Abstract
A combination of fluorescence spectroscopy and molecular dynamics (MD) is applied to assess the conformational dynamics of a peptide making up the outermost ring of the nicotinic acetylcholine receptor (AChR) transmembrane region and the effect of membrane thickness and cholesterol on the hydrophobic matching of this peptide. The fluorescence studies exploit the intrinsic fluorescence of the only tryptophan residue in a synthetic peptide corresponding to the fourth transmembrane domain of the AChR γ subunit (γM4-Trp6) reconstituted in lipid bilayers of varying thickness, and combine this information with quenching studies using depth-sensitive phosphatidylcholine spin-labeled probes and acrylamide, polarization of fluorescence, and generalized polarization of Laurdan. A direct correlation was found between bilayer width and the depth of insertion of Trp6. We further extend our recent MD study of the conformational dynamics of the AChR channel to focus on the crosstalk between M4 and the lipid-belt region. The isolated γM4 peptide is shown to possess considerable orientational flexibility while maintaining a linear α-helical structure, and to vary its tilt depending on bilayer width and cholesterol (Chol) content. MD studies also show that γM4 also establishes contacts with the other TM peptides on its inner face, stabilizing a shorter TM length that is still highly sensitive to the lipid environment. In the native membrane the topology of the M4 ring is likely to exhibit a similar behavior, dynamically modifying its tilt to match the hydrophobic thickness of the bilayer.
Introduction
The nicotinic acetylcholine receptor (AChR; Karlin [Citation2002]) belongs to the superfamily of ligand-gated ion channels (LGIC; Lester et al. [Citation2004]), together with the 5-HT3 (a subtype of serotonin receptor), GABA, and glycine receptors. The AChR, the best characterized member of the LGIC superfamily, is an integral membrane protein composed of homologous subunits organized pseudo-symmetrically as a pentamer around a central pore, most typically with an α2βγδ stoichiometry. Each AChR subunit contains a relatively large amino-terminal extracellular domain and four membrane-embedded hydrophobic domains of 20–30 amino acids in length (M1–M4) connected by loops of varying length and ending with a very short extracellular carboxyl terminus (Karlin [Citation2002]).
The topology and lipid–protein interactions of individual AChR transmembrane segments have been studied using isolated or synthetic peptides. According to NMR data M2 inserts in the lipid bilayer at an angle of 12° relative to the bilayer normal (Opella et al. [Citation1999]), and is either straight (Opella et al. [Citation1999]) or presents a slight kink half-way through its length (Miyazawa et al. [Citation2003]). MD simulation studies of the M2 helix bundle suggest kinking motions in this segment (Hung et al. [Citation2005]). For M1, fluorescence experiments (Barrantes et al. [Citation2000]) and more recently NMR data (de Planque et al. [Citation2004a], Citation[b]), indicate that M1 is kinked – or contains non-helical elements – in the region of the proline residues. Two-dimensional 1H NMR spectroscopy (Lugovskoy et al. [Citation1998]) and CD and FTIR spectroscopy indicated α-helical structure for M3 and M4 segments (Corbin et al. [Citation1998]). More recently, the availability of the cryoelectron microscopy data has prompted MD simulations/modelling exercises of individual TM peptides, such as δM2 (Kessel et al. [Citation2003]; Law et al. [Citation2003]; Kim et al. [Citation2004]; Saiz & Klein [Citation2005]). We have recently undertaken a 35-ns MD simulation to explore the conformational dynamics of the AChR channel, in a large dipalmitoyl-phosphatidylcholine (DPPC) bilayer (Xu et al. [Citation2005]).
In the present work we focus on the outer ring (Barrantes [Citation2003], [Citation2004]) of the AChR TM region, conformed by M4 TM segments, which establishes close interactions with neighboring lipid molecules, and is totally secluded from contact with the AChR pore region. We apply a combination of experimental and in silico methods – fluorescence spectroscopy and MD simulations – to study the γM4 peptide alone or with the rest of the helix bundle (γM1-3), embedded in different host lipid membranes, with or without cholesterol (Chol), in which the degree of hydrophobic peptide/bilayer mismatch was varied. We establish the topology of the only tryptophan (Trp) residue in the γM4-Trp6 28-mer peptide by differential fluorescence quenching with spin-labeled PCs and with acrylamide. We identify bilayer forces that influence the orientation of γM4: an increase in the degree of mismatch between the peptide length and the bilayer width caused an increment in the tilt of γM4-Trp6 with respect to the bilayer normal. Our results indicate that the orientation in the membrane of γM4 – a straight α-helix (Xu et al. [Citation2005]) – is sensitive to bilayer thickness and Chol content.
Materials and methods
Materials
A peptide corresponding to the Torpedo californica AChR TM segment γM4 (γM4-Trp6) and the two extramembranous regions, and having the sequence DKACFWIALLLFSIGTLAIFLTGHFNQV (28-mer peptide) was purchased from Biosynthesis Inc., Lewisville, TX. The resulting peptide was deemed to be over 90% pure as determined by mass spectroscopy and analytical HPLC. M4 was kept lyophilized at −80°C until use. The spin labeled PCs, derived from the 1-palmitoyl-2-stearoyl-phosphatidylcholine substituted at positions 5 (5-SLPC), 7 (7-SLPC), 10 (10-SLPC), and 12 (12-SLPC), were obtained from Avanti Polar Lipids, Birmingham, AL. All other drugs were obtained from Sigma Chem. Co., St Louis.
Methods
Sample preparation
Mixtures of lipids dissolved in chloroform: methanol (2:1 v/v), and γM4-Trp6 peptide dissolved in ethanol, were first dried under nitrogen (1 h) and then multilamellar vesicles (MVL) were obtained following conventional procedures. Briefly, 300 µl of buffer A (150 mM NaCl, 0.25 mM MgCl2, and 20 mM Hepes buffer, pH 7.4) were added to the dried sample (0.5 mg/ml lipid:buffer relationship), vortexed for 2 min and finally bath sonicated (Voglino et al. [Citation1999]). Final peptide and lipid concentrations were 3.75 µM and 150 µM, respectively. For parallax measurements, vesicles with the addition of 10% quencher-(nitroxide)-labeled PCs were prepared. Different lipids were used for this purpose: dimyristoylphosphatidylcholine (DMPC) (phase transition temperature [Tt] = 23°C; Ladbrooke & Chapman [Citation1969]), dipalmitoylphosphatidylcholine (DPPC) (Tt=41.2°C; Füldner [Citation1981]), dioleoylphosphatidylcholine (DOPC) (Tt= − 21°C; Ladbrooke & Chapman [Citation1969]) and dilaurylphosphatidylcholine (DLPC) (Tt= − 1.8°C; Mabrey & Sturtevant [Citation1976]).
The thickness of the hydrocarbon core of a gel and a liquid-crystalline phase bilayer (DG and DLC, respectively) was calculated according to Sperotto and Mouritsen ([Citation1988]) and Lewis and Engelman ([Citation1983]) as follows:1
2 in which NC is the number of carbon atoms in the fatty acyl chains.
Fluorescence measurements
All fluorimetric measurements were performed in an SLM model 4800 fluorimeter (SLM Instruments, Urbana, IL) using the vertically polarized light beam from a Hannovia 200 W Hg/Xe arc obtained with a Glan–Thompson polarizer (4 nm excitation and emission slits) and 5×5 mm quartz cuvettes. The temperature was set with a thermostated circulating water bath (Haake, Darmstadt, Germany).
Parallax measurements
Measurements were performed with an excitation wavelength of 290 nm and an emission wavelength of 326 mn in the presence (F) and absence (Fo) of 10 mol% doxyl PCs. Parallax analysis employed the following formalism (Chattopadhyay & London [Citation1987]):3 where ZCF is the distance of the Trp residue from the bilayer center, LC1 represents the distance from the bilayer center to the shallow quencher, F1 and F2 are the relative fluorescence intensities in the presence of the shallower and deeper quenchers, respectively, C is the mole fraction of quencher divided by the lipid area (70 Å2), and L21 is the difference in the depth of the two quenchers. When the Trp residue was deeply buried in the membrane (indicated by a ZCF value <5 Å) the following equation was used for the calculation of ZCF (Ren et al. [Citation1997]):
4 where LC2 is the distance from the bilayer center to the deeper quencher.
Upon obtention of the ZCF, the probe insertion depth (SHF) was calculated as follows:5
When experiments were carried out in the presence of Chol, it was not possible to estimate the exact thickness of the bilayer. Thus, instead of using Equation 5 to calculate SHF, we modified Equation 3 in order to directly calculate the probe's insertion depth (SHF) in the bilayer as follows:6 where SH1 is the distance from the hemilayer “hydrocarbon” surface to the shallow quencher. This is equal to the hemilayer thickness minus the LCi of each spin-labeled probe. Data of Chattopadhyay and London ([Citation1987]) was used for these calculations (a hemilayer thickness of 15 Å, and LCi values of 12.15, 10.35, 7.65 and 5.85 for 5-SLPC, 7-SLPC, 10-SLPC and 12-SLPC, respectively).
Quenching of Trp6 emission by acrylamide
Aliquots of a 9.0 M acrylamide solution were added to the samples. The scatter contribution was derived from acrylamide titration of lipid vesicles without the Trp6 peptide. The excitation wavelength was 295 nm, chosen in order to reduce acrylamide absorbance and the inner filter effect. Inner filter effects were corrected according to the following formula:7 where Fobs is the measured fluorescence intensity of the Trp and ODex and ODem are the optical densities of the sample at the excitation and emission wavelength, respectively (Lakowicz [Citation1999]). The data were analysed according to the Stern–Volmer equation,
8 where Fo and F correspond to the fluorescence emission of Trp in the absence and presence of acrylamide, and [Q] is the concentration of the quencher. Plots of Fo/F versus [Q] yield a slope equal to KSV, the Stern–Volmer constant. In order to discard the possibility that acrylamide failed to quench the Trp fluorescence because of the multilamellar nature of the preparation, control experiments were undertaken by reconstituting individual samples of the peptide with varying acrylamide concentrations, such that the interbilayer concentration of acrylamide matched that of the bulk solution. No differences were observed in Fo/F between these and the conventional multilamellar vesicles (MLV) samples.
Q-ratio*
The Q-ratio* values were calculated as in Caputo and London ([Citation2003]) from the formula,9 where Fo is the fluorescence of a sample lacking quencher and Facrylamide and F10-SLPC are the fluorescence intensities in the presence of acrylamide or 10-SLPC, respectively.
Laurdan measurements
Laurdan was added to the liposomes from an ethanol solution to give a final probe concentration of 0.6 µM. The amount of organic solvent was kept below 0.1%. The samples were incubated in the dark for 60 min at room temperature.
a) Generalized polarization (GP) measurements of Laurdan employed the following algorithm (Parasassi et al. [Citation1990], [Citation1991]):10 where I434 and I490 are the emission intensities at the characteristic wavelength of the gel phase (434 nm) and the liquid-crystalline phase (490 nm), respectively. The excitation wavelength used was 360 nm.
b) Anisotropy measurements were carried out in the T format with Schott KV418 filters in the emission channels and corrected for optical inaccuracies and for background signals. The anisotropy value (r) was obtained as follows:11 where (Iv/Ih)v and (Iv/Ih)h are the ratios of the emitted vertical or horizontally polarized light to the exciting, vertical or horizontally polarized light, respectively. The excitation wavelength used was 360 nm.
Data analysis
Intergroup comparisons were carried out using the paired Student's t-test.
Molecular dynamics (MD) simulations
Structures of the single γM4 peptide and of γM4 together with the rest of the helix bundle (γM1-3) were taken from the structure determined by Miyazawa et al. ([Citation2003]) (PDB code 1OED) as in Xu et al. ([Citation2005]). Three models were developed in the present study: simulation I (isolated γM4 peptide in POPC), simulation II (isolated γM4 peptide in DPPC) and simulation III γM4 peptide together with γM1-3 in DPPC). In simulations I and II, the γM4 peptide was built with 33 residues: (VIDKACFWIALLLFSIGTLAIFLTGHFNQVPEF). In simulation III the sequence of γM4 (DKACFWIALLLFSIGTLAIFLTGHFNQV, 28 residues) was the same as that of the synthetic γM4-Trp6 peptide used in the fluorescence experiments, with no connecting loop between γM4 and the rest of the γM1-3 helix bundle. The set-up of the peptide/bilayer/water system for MD simulations was essentially as described previously (Xu et al. [Citation2005]; Tieleman & Berendsen [Citation1996]; Tieleman et al. [Citation1999a]; Law et al. [Citation2000]). Two lipid molecules and 17 lipid molecules were removed from the bilayer to fit in the single γM4 peptide and the γM4 peptide together with the γM1-3 bundle, respectively. The protein/DPPC system was then solvated in a bath of SPC water molecules (Berendsen et al. [Citation1981]). The resulting system was submitted to energy minimization to remove unfavorable contacts and equilibrated for 1 ns with positional restraints on the protein atoms. Cl− or Na+ counterions were then added to the system to provide a neutral simulation cell and the whole system was minimized once more.
MD simulations were carried out with a GROMACS package (Berendsen et al. [Citation1995]; Lindahl et al. [Citation2001]) using NPT and periodic boundary conditions. The modified GROMOS87 force field (van Gunsteren & Berendsen [Citation1987]) was applied for protein and the lipid parameters adopted were those used in previous MD studies of lipid bilayers (Berger et al. [Citation1997]; Marrink et al. [Citation1998]; Tieleman et al. [Citation1999a], Citation[b]). The LINCS method (Hess et al. [Citation1997]) was used to constrain bond lengths, allowing an integration step of 2 fs. The structures for analysis were saved every 500 steps (1ps). The electrostatic interactions were calculated using the Particle-Mesh Ewald (PME) algorithm (Essmann et al. [Citation1995]). The cut-off for van der Waals interactions was 0.9 nm. A constant pressure of 1 bar was applied independently on X, Y and Z directions of the whole system with a coupling constant of 1.0 ps (Berendsen et al. [Citation1984]). Water, lipids and peptide were coupled separately to a temperature bath (300K, 27°C, for POPC and 323K, 50°C, for DPPC) using a coupling time of 0.1 ps (Berendsen et al. [Citation1984]). Afterwards, the peptide/lipids/water systems were submitted to a 30 ns equilibrium simulation. Throughout the entire duration of simulation III, the main-chain atoms of γM1, γM2 and γM3 were fixed with force constant 1000. MD simulations were run on a 128-CPU Silicon Graphics (Mountainview, CA) Origin3800 server. Analyses were performed using facilities within the GROMACS package. Secondary structure analysis employed the DSSP (define secondary structure of proteins) definitions (Kabsch & Sander [Citation1983]).
Results
Intrinsic fluorescence and partitioning of the γM4-Trp6 peptide in different bilayer systems
γM4-Trp6 is a 28-mer peptide obtained by solid-state synthesis and having the following sequence: DKACFW6IALLLFSIGTLAIFLTGHFNQV, predicted by hydropathy plots to be the bilayer-embedded fourth TM segment of the Torpedo californica AChR γ subunit. It possesses charged/polar residues at its two extremes and a single Trp residue at position 6, which constitutes a convenient intrinsic probe for studying peptide topology with respect to the membrane and a useful landmark for secondary structure studies using fluorescence spectroscopy.
In order to determine peptide orientation with respect to the membrane bilayer, γM4-Trp6 was reconstituted in different pure lipid systems (DLPC, DOPC, DMPC and DPPC, for details see Materials and methods), which vary in bilayer width (see ). In all the reconstituted systems analysed, the fluorescence spectra showed one main emission band with similar intensity. The emission maximum varied between 326 and 328 nm, indicating a hydrophobic environment for the Trp6 residue, and the effective insertion of the peptide into the bilayer. The fluorescence emission spectrum for Trp in water is centered at about 360 nm, whereas that of a Trp residue of a peptide located in the hydrophobic region of a lipid bilayer is centered at 323–330 nm, depending on bilayer thickness and peptide length (Webb et al. [Citation1998]).
Table I. Bilayer width (BW) of different synthetic PCs and variation in the distance of Trp6 in γM4-Trp6 to the membrane interface (i.e., insertion depth, SHF) as a function of bilayer thickness.
In order to corroborate that γM4-Trp6 was indeed inserted into the hydrophobic region of the bilayer and to assess its sensitivity to bilayer thickness and phase state, we used the effective water-soluble quencher, acrylamide, to perform quenching experiments in different reconstituted lipid systems (two in the liquid/crystalline phase, DLPC and DOPC, and two in the gel phase, DMPC and DPPC at 15°C). Quenching by acrylamide obeys a collisional mechanism, in which fluorophore molecules need to be accessible to the quencher for quenching to occur; if the Trp residue(s) is(are) protected in the protein core or embedded in the membrane, fluorescence is not expected to be quenched (see e.g., Lakowicz [Citation1999]). The fluorescence intensity of γM4-Trp6 exhibited only a negligible concentration-dependent decrease upon addition of acrylamide. Stern–Volmer plots of γM4-Trp6 fluorescence quenching in different reconstituted lipid systems yielded similar values of Ksv (2.7±0.5 M−1 for DLPC, 2.0±0.1 M−1 for DMPC at 15°C, 2.0±0.1 M−1 for DPPC at 15°C, and 3.1±0.5 M−1 for DOPC). Comparison of the experimental Ksv values with those reported for Trp quenching in water (Ksv=13.7; Coutinho & Prieto [Citation1993]) suggests that γM4-Trp6 is poorly accessible to the quencher. As the Trp is located in the 6th position in the peptide sequence, this result indicates that the peptide is effectively inserted into the hydrophobic region of all the bilayer systems tested. Also, addition of the peptide to only buffer caused its precipitation and no fluorescence signal. This, together with the fact that all the reconstituted samples exhibited similar fluorescence intensity values, suggests that the incorporation was efficient and quantitatively similar for the different reconstitution conditions.
Depth of the single Trp residue in γM4-Trp6
The topography of the only Trp residue in γM4-Trp6 with respect to the membrane bilayer was studied directly using fluorescence quenching in combination with the parallax analytical method (Chattopadhyay & London [Citation1987]). In this method, the depth of a fluorophore is calculated from the ratio of its quenching by lipids carrying a nitroxide spin-label at different depths. shows the correlation between Trp fluorescence quenching and bilayer width for γM4-Trp6 reconstituted in three lipid systems. As the bilayer width increased (DLPC < DOPC < DMPCgel) γM4-Trp6 was quenched more efficiently by probes having their nitroxide groups deeper in the bilayer.
Figure 1. Normalized fluorescence intensity of γM4-Trp6 at 328 nm in reconstituted liposomes of different bilayer width with SLPCs. Trp6 in reconstituted liposomes of DLPC (____), DOPC (_ _ _ _) and DMPC at 11°C (......) with or without SLPCs having a nitroxide group at different depths (positions 5, 7, 10 and 12 of the phospholipid acyl chain). The fluorescence intensity values obtained in the presence of SLPC were normalized with respect to the corresponding fluorescence intensity value in the absence of spin probe (100%).
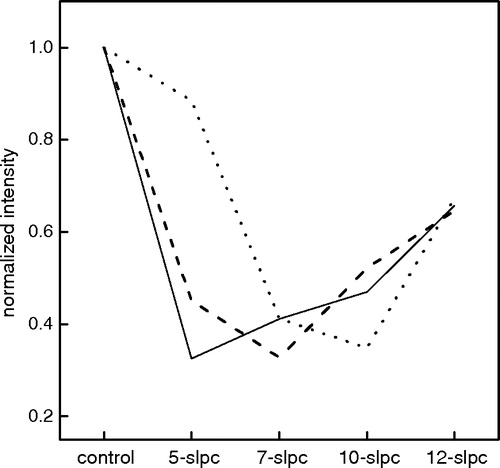
The depth of Trp insertion in the bilayer (SHF) was defined as the distance (perpendicular to the plane of the membrane) from the polar head region/ hydrophobic region interface to the only Trp residue, Trp6. SHF increased in parallel with the increment in bilayer thickness, clearly indicating that Trp6 penetrates deeper into the membrane as the bilayer width increases (see ).
Figure 2. (a) γ-M4-Trp6 insertion depth (SHF) as a function of hemilayer width. From left to right the points correspond to the experimentally measured position of γM4-Trp6 peptide reconstituted in DLPC, DMPC at 37°C, DPPC at 52°C, DOPC, DMPC at 12°C, and DPPC at 30°C. Inset: Q-ratio* values as a function of hemilayer width. Each point corresponds to the average±SD of at least four independent determinations. (b) Laurdan GP (♦, ▪, •, ▴) and anisotropy (⋄, □, ○, ▵) as a function of the depth of insertion of Trp6 (SHF) in liposomes of DMPC with different amounts of Chol. From left to right: 0% (♦, ⋄), 15% (▪,□) and 30% (•,○) Chol in DMPC liposomes. Experiments were carried out at 37°C to ensure that DMPC was in the liquid-crystalline phase. The same parameters are presented for DMPC without Chol at 11°C (gel phase, ▴, ▵) for comparison (SHF=6.04).
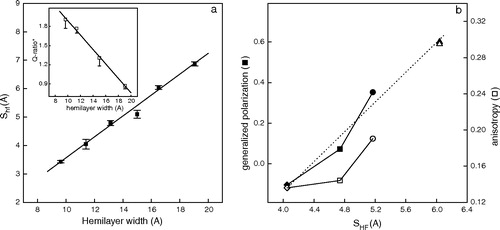
The ratio of acrylamide quenching to that of 10-DN quenching (Q-ratio) was found to be linearly dependent on the depth of the Trp in the membrane (Caputo & London [Citation2003]). We calculated a similar ratio value (Q-ratio*) by comparing the quenching of acrylamide to that of 10-SLPC (see , inset). Q-ratio* values decreased alongside the increment in bilayer width. The inverse relationship between SHF and the Q-ratio* confirms our results: as the bilayer becomes wider the Trp localizes deeper in the hydrocarbon region.
Native AChR-rich membranes are particularly rich in Chol, and Chol is known to stabilize the structure and function of the AChR protein (see review in Barrantes [[Citation2003]). Baenziger et al. (2000) showed that AChR reconstituted in PC alone is stabilized in a desensitized-like state, whereas reconstitution in the presence of either Chol or phosphatidic acid stabilizes the AChR in the resting state. These authors postulated a model in which the equilibrium between the two states is adjusted by bulk physical properties of the lipid bilayer. One such bulk physical property is bilayer width, which is modified (increased) by Chol levels. Bilayer width, in turn, influences the behavior of TM peptides (Bretscher & Munro [Citation1993]). We therefore studied the influence of Chol on the γM4-Trp6 peptide reconstituted in synthetic bilayers of DMPC at 38°C (i.e., in the liquid-crystalline phase). First we studied the relationship between the changes produced by the presence of increasing Chol concentrations on the physical characteristics of the bilayer, using the fluorescent probe Laurdan. shows the correlation between the increase in SHF and the so-called GP and anisotropy values of Laurdan, respectively. Upon increasing Chol concentration the phase state of the membrane changed from liquid-crystalline (0% Chol) to liquid-ordered (30% Chol), with a concomitant increase in both parameters. This shows that Chol caused lateral mobility (as judged by the increment in fluorescence anisotropy values) to decrease and the order of the liquid phase (as judged by the increment in Laurdan GP) to increase. It is well known that Chol content is one of the key physiological factors controlling bilayer thickness (Nezil & Bloom [Citation1992]). This is corroborated here with the increase in acyl-chain order, rendering the bilayer thicker. One can also observe that the position of Trp6 in the bilayer varies as a function of Chol content (see ): the SHF value increased as Chol content increased. Thus, the γM4-Trp peptide is not only sensitive to bilayer thickness in a pure phospholipid system () but also to the bilayer width and phase state in a more realistic, Chol-containing bilayer ().
In a previous work the γM4 peptide obtained by controlled proteolysis of the AChR was shown to exhibit α-helical secondary structure in a reconstituted lipid (asolectin) system of mixed composition (Barrantes et al. [Citation2000]). Here we used pure synthetic lipid bilayers to analyse the possible helix-tilting dependence of a synthetic γM4-Trp6 on bilayer thickness. We assumed the helix tilt to be proportional to the ratio of bilayer width to the length of the hydrophobic peptide, as this ratio has been suggested to be a major determinant of TM segment orientation (Bretscher & Munro [Citation1993]). As the same peptide was used in all tested samples, the bilayer width was the major, if not the only variable. The bilayer width was changed using various strategies: (a) using PCs of different acyl chain lengths; (b) using the same PC in either the gel or the liquid-crystalline state, respectively; or (c) using the same PC in the liquid phase but containing varying amounts of Chol. It is important to stress that despite possible differences in membrane biophysical properties (e.g., lipid order, lateral mobility) among the different experimental paradigms, all the combinations resulted in changes in bilayer width.
We calculated a “helix chain tilt parameter” (HCTPexp) as follows (see ):12 where Trph is the distance from the first hydrophobic amino acid in γM4-Trp6 peptide (Ala3) to the Trp6 residue, measured for an α-helical conformation obtained by molecular rendering of the γM4 segment. We assumed one fixed entry point for γM4 at residue Lys2, anchored at the interface polar headgroup region of the bilayer. We followed the criterion that the hydrophobic thickness of membrane polypeptides can be estimated by the location of charged residues – as charged residues often mark the ends of transmembrane α-helices (reviewed in Lee [[Citation2003] and references therein). The cost of burying a charged residue within the hydrocarbon core of a lipid bilayer is so high (about 37 kJ mol−1 for a Lys residue; Engelman et al. [Citation1986]) that unpaired charged residues at the ends of transmembrane α-helices are much more likely to be located in the headgroup region than in the hydrophobic core of the bilayer. The criterion that we followed is well-established: Lee ([Citation2003]) listed the hydrophobic thickness of almost 30 intrinsic membrane proteins with known high-resolution structure; in 21 of them the hydrophobic thickness was set as the (minimal) distance between charged residues and/or Trp residues at the two sides of the bilayer. Furthermore, in our study the bilayer thickness was always similar to or smaller than the hydrophobic length of the peptide, discarding the possibility that Lys residues resided within the hydrophobic core of the bilayer and snorkeled up to the surface to expose their charged group in the headgroup region of the bilayer. This possibility would apply if the bilayer were thicker than the hydrophobic length of the peptide. Finally, the cryoelectron microscopy study of Miyazawa et al. ([Citation2003]) at a resolution of 4 Å also defined residue Lys2 as the first amino acid of the γM4 TM peptide, and positioned it at the polar headgroup interface region.
Figure 3. (a) Schematic diagram indicating the different parameters taken into account to calculate the helix chain tilt parameters (HCTP) for the γ-M4-Trp6 peptide as a linear α-helix in the bilayer (see text for details). (b) Experimental and calculated tilt parameters as a function of bilayer width. From left to right the experimental (HCTPexp, ▪) and calculated (HCTP25, □; HCTP21, ○) tilt parameter points correspond to values of γ-M4 peptide reconstituted in DLPC, DMPC at 37°C, DPPC at 52°C, DOPC, DMPC at 12°C, and DPPC at 30°C, respectively. HCTPsim corresponds to the MD simulations: I) γM4 in POPC at 27°C (▾), II) γM4 in DPPC at 50°C (▴), and III) γM4 helix modeled with the rest (γM1-3) of the helix bundle in DPPC at 50°C (♦). HCTP obtained for the γM4 of the whole AChR in native membranes of T. californica (•, arrow). This Figure is reproduced in colour in Molecular Membrane Biology online.
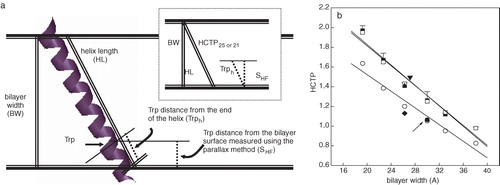
From it is clear that the helix is aligned more perpendicular (i.e., less tilted, lower HCTPexp values) to the bilayer normal as the bilayer becomes wider, reaching HCTPexp values near unity at high bilayer widths (implying that the bilayer width matches the length of the hydrophobic peptide).
To further test whether an α-helical structure was compatible with the experimental data, we also calculated the HCTP for membrane-embedded γM4 peptides considering two different transmembrane lengths: A 25-mer hydrophobic TM domain (HCTP25, which comprises the TM region formed by all the hydrophobic amino acids of the sequence starting with Ala3), and a 21-mer (following the recent work of Miyazawa et al. ([Citation2003]), in which they depict all M4 segments, including γM4, as linear α-helices in which only 21 amino acids conform the hydrophobic TM domain (HCTP21)), starting at Lys2 as an interfacial amino acid and having Ala3 as the first hydrophobic amino acid. 13 where BW is the bilayer width and HL the helix length, calculated assuming a length of 1.5 Å per hydrophobic amino acid (Harzer & Bechinger [Citation2000]). compares HCTPexp with the HCTP25 and HCTP21 as a function of bilayer width. HCTP25 corresponds to a theoretical straight α-helical peptide whose membrane-embedded domain is equivalent to the hydrophobic amino acid sequence of the segment. HCTP25 presented a tendency similar to that of HCTPexp as a function of bilayer width, which is a solid indication that: (a) the experimentally used γM4 peptide is a straight α-helical structure; and (b) γM4 adapts to its membrane environment by displaying orientational flexibility (i.e., by tilting in order to minimize the hydrophobic mismatch).
The HCTP21 absolute values differed from those of the other HCTP values. This indicates that in the case of γM4-Trp6 it is most probable that more than 21 amino acids are inserted inside the hydrophobic core, which is highly likely considering that residue 22 and the subsequent ones are also hydrophobic, and that in the presence of the rest of the AChR protein-protein interactions stabilize a shorter transmembrane M4 segment.
The question arose next as to the orientation of γM4 in the intact AChR molecule at the postsynaptic membrane. To address this question we calculated an HCTP value from (a) the tilt angle distended by the corresponding TM segment in the cryoelectron micrographs of Miyazawa et al. ([Citation2003]), which turned out to be ∼20°; and (b) the bilayer thickness in Torpedo membranes. The latter could be calculated from the X-ray diffraction study of Ross et al. ([Citation1977]). From their value of 40±3 Å for the distance between the highest electron-dense peaks on either side of the negative density region, interpreted as the center of the bilayer, we subtracted twice the length of the glycerol-group region (∼5 Å) from the phosphate to the first methylene of the hydrocarbon chain (Harroun et al. [Citation1999]), resulting in a value of ∼30 Å for the hydrophobic width of the bilayer in Torpedo californica membranes. The calculated HCTP using this information falls exactly in the curve generated with the HCTP21 values (, arrow), suggesting that the tilt angle of γM4 in the intact AChR macromolecule differs from that of the isolated γM4 in a model system (but see below).
Molecular dynamics (MD) simulations
MD simulations were performed to determine the conformational stability and orientation of the γM4 peptide in a lipid bilayer. In simulations I and II only the single γM4 helix was inserted in a bilayer (POPC or DPPC), while in simulation III the γM4 helix was modeled together with the rest of the helix bundle (γM1-3) in a DPPC bilayer, with the aim of resolving the discrepancy between our fluorescence experimental results and the postulated membrane-embedded length of γM4 in Miyazawa et al. ([Citation2003]), obtained using AChR-rich tubules prepared from Torpedo membranes. The main-chain root mean square deviation (RMSD) of the main-chain atoms of γM4 (Supplementary material) showed that γM4 in DPPC (simulation II) is more flexible in this lipid environment in comparison to the other two bilayer systems, whereas γM4 is most stable in the presence of the rest of the γM1-3 helix bundle (simulation III). No large increase or decrease in the RMSD was apparent in any of the three simulations, which remained in all cases below 2.5 Å, suggesting that the conformation of the peptide is stable in all the bilayer systems studied. A more fine-grained description of the conformational dynamics of γM4 in bilayer lipids was provided by analysis of the time-dependent secondary structure using the DSSP algorithm (Kabsch & Sander [Citation1983]) for the three different simulations (data not shown). From this analysis it was apparent that γM4 always maintained its α-helicity in the bilayer lipids (POPC and DPPC), either as an isolated peptide or together with the γM1-3 helix bundle. In addition, there were no kinks or breaks in the α-helix stretch. Therefore the results of the three simulations concurred with the fluorescence experimental data in that γM4 is a linear membrane-spanning α-helix. It is important to bear in mind that in the case of the 33-mer peptide, the five terminal amino acid residues are polar and therefore most likely reside outside the hydrophobic core of the membrane. In both simulations I and II such terminal amino acids were excluded from the membrane-spanning region, whereas in simulation III all amino acids were included in the membrane-spanning α-helix. It should further be noted that the hydrophobic length was the same for the two peptides, and also similar to that of the γM4-Trp6 used in the fluorescence experiments.
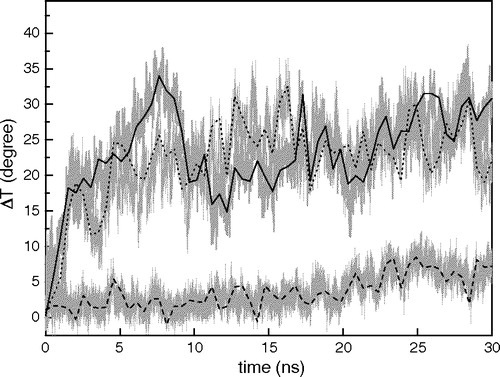
We regarded the angle between the axis of the γM4 peptide and the Z-axis (i.e., the axis perpendicular to the plane of the bilayer) as the tilt angle of the peptide with respect to the bilayer, whereas the X-axis is parallel to the plane of bilayer. shows the change in the tilt angle resulting from each MD simulation as a function of time. At time zero the peptide was positioned at 20°, as in Miyazawa et al. ([Citation2003]). The α values for the isolated γM4 peptide increased during the first 10 ns and subsequently reached equilibrium at ∼30 ns, both in POPC and DPPC. In contrast, when γM4 was modeled together with the γM1-3 helix bundle, the tilt angle reached equilibrium immediately, and maintained its position during the rest of the time window of the MD simulation.
Figure 4. Changes of the angle (ΔT) between the axis of γM4 and the Z-axis along each 30 ns trajectory. Three MD simulations were analyzed: I)? γM4 in POPC at 27°C (____), II) γM4 in DPPC at 50°C (......), and III) γM4 helix modeled with the γM1-3 bundle in DPPC at 50°C (---). The centers of mass of residues 5–8 and 20–23 are set as the top and bottom of the axis of the γM4 helix, respectively.
An equilibrated tilt angle (α) was attained for each condition, and an HCTPsim was calculated from the MD data as follows (see ):14
The HCTPsim values obtained for γM4 in DPPC and POPC fell well within the curve described by HCTP25 (and HCTPexp), which corresponds to the γM4 peptide with its hydrophobic portion totally inserted in the membrane. The HCTPsim value obtained for γM4 modeled together with the γM1-3 helix bundle came close to the values of HCTP21, in which only 21 residues were considered in the membrane-embedded domain (Miyazawa et al. [Citation2003]), as was the case with the value obtained for γM4 in the whole AChR. Thus γM4 adopts two possible arrangements depending on the presence or absence of the rest of the bundle. The two curves have similar slopes, indicating that in spite of the difference in hydrophobic length between the two γM4 peptides, γM4 adapts in a similar manner to its lipid environment.
Discussion
The recent cryoelectron microscopy studies of Unwin and coworkers (Miyazawa et al. [Citation2003]; Unwin [Citation2005]) represent the highest resolution structural work on the AChR produced to date. The present work constitutes the first successful attempt to test the solid-state structural information using spectroscopic techniques with sub-nanometer resolution in solution, and correlate the experimental data with in silico molecular dynamics studies.
Intrinsic fluorescence spectra and acrylamide quenching experiments demonstrated that γM4-Trp6 effectively partitioned into different bilayer lipid systems (γM4-Trp6 exhibiting wavelength emission maxima near 328 nm, and low Ksv values in the presence of acrylamide), irrespective of which lipids were used in the host bilayer. In order to address the issue of γM4-Trp6 secondary structure in the membrane we studied the transbilayer location of the single Trp6 in γM4-Trp6. Trp6 should localize at different bilayer positions depending on whether the peptide adopts an α-helical or a non-helical, e.g., a pleated β-structure (the only possible TM structural configurations of membrane proteins to minimize the high energetic cost of dehydrating the peptide bond; White et al. [Citation2001]). Spin-labeled PCs, having nitroxide groups at different positions, were used as molecular rulers that enabled us to determine the position of Trp6 in different bilayer systems. As the bilayer became thicker, Trp6 was quenched more efficiently by nitroxide probes located deeper in the bilayer (see ). Further analysis of the data enabled us to calculate the depth of insertion of Trp6 in the bilayer (). Using differential quenching of pyrene-labeled whole AChR and constituent peptides by fatty acid, phospholipid, and steroid spin labels we determined that pyrene-labeled Cys residues in the AChR were located in shallow positions in the membrane (Barrantes et al. [Citation2000]). In particular, the pyrene-labeled Cys451 residue, separated by only two residues from Trp453 in γM4 (Cys4 and Trp6, respectively, in the nomenclature adopted here for γM4-Trp6), was found to lie near the lipid–water interface. The same pattern was obtained for reconstituted whole AChR and individual, isolated AChR TM peptides, thus reinforcing the view that studies with isolated membrane peptides can provide faithful information on the topography of the naturally occurring segment in the native protein (Barrantes et al. [Citation2000]).
Upon analysing the location of Trp6 in the membrane, a direct relationship between bilayer width and Trp6 depth was apparent (). Comparison between acrylamide and spin-labeled PC quenching further strengthened this view (, inset). Using liposomes with increasing Chol content, which augmented bilayer width (), the thicker the bilayer, the deeper the position of Trp6. Mismatch between the helix length and the bilayer width may be compensated for by peptide tilting and/or bilayer distortion (Ren et al. [Citation1997]). The present results suggest that γM4 adapts its tilt angle to bilayer width in order to minimize the hydrophobic mismatch with surrounding lipids. When γM4 is incorporated in a liquid-disordered lipid phase, peptide-peptide interactions are not favorable, leading to the suggestion that γM4 induces a local disordering on its surrounding lipids in order to solve the hydrophobic mismatch problem (de Almeida et al. [Citation2004]).
We challenged the cryoelectron microscopy data (Miyazawa et al. [Citation2003]) and our own experimentally observed variations in the tilt of γM4 as a response to hydrophobic mismatch with MD simulations using γM4 peptides in two host lipid systems. As observed using the whole AChR TM region (Xu et al. [Citation2005]) in a large DPPC bilayer, the present modeling studies show that the γM4 peptide retains its straight α-helical conformation in either DPPC and POPC bilayer lipids, in full agreement with the experimental fluorescence data on the synthetic γM4-Trp6 peptide, and also in agreement with magic-angle and wideline NMR, and CD spectroscopy data of γM4 in reconstituted, magnetically aligned lipid bilayers (Williamson et al. this issue). The γM4 peptide changed its orientation with respect to the membrane bilayer during the simulation process, a phenomenon that reduced the peptide/bilayer hydrophobic mismatch. We calculated four different “helix chain tilt parameter” (HCTP) values for the γM4 peptide inserted in bilayers of varying widths (), and a fifth value derived from the cryoelectron microscopy data of Miyazawa et al. ([Citation2003]). Clear differences in the absolute values were found between the HCTP values obtained for the isolated γM4 peptide (HCTPexp, HCTP25, and HCTPsim; corresponding to experimental, theoretical, and MD simulations I and II, respectively) and the HCTP values obtained for the γM4 in the presence of the γM1-3 bundle (HCTPsim, corresponding to MD simulation III) and the theoretical HCTP21. This large difference in HCTP values is clearly depicted in the snapshots of the start and end points of each simulation (see ). The smaller HCTPsim of γM4 in the latter case is most likely a consequence of the presence of the rest of the helix bundle, which stabilizes a shorter transmembrane region (21 amino acids instead of 25) (). Thus M4 adapts in a similar manner to different lipid environments. These results reinforce the view that isolated, reconstituted membrane peptides can provide faithful information on the behavior of the relevant peptide in the whole protein.
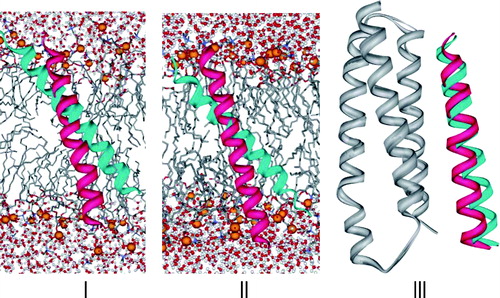
Figure 5. Ribbon snapshots of γM4 along the 30 ns trajectory. Two snapshots are presented: 0 ns (red) and 30 ns (cyan) for the three simulations: I) γM4 in POPC at 27°C, II) γM4 in DPPC at 50°C, and III) γM4 with the γM1-3 bundle (grey) in DOPC at 50°C. In I and II the phosphorus atoms of the phospholipid polar headgroups are shown in space-filling format (orange) the other atoms of the lipid are represented as sticks, water molecules are displayed as balls (red) and sticks, and the front half of the bilayer lipids is not shown to facilitate the visualization of γM4 within the core of the lipid bilayer. The lipid moiety was not included in III for the sake of clarity.
The fourth segment of each subunit, M4, located at the outermost layer of the TM region, is particularly important since it establishes more interactions with neighboring lipid molecules than any other TM segment, and is totally secluded from contact with the AChR pore region proper. Here, we demonstrate that γM4-Trp6, a synthetic γM4 peptide, possesses α-helical structure, and an extraordinary sensitivity to its lipid environment, changing its tilt angle relative to the bilayer normal in order to solve the hydrophobic mismatch resulting from insertion of the peptide in different lipid phases, changes in lipid composition, and/or Chol content. The varying tilt of the isolated γM4-Trp6 in bilayers of different width suggests that the peptide maximizes contact with lipids to optimize hydrophobic matching. The γM4 peptide also establishes contacts with itself at high concentrations (de Almeida et al. [Citation2004]) or with the other TM peptides – the γM1-3 helix bundle – predominantly on its inner face, a condition that is also likely to prevail in the intact AChR molecule, stabilizing a shorter TM length that is still highly sensitive to the lipid environment. The importance of peptide–peptide interactions became apparent in our recent MD study of the whole TM region, in which each M4 helix was found to deviate with a different degree of tilt from its initial position within the time frame of the 35-ns simulation, but the ensemble of five M4 peptides – the whole outer ring – tilted concertedly in a clockwise direction (Xu et al. [Citation2005]), thus maximizing its hydrophobic interactions with the surrounding lipid molecules by burying itself deeper into the hydrophobic region of the bilayer. The van der Waals interactions between the five M4 segments and DPPC reflected by the Lennard-Jones interaction energy tended to be stronger along the simulation time and fluctuated slightly around −6500 kJ/mol after ∼30 ns (Xu et al. [Citation2005]).
The present study provides new insights on the modulation exerted by the lipid microenvironment on AChR function in particular, and other membrane proteins in general. Individual amino acid mutations in the M4 AChR TM domain modulate AChR function (Lee et al. [Citation1994]; Lasalde et al. [Citation1996]; Ortiz-Miranda et al. [Citation1997]; Bouzat et al. [Citation1998]; Tamamizu et al. [Citation1999], [Citation2000]), a clear indication that AChR function is effectively influenced by M4 (reviewed in Barrantes [Citation2004]), the only member of the outermost TM “ring” (Barrantes [Citation2003]). Furthermore, changes in membrane lipid composition (Criado et al. [Citation1984]; Sunshine & McNamee [Citation1994]; Baenziger et al. [Citation2000]; daCosta et al. [Citation2002]) and/or presence of exogenous hydrophobic molecules (Andreasen & McNamee [Citation1980]; Villar et al. [Citation1988]; Bouzat & Barrantes [Citation1993a], Citation[b], [Citation1996]; Lasalde et al. [Citation1995]; Nurowska & Ruzzier [Citation1996]; Blanton et al. [Citation1999]; Santiago et al. [Citation2001]; Garbus et al. [Citation2001], [Citation2002]) also affect AChR function. Changes in AChR lipid microenvironment could induce hydrophobic mismatch between the hydrophobic TM segments and the hydrophobic width of the bilayer, as revealed here in the series of experiments on Chol addition. A representative AChR TM peptide, γM4, is able to solve this problem by exhibiting considerable orientational flexibility, which allows it to tilt in order to decrease its hydrophobic mismatch with the bilayer width. Such orientational flexibility is also observed in the presence of the rest of the bundle. Topological changes in the AChR protein in response to hydrophobic mismatch caused by hydrophobic compounds might thus constitute an alternative explanation for the modulatory effects caused by such ligands. Thus, in the native membrane the topology of the outermost M4 ring could dynamically modify its tilt to match the hydrophobic thickness of the bilayer, in particular in the presence of hydrophobic compounds favorably partitioning in the lipid-belt region of the AChR, as a variety of pharmacologically active compounds do. Future studies using the successful combination of spectroscopic and MD approaches should provide further insight into this hypothesis.
Thanks are due to Prof. M. Prieto for valuable discussions. This work was supported in part by grants from the Universidad Nacional del Sur, the Agencia Nacional de Promoción Científica (FONCYT), Argentina, and Antorchas/British Council grants to F.J.B.
References
- Andreasen TJ, McNamee MG. Inhibition of ion permeability control properties of acetylcholine receptor from Torpedo californica by long-chain fatty acids. Biochemistry 1980; 19: 4719–4726
- Baenziger JE, Morris ML, Darsaut TE, Ryan SE. Effect of membrane lipid composition on the conformational equilibria of the nicotinic acetylcholine receptor. J Biol Chem 2000; 275: 777–784
- Barrantes FJ. Transmembrane modulation of nicotinic acetylcholine receptor function. Curr Opin Drug Disc Develop 2003; 6: 620–632
- Barrantes FJ. Structural basis for lipid modulation of nicotinic acetylcholine receptor function. Brain Res Brain Res Rev 2004; 47: 71–95
- Barrantes FJ, Antollini SS, Blanton MP, Prieto M. Topography of nicotinic acetylcholine receptor membrane-embedded domains. J Biol Chem 2000; 275: 37333–37339
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys 1984; 81: 3684–3690
- Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. Interaction models for water in relation to protein hydration. Intermolecular forces, B Pullman. Reidel, DordrechtThe Netherlands 1981; 331–342
- Berendsen HJC, van der Spoel D, van Drunen R. GROMACS: A message-passing parallel molecular dynamics implementation. Comp Phys Comm 1995; 91: 43–56
- Berger O, Edholm O, Jahnig F. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys J 1997; 72: 2002–2013
- Blanton MP, Xie Y, Dangott LJ, Cohen JB. The steroid promegestone is a noncompetitive antagonist of the Torpedo nicotinic acetylcholine receptor that interacts with the lipid-protein interface. Mol Pharmacol 1999; 55: 269–278
- Bouzat C, Barrantes FJ. Hydrocortisone and 11-desoxycortisone modify acetylcholine receptor channel gating. Neuroreport 1993a; 4: 143–146
- Bouzat CB, Barrantes FJ. Effects of long-chain fatty acids on the channel activity of the nicotinic acetylcholine receptor. Recept Channels 1993b; 1: 251–258
- Bouzat C, Barrantes FJ. Modulation of muscle nicotinic acetylcholine receptors by the glucocorticoid hydrocortisone. Possible allosteric mechanism of channel blockade. J Biol Chem 1996; 271: 25835–25841
- Bouzat C, Roccamo AM, Garbus I, Barrantes FJ. Mutations at lipid-exposed residues of the acetylcholine receptor affect its gating kinetics. Mol Pharmacol 1998; 54: 146–153
- Bretscher M, Munro S. Cholesterol and the Golgi apparatus. Science 1993; 261: 1280–1281
- Caputo GA, London E. Using a novel dual fluorescence quenching assay for measurement of tryptophan depth within lipid bilayers to determine hydrophobic alpha-helix locations within membranes. Biochemistry 2003; 42: 3265–3274
- Chattopadhyay A, London E. Parallax method for direct measurement of membrane penetration depth utilizing fluorescence quenching by spin-labeled phospholipids. Biochemistry 1987; 26: 39–45
- Corbin J, Wang HH, Blanton MP. Identifying the cholesterol binding domain in the nicotinic acetylcholine receptor with [125I]azido-cholesterol. Biochim Biophys Acta 1998; 1414: 65–74
- Coutinho A, Prieto M. Ribonuclease TI and alcohol dehydrogenase fluorescence quenching by acrylamide: A laboratory experiment for undergraduate students. J Chem Educ 1993; 70: 425–428
- Criado M, Eibl H, Barrantes FJ. Functional properties of the acetylcholine receptor incorporated in model lipid membranes. Differential effects of chain length and head group of phospholipids on receptor affinity states and receptor-mediated ion translocation. J Biol Chem 1984; 259: 9188–9198
- daCosta CJ, Ogrel AA, McCardy EA, Blanton MP, Baenziger JE. Lipid–protein interactions at the nicotinic acetylcholine receptor. A functional coupling between nicotinic receptors and phosphatidic acid-containing lipid bilayers. J Biol Chem 2002; 277: 201–208
- de Almeida RFM, Loura LM, Prieto M, Watts A, Fedorov A, Barrantes FJ. Cholesterol modulates the organization of the γM4 transmembrane domain of the muscle nicotinic acetylcholine receptor. Biophys J 2004; 86: 2261–2272
- de Planque MRR, Rijkers DTS, Liskamp RMJ, Separovic F. The alphaM1 transmembrane segment of the nicotinic acetylcholine receptor interacts strongly with model membranes. Magn Reson Chem 2004a; 42: 148–154
- de Planque MR, Rijkers DT, Fletcher JI, Liskamp RM, Separovic F. The alphaM1 segment of the nicotinic acetylcholine receptor exhibits conformational flexibility in a membrane environment. Biochim Biophys Acta 2004b; 1665: 40–47
- Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Ann Rev Biophys Biophys Chem 1986; 15: 321–353
- Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LGA. A smooth particle mesh Ewald potential. J Chem Phys 1995; 103: 8577–8592
- Füldner HH. Characterization of a third phase transition in multilamellar dipalmitoyllecithin liposomes. Biochemistry 1981; 20: 5707–5710
- Garbus I, Bouzat C, Barrantes FJ. Steroids differentially inhibit the nicotinic acetylcholine receptor. Neuroreport 2001; 12: 227–231
- Garbus I, Roccamo AM, Barrantes FJ. Identification of threonine 422 in transmembrane domain alpha M4 of the nicotinic acetylcholine receptor as a possible site of interaction with hydrocortisone. Neuropharmacology 2002; 43: 65–73
- Harroun TA, Heller WT, Weiss TM, Yang L, Huang HW. Theoretical analysis of hydrophobic matching and membrane-mediated interactions in lipid bilayers containing gramicidin. Biophys J 1999; 76: 3176–3185
- Harzer U, Bechinger B. Alignment of lysine-anchored membrane peptides under conditions of hydrophobic mismatch: a CD, 15N and 31P solid-state NMR spectroscopy investigation. Biochemistry 2000; 39: 13106–13114
- Hess B, Bekker B, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comp Chem 1997; 18: 1463–1472
- Hung A, Tai K, Sansom MS. Molecular dynamics simulation of the M2 helices within the nicotinic acetylcholine receptor transmembrane domain: structure and collective motions. Biophys J 2005; 88: 3321–3333
- Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983; 22: 2577–2637
- Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nat Rev Neurosci 2002; 3: 102–114
- Kessel A, Shental-Bechor D, Haliloglu T, Ben-Tal N. Interactions of hydrophobic peptides with lipid bilayers: Monte Carlo simulations with M2delta. Biophys J 2003; 85: 3431–3444
- Kim S, Chamberlain AK, Bowie JU. A model of the closed form of the nicotinic acetylcholine receptor M2 channel pore. Biophys J 2004; 87: 792–799
- Ladbrooke BD, Chapman D. Thermal analysis of lipids, proteins and biological membranes. A review and summary of some recent studies. Chem Phys Lipids 1969; 3: 304–356
- Lakowicz JR. Principles of fluorescence spectroscopy. Kluwer Academic/Plenum Publishers, New York 1999
- Lasalde JA, Colom A, Resto E, Zuazaga C. Heterogeneous distribution of acetylcholine receptors in chick myocytes induced by cholesterol enrichment. Biochim Biophys Acta 1995; 1235: 361–368
- Lasalde JA, Tamamizu S, Butler DH, Vibat CR, Hung B, McNamee MG. Tryptophan substitutions at the lipid-exposed transmembrane segment M4 of Torpedo californica acetylcholine receptor govern channel gating. Biochemistry 1996; 35: 14139–14148
- Law RJ, Forrest LR, Ranatunga KM, La Rocca P, Tieleman DP, Sansom MS. Structure and dynamics of the pore-lining helix of the nicotinic receptor: MD simulations in water, lipid bilayers, and transbilayer bundles. Proteins Struc Funct Genet 2000; 39: 47–55
- Law RJ, Tieleman DP, Sansom MS. Pores formed by the nicotinic receptor M2delta Peptide: A molecular dynamics simulation study. Biophys J 2003; 84: 14–27
- Lee AG. Lipid-protein interactions in biological membranes: A structural perspective. Biochim Biophys Acta 2003; 1612: 1–40
- Lee YH, Li L, Lasalde J, Rojas L, McNamee MG, Ortiz-Miranda SI, Pappone P. Mutations in the M4 domain of Torpedo californica acetylcholine receptor dramatically alter ion channel function. Biophys J 1994; 66: 646–653
- Lester HA, Dibas MI, Dahan DS, Leite JF, Dougherty DA. Cys-loop receptors: new twists and turns. Trends Neurosci 2004; 27: 329–336
- Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J Mol Biol 1983; 166: 211–217
- Lindahl E, Hess B, van der Spoe Dl. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J Mol Model 2001; 7: 306–317
- Lugovskoy AA, Maslennikov IV, Utkin YN, Tsetlin V, Cohen JB, Arseniev AS. Spatial structure of the M3 transmembrane segment of the nicotinic acetylcholine receptor alpha subunit. Eur J Biochem 1998; 255: 455–461
- Mabrey S, Sturtevant JM. Investigation of phase transitions of lipids and lipid mixtures by sensitivity differential scanning calorimetry. Proc Natl Acad Sci USA 1976; 73: 3862–37866
- Marrink SJ, Berger O, Tieleman P, Jahnig F. Adhesion forces of lipids in a phospholipid membrane studied by molecular dynamics simulations. Biophys J 1998; 74: 931–943
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature 2003; 423: 949–955
- Nezil FA, Bloom M. Combined influence of cholesterol and synthetic amphiphillic peptides upon bilayer thickness in model membranes. Biophys J 1992; 61: 1176–1183
- Nurowska E, Ruzzier F. Corticosterone modifies the murine muscle acetylcholine receptor channel kinetics. Neuroreport 1996; 8: 77–80
- Opella SJ, Marassi FM, Gesell JJ, Valente AP, Kim Y, Oblatt-Montal M, Montal M. Structures of the M2 channel-lining segments from nicotinic acetylcholine and NMDA receptors by NMR spectroscopy. Nat Struct Biol 1999; 6: 374–379
- Ortiz-Miranda SI, Lasalde JA, Pappone PA, McNamee MG. Mutations in the M4 domain of the Torpedo californica nicotinic acetylcholine receptor alter channel opening and closing. J Membr Biol 1997; 158: 17–30
- Parasassi T, De Stasio G, d'Ubaldo A, Gratton E. Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence. Biophys J 1990; 57: 1179–1186
- Parasassi T, De Stasio G, Ravagnan G, Rusch RM, Gratton E. Quantitation of lipid phases in phospholipid vesicles by the generalized polarization of Laurdan fluorescence. Biophys J 1991; 60: 179–189
- Ren J, Lew S, Wang Z, London E. Transmembrane orientation of hydrophobic α-helices is regulated both by the relationship of helix length to bilayer thickness and by the cholesterol concentration. Biochemistry 1997; 36: 10213–10220
- Ross MJ, Klymkowsky MW, Agard DA, Stroud RM. Structural studies of a membrane-bound acetylcholine receptor from Torpedo californica. J Mol Biol 1977; 116: 635–659
- Saiz L, Klein ML. The transmembrane domain of the acetylcholine receptor: Insights from simulations on synthetic peptide models. Biophys J 2005; 88: 959–970
- Santiago J, Guzman GR, Rojas LV, Marti R, Asmar-Rovira GA, Santana LF, McNamee M, Lasalde-Dominicci JA. Probing the effects of membrane cholesterol in the Torpedo californica acetylcholine receptor and the novel lipid-exposed mutation alpha C418W in Xenopus oocytes. J Biol Chem 2001; 276: 46523–46532
- Sperotto MM, Mouritsen OG. Dependence of lipid membrane phase transition temperature on the mismatch of protein and lipid hydrophobic thickness. Eur Biophys J 1988; 16: 1–10
- Sunshine C, McNamee MG. Lipid modulation of nicotinic acetylcholine receptor function: The role of membrane lipid composition and fluidity. Biochim Biophys Acta 1994; 1191: 59–64
- Tamamizu S, Guzman GR, Santiago J, Rojas LV, McNamee MG, Lasalde-Dominicci JA. Functional effects of periodic tryptophan substitutions in the alpha M4 transmembrane domain of the Torpedo californica nicotinic acetylcholine receptor. Biochemistry 2000; 39: 4666–4673
- Tamamizu S, Lee Y, Hung B, McNamee MG, Lasalde-Dominicci JA. Alteration in ion channel function of mouse nicotinic acetylcholine receptor by mutations in the M4 transmembrane domain. J Membr Biol 1999; 170: 157–164
- Tieleman DP, Berendsen HJC. Molecular dynamics simulations of fully hydrated DPPC with different macroscopic boundary conditions and parameters. J Chem Phys 1996; 105: 4871–4880
- Tieleman DP, Sansom MS, Berendsen HJC. Alamethicin helices in a bilayer and in solution: molecular dynamics simulations. Biophys J 1999a; 76: 40–49
- Tieleman DP, Berendsen HJ, Sansom MS. An alamethicin channel in a lipid bilayer: Molecular dynamics simulations. Biophys J 1999b; 76: 1757–1769
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4 Å resolution. J Mol Biol 2005; 346: 967–989
- van Gunsteren WF, Berendsen HJC. 1987. Gromos-87 manual. GroningenThe Netherlands: Biomos BV Nijenborgh.
- Villar MT, Artigues A, Ferragut JA, González-Ros JM. Phospholipase A2 hydrolysis of membrane phospholipids causes structural alteration of the nicotinic acetylcholine receptor. Biochim Biophys Acta 1988; 938: 35–43
- Voglino L, Simon SA, McIntosh TL. Orientation of LamB signal peptides in bilayers: Influence of lipid probes on peptide binding and interpretation of fluorescence quenching data. Biochemistry 1999; 38: 7509–7516
- Webb RJ, East JM, Shanna RP, Lee AG. Hydrophobic mismatch and the incorporation of peptides into lipid bilayers: A possible mechanism for retention in the Golgi. Biochemistry 1998; 37: 673–679
- White SH, Ladokhin AS, Jayasinghe S, Hristova K. How membranes shape protein structure. J Biol Chem 2001; 276: 32395–32398
- Xu Y, Barrantes FJ, Luo X, Chen K, Shen J, Jiang H. Conformational dynamics of the nicotinic acetylcholine receptor channel: A 35-ns molecular dynamics simulation study. J Am Chem Soc 2005; 127: 1291–1299
