with Crohn's disease - A preliminary report. Bosn J Basic Med Sci 17:323–327.
- Buchan BW, Peterson JF, Cogbill CH, Anderson DK, Ledford JS, White MN, et al. 2011. Evaluation of a microarray-based genotyping assay for the rapid detection of cytochrome P450 2C19 *2 and *3 polymorphisms from whole blood using nanoparticle probes. Am J Clin Pathol 136:604–608.
- Carlquist JF, Knight S, Horne BD, Huntinghouse JA, Rollo JS, Muhlestein JB, et al. 2013. Cardiovascular risk among patients on clopidogrel anti-platelet therapy after placement of drug-eluting stents is modified by genetic variants in both the CYP2C19 and ABCB1 genes. Thromb Haemost 109:744–754.
- Cayla G, Hulot JS, O’Connor SA, Pathak A, Scott SA, Gruel Y, et al. 2011. Clinical, angiographic, and genetic factors associated with early coronary stent thrombosis. JAMA 306:1765–1774.
- Cervinski MA, Schwab MC, Lefferts JA, Lewis LD, Lebel KA, Tyropolis AM, et al. 2013. Establishment of a CYP2C19 genotyping assay for clinical use. Am J Clin Pathol 139:202–207.
- Chae H, Kim M, Koh YS, Hwang BH, Kang MK, Kim Y, et al. 2013. Feasibility of a microarray-based point-of-care CYP2C19 genotyping test for predicting clopidogrel on-treatment platelet reactivity. Biomed Res Int 2013:1.
- Choi JL, Kim BR, Kim JE, Woo KS, Kim KH, Kim JM, et al. 2014. Association between the microarray-based CYP2C19 genotyping assay and the platelet function test in cardiovascular patients receiving clopidogrel. Int J Lab Hematol 36:e80–e83.
- Dai DP, Xu RA, Hu LM, Wang SH, Geng PW, Yang JF, et al. 2014. CYP2C9 polymorphism analysis in Han Chinese populations: building the largest allele frequency database. Pharmacogenomics J 14:85–92.
- Ellis JA, Ong B. 2017. The MassARRAY® System for Targeted SNP Genotyping. Methods Mol Biol 1492:77–94.
- Ferguson RJ, De Morais SM, Benhamou S, Bouchardy C, Blaisdell J, Ibeanu G, et al. 1998. A new genetic defect in human CYP2C19: mutation of the initiation codon is responsible for poor metabolism of S-mephenytoin. J Pharmacol Exp Ther 284:356–361.
- Fleitas T, Ibarrola-Villava M, Ribas G, Cervantes A. 2016. MassARRAY determination of somatic oncogenic mutations in solid tumors: Moving forward to personalized medicine. Cancer Treat Rev 49:57–64.
- Fung KL, Gottesman MM. 2009. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta 1794:860–871.
- Gabriel S, Ziaugra L, Tabbaa D. 2009. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet Chapter 2:Unit 2 12. DOI: 10.1002/0471142905.hg0212s60
- Ganoci L, Bozina T, Mirosevic Skvrce N, Lovric M, Mas P, Bozina N. 2017. Genetic polymorphisms of cytochrome P450 enzymes: CYP2C9, CYP2C19, CYP2D6, CYP3A4, and CYP3A5 in the Croatian population. Drug Metab Pers Ther 32:11–21.
- Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. 2015. A global reference for human genetic variation. Nature 526:68–74.
- Ghasemi Z, Hashemi M, Ejabati M, Ebrahimi SM, Kheiri Manjili H, Sharafi A, et al. 2016. Development of a high-resolution melting analysis method for CYP2C19*17 genotyping in healthy volunteers. Avicenna J Med Biotechnol 8:193–199.
- Hu LM, Dai DP, Hu GX, Yang JF, Xu RA, Yang LP, et al. 2012. Genetic polymorphisms and novel allelic variants of CYP2C19 in the Chinese Han population. Pharmacogenomics 13:1571–1581.
- Jani M, Barton A, Ho P. 2015. Pharmacogenetics of treatment response in psoriatic arthritis. Curr Rheumatol Rep 17:44.
- Kazui M, Nishiya Y, Ishizuka T, Hagihara K, Farid NA, Okazaki O, et al. 2010. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos 38:92–99.
- Kubica A, Kasprzak M, Siller-Matula J, Koziński M, Pio Navarese E, Obońska K, et al. 2014. Time-related changes in determinants of antiplatelet effect of clopidogrel in patients after myocardial infarction. Eur J Pharmacol 742:47–54.
- Kurose K, Sugiyama E, Saito Y. 2012. Population differences in major functional polymorphisms of pharmacokinetics/pharmacodynamics-related genes in Eastern Asians and Europeans: implications in the clinical trials for novel drug development. Drug Metab Pharmacokinet 27:9–54.
- Li Y, Tang HL, Hu YF, Xie HG. 2012. The gain-of-function variant allele CYP2C19*17: a double-edged sword between thrombosis and bleeding in clopidogrel-treated patients. J Thromb Haemost 10:199–206.
- Masimirembwa C, Bertilsson L, Johansson I, Hasler JA, Ingelman-Sundberg M. 1995. Phenotyping and genotyping of S-mephenytoin hydroxylase (cytochrome P450 2C19) in a Shona population of Zimbabwe. Clin Pharmacol Ther 57:656–661.
- Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. 2009. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 360:354–362.
- Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, et al. 2010. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA 304:1821–1830.
- Min KW, Kim WS, Jang SJ, Choi YD, Chang S, Jung SH, et al. 2016. MassARRAY, pyrosequencing, and PNA clamping for EGFR mutation detection in lung cancer tissue and cytological samples: a multicenter study. J Cancer Res Clin Oncol 142:2209–2216.
- Nguyen TA, Diodati JG, Pharand C. 2005. Resistance to clopidogrel: a review of the evidence. J Am Coll Cardiol 45:1157–1164.
- Oeth P, del Mistro G, Marnellos G, Shi T, van den Boom D. 2009. Qualitative and quantitative genotyping using single base primer extension coupled with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MassARRAY). Methods Mol Biol 578:307–343.
- Rytkin E, Mirzaev KB, Grishina EA, Smirnov VV, Ryzhikova KA, Sozaeva ZA, et al. 2017. Do CYP2C19 and ABCB1 gene polymorphisms and low CYP3A4 isoenzyme activity have an impact on stent implantation complications in acute coronary syndrome patients? Pharmgenomics Pers Med 10:243–245.
- Santos PC, Soares RA, Santos DB, Nascimento RM, Coelho GL, Nicolau JC, et al. 2011. CYP2C19 and ABCB1 gene polymorphisms are differently distributed according to ethnicity in the Brazilian general population. BMC Med Genet 12: DOI:10.1186/1471-2350-12-13
- Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, et al. 2013. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94:317–323.
- Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Meneveau N, et al. 2009. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 360:363–375.
- Svidnicki MC, Silva-Costa SM, Ramos PZ, dos Santos NZ, Martins FT, Castilho AM, et al. 2015. Screening of genetic alterations related to non-syndromic hearing loss using MassARRAY iPLEX(R) technology. BMC Med Genet 16: DOI:10.1186/s12881-015-0232-8
- Udell JA, Bonaca MP, Collet JP, Lincoff AM, Kereiakes DJ, Costa F, et al. 2016. Long-term dual antiplatelet therapy for secondary prevention of cardiovascular events in the subgroup of patients with previous myocardial infarction: a collaborative meta-analysis of randomized trials. Eur Heart J 37:390–399.
- Wiviott SD, Braunwald E, McCabe CH, Montalescot G, Ruzyllo W, Gottlieb S, et al. 2007. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med 357:2001–2015.
- Yip VL, Hawcutt DB, Pirmohamed M. 2015. Pharmacogenetic Markers of Drug Efficacy and Toxicity. Clin Pharmacol Ther 98:61–70.
- Zhai Y, He H, Ma X, Xie J, Meng T, Dong Y, et al. 2017. Meta-analysis of effects of ABCB1 polymorphisms on clopidogrel response among patients with coronary artery disease. Eur J Clin Pharmacol 73:843–854.
- Zhang L, Cui G, Li Z, Wang H, Ding H, Wang DW. 2013. Comparison of high-resolution melting analysis, TaqMan Allelic discrimination assay, and sanger sequencing for Clopidogrel efficacy genotyping in routine molecular diagnostics. J Mol Diagn 15:600–606.
- Zhou Q, Yu XM, Lin HB, Wang L, Yun QZ, Hu SN, et al. 2009. Genetic polymorphism, linkage disequilibrium, haplotype structure and novel allele analysis of CYP2C19 and CYP2D6 in Han Chinese. Pharmacogenomics J 9:380–394.
- Zhuo ZL, Xian HP, Long Y, Liu C, Sun YY, Ma YT, et al. 2018. Association between CYP2C19 and ABCB1 polymorphisms and clopidogrel resistance in clopidogrel-treated Chinese patients. Anatol J Cardiol 19(2):123–129.