Abstract
Candida albicans is a common fungus of the human microbiota. While generally a harmless commensal in healthy individuals, several factors can lead to its overgrowth and cause a range of complications within the host, from localized superficial infections to systemic life-threatening disseminated candidiasis. A major virulence factor of C. albicans is its ability to form biofilms, a closely packed community of cells that can grow on both abiotic and biotic substrates, including implanted medical devices and mucosal surfaces. These biofilms are extremely hard to eradicate, are resistant to conventional antifungal treatment and are associated with high morbidity and mortality rates, making biofilm-associated infections a major clinical challenge. Here, we review the current knowledge of the processes involved in C. albicans biofilm formation and development, including the central processes of adhesion, extracellular matrix production and the transcriptional network that regulates biofilm development. We also consider the advantages of the biofilm lifestyle and explore polymicrobial interactions within multispecies biofilms that are formed by C. albicans and selected microbial species.
Introduction
A biofilm is a consortium of microbes attached to a biotic or abiotic substrate embedded within a matrix of extracellular polymeric substances. For many microbes, the ability to form biofilms is an important virulence factor. Biofilms are found on natural substrates including plant and mammalian tissues and form readily on synthetic polymers such as plastics found in catheters, prostheses and implanted heart valves (Nobile and Johnson Citation2015). Rapid medical intervention, high concentrations of antimicrobials and replacement of the infected device are required to manage a biofilm infection. However, such procedures are not without significant patient risk, as toxicity issues may arise from antimicrobial therapy and replacement of infected devices may sometimes require major surgery (for instance, when replacing coronary stents or heart valves). Thus, biofilm-related infections contribute significantly to the economic burden of global healthcare (Pittet Citation1994; Crump and Collignon Citation2000; Schierholz and Beuth Citation2001; Orsi et al. Citation2002; Yousif et al. Citation2015) and are associated with poor clinical outcome.
Candida species, which include the opportunistic pathogen Candida albicans, are one of the main causes of nosocomial infections worldwide, particularly in immunocompromised individuals (Hajjeh et al. Citation2004). Candida albicans can form complex biofilms on medical devices and, often, device removal is recommended (Kojic and Darouiche Citation2004; Pappas et al. Citation2016; Cavalheiro and Teixeira Citation2018; Nett and Andes Citation2020). Biofilms are also associated with recurrent candidaemia, which affects more than 400 000 people per year and has an alarming mortality rate of approximately 50% (Tumbarello et al. Citation2012; Bongomin et al. Citation2017; Li et al. Citation2018).
Candida albicans forms biofilms comprised of yeast cells, pseudohyphae and hyphae. The transition from planktonic growth to biofilm is accompanied by a complex remodelling of phenotypic behaviour underpinned by myriad changes in gene expression. The development of C. albicans biofilm occurs progressively, and is typically classified into four stages; (i) adsorption and adhesion of C. albicans yeast cells to a substrate, (ii) formation of microcolonies and production of the extracellular matrix, (iii) maturation and (iv) dispersal of cells from the mature biofilm. Once established, C. albicans biofilms are highly tolerant to antifungal therapy and can serve as a reservoir for recurrent infection. Indeed, the medical impact of C. albicans biofilm is underscored by a positive correlation between biofilm formation, increased virulence and higher patient mortality (Tumbarello et al. Citation2012; Rajendran, May, et al. Citation2016).
In this review, we summarise the current understanding of the processes involved in C. albicans biofilm formation and the regulatory circuits that are pivotal to C. albicans biofilm development. We explore multispecies biofilms, the advantages conferred to microbes during biofilm growth, persister cells and resistance to host immune clearance.
Biofilm formation
Biofilm formation begins with the adhesion of C. albicans yeast cells to a suitable surface. These early contact events comprise several non-specific factors including attractive and repulsive forces (such as van der Waals forces, hydrophobic interactions and Brownian movement forces), and electrostatic interactions that facilitate initial adherence (Douglas Citation1987; Hazen Citation1989; Hazen et al. Citation1990; Hobden et al. Citation1995; Jones et al. Citation1995; Williams et al. Citation2013). While these factors may not be the predominant mechanism for yeast cells to adhere, they are nevertheless an important prerequisite for biofilm formation (Park et al. Citation2008; Silva-Dias et al. Citation2015).
Following initial contact, cell wall-associated adhesion molecules are expressed to further strengthen fungal adhesion. There are three key families of C. albicans adhesins that play a major role in mediating adherence during biofilm formation: the agglutinin-like sequence (Als) family, the hyphal wall protein (Hwp) family, and the individual protein file family F/hyphally regulated (Iff/Hyr) family (de Groot et al. Citation2013). The most widely studied are the Als adhesins (Hoyer and Cota Citation2016) comprising eight members (Als1p-Als7p and Als9p), which are glycosylphosphatidylinositol (GPI)-linked to β-1,6 glucans in the fungal cell wall. The Als family of adhesins is key in both yeast and hyphal adhesion during biofilm formation and development.
During the initial adhesion of yeast cells to surfaces, Als1p and Als5p facilitate adhesion via their amyloid-forming region that is critical for yeast cell–cell aggregation and cell–substrate adhesion (Rauceo et al. Citation2004; Ramsook et al. Citation2010; Garcia et al. Citation2011). These yeast cell aggregates form quickly and are key to the establishment of mixed-species biofilms with other fungal species and bacteria (Klotz et al. Citation2007; Alsteens et al. Citation2010; Ramsook et al. Citation2010; Lipke et al. Citation2012, Citation2014). In addition to the Als family, other adhesins are important in mediating the initial attachment of C. albicans yeast cells during biofilm formation (). The enhanced adhesion to polystyrene protein (Eap1p) is significant in the clinical setting due to its ability to mediate attachment of C. albicans to plastics found in medical devices (Chong et al. Citation2018; McCall et al. Citation2019). Deletion of EAP1 renders C. albicans unable to attach to plastic surfaces and epithelial cells in vitro. In addition, Eap1p is required for biofilm formation in a rat central venous catheter model (Li et al. Citation2007).
Figure 1. Stages of Candida albicans biofilm formation and development. Candida albicans biofilm formation is a multifactorial process that consists of four main stages. (A) Initial attachment of planktonic cells: C. albicans yeasts attach to a surface (e.g. epithelia, biomaterials or cellular aggregates) through adhesins such as Als family members. (B) Proliferation and filamentation: yeasts transition to hyphae and this process is regulated by many transcription factors (TFs) including Tec1p and Efg1p. Hyphae express specific adhesins such as Hwp1p and Hyr1p. (C) Biofilm maturation and extracellular matrix formation: the matrix forms around the C. albicans cells, positively regulated by the TF Rlm1p, providing structural support and protection against antifungals and the host immune system. Adhesion is maintained and amino acid metabolism is increased in the biofilm. (D) Biofilm dispersion: yeast cells disperse from the biofilm to colonise other parts of the body. These cells differ from initial planktonic cells as they are more virulent and more likely to form biofilms. Figure created with Adobe Illustrator.
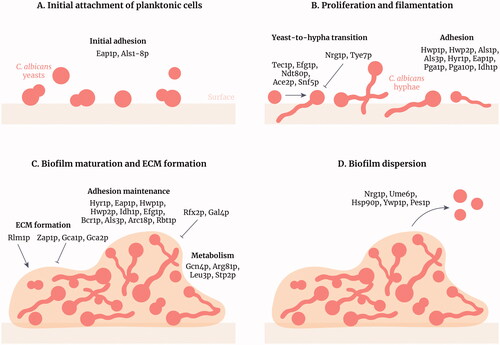
Upon contact with a suitable surface, yeast cells germinate to form hyphae and pseudohyphae. The adhesion of these distinct morphological forms is crucial during the later stages of biofilm development. A vast array of adhesins orchestrate the adherence of yeast, pseudohyphae and hyphae, thereby contributing to biofilm maintenance. Als1p and Als3p have critical roles in facilitating hyphal adhesion during biofilm formation (Nobile et al. Citation2008). A C. albicans mutant unable to express ALS3 forms weaker and more disorganised biofilms compared to wild-type fungi in vitro (Zhao et al. Citation2006). Furthermore, disruption of both ALS3 and ALS1 (als3ΔΔ/als1ΔΔ) reduces the biofilm biomass when compared with controls (McCall et al. Citation2019).
Other Als family members also make significant contributions to biofilm development. Increased expression of ALS6, ALS7, and ALS9 rescues the biofilm defect of an als3ΔΔ/als1ΔΔ mutant (Nobile et al. Citation2008), while mutations in the amyloid-forming region of Als5p prevent the formation of adhesion nanodomains, cellular aggregates and biofilm development on polystyrene surfaces in vitro (Garcia et al. Citation2011). Conversely, decreased expression of ALS2 but not ALS4 results in impaired biofilm formation in a catheter biofilm model (Zhao et al. Citation2005). However, overexpression of ALS2 or ALS4 fails to rescue biofilm formation in an als3ΔΔ/als1ΔΔ null mutant in vivo (Nobile et al. Citation2008). This suggests that only certain Als proteins may function equivalently to Als1p and Als3p in an in vivo model of biofilm formation (Nobile et al. Citation2008).
Hyphal wall protein 1 (Hwp1p) is the most thoroughly characterised member of the Hwp family. Hwp1p is a molecular mimic of the substrate recognised by mammalian transglutaminase enzymes which cross-link Hwp1p to the surface of the host cell (Staab Citation1999), facilitating adhesion. A C. albicans hwp1ΔΔ mutant is unable to produce hyphae or form biofilms (Nobile, Nett, et al. Citation2006), underscoring the critical role of Hwp1p in these processes. Interestingly, overexpression of HWP1 is unable to promote biofilm formation in an als3ΔΔ/als1ΔΔ mutant in vitro or in vivo (Nobile et al. Citation2008), though a mixture of biofilm defective hwp1ΔΔ and als3ΔΔ/als1ΔΔ strains can form a hybrid biofilm. This suggests that while Hwp1p has a distinct function to the Als proteins, the three adhesins may have complementary roles in mediating adherence of C. albicans cells during biofilm formation (Nobile et al. Citation2008). Other Hwp family members including Hwp2p and Rbt1p contribute to initial cell attachment and the maintenance of adhesion (Li et al. Citation2007; Ene and Bennett Citation2009; Younes et al. Citation2011; Desai and Mitchell Citation2015; McCall et al. Citation2019). Mutants defective in HWP2 are unable to adhere efficiently to epithelial cells and present a decrease in biofilm mass (Younes et al. Citation2011), while RBT1 overexpressing strains exhibit an increase in cell surface hydrophobicity, which is essential for C. albicans adherence to polystyrene (Monniot et al. Citation2013).
The Iff/Hyr family of adhesins includes the hyphae specific Hyr1p (Bailey et al. Citation1996), which appears to be the only member of this family associated with biofilm formation and development. Mutants defective in HYR1 exhibit reduced adhesion in a flow system during the initial stages of biofilm development, as well as a reduction in biomass due to defective attachment (McCall et al. Citation2019). Furthermore, a significant reduction in both initial attachment and subsequent adhesion was observed following the loss of HYR1, while the loss of ALS3/ALS1 only affected the maintenance of adhesion, suggesting that Hyr1p is key in mediating adhesion during both early and late stages of biofilm development (McCall et al. Citation2019).
While research has focussed predominantly on the three main C. albicans adhesin families, several additional factors contribute to biofilm adhesion, including the putative GPI-anchored proteins Pga1p and Pga10p (de Groot et al. Citation2003; Pérez et al. Citation2006; Hashash et al. Citation2011; Awad et al. Citation2018), as well as Ecm33p, Mp65p, Msb2p, Pbr1p, Arc18p, Pmt1p, Mnn9p, Spt7p, orf19.831 and Ihd1p (Sahni et al. Citation2009; Sandini et al. Citation2011; Puri et al. Citation2012; Rouabhia et al. Citation2012; Lee et al. Citation2016; McCall et al. Citation2019). Gene deletion studies have demonstrated decreased or impaired adhesion and fragile biofilms. Collectively, these observations highlight a clear role for adhesins and adhesin-like molecules during the initial stages of biofilm formation.
Biofilm development
Following adhesion, biofilm development is accompanied by changes in morphology, cell number, the secretion of extracellular polymeric substances and production of an extracellular matrix (ECM). Once adhered, yeast cells proliferate to form microcolonies, forming the basal layer of the biofilm. As the biofilm matures, the biomass increases with yeast cells, hyphae, pseudohyphae and the ECM accounting for a large proportion of the biofilm (). Hyphae are fundamental components that support the structural integrity of the biofilm and provide a scaffold for the attachment of additional yeast cells, pseudohyphae, other hyphae, as well as bacteria in multispecies biofilms (Chandra et al. Citation2001; Harriott and Noverr Citation2009). Mutants defective in the production of hyphae or hypha-specific adhesins are also defective in the production of substantial biofilms (Baillie and Douglas Citation1999; Ramage, VandeWalle, et al. Citation2002; Richard et al. Citation2005). Accordingly, the ability to form hyphae is critical for the normal development and maintenance of C. albicans biofilms. In a typical biofilm, an adherent layer of yeast cells is covered by an entangled mesh of hyphal filaments and pseudohyphae and encased in an ECM (Baillie and Douglas Citation1999; Chandra et al. Citation2001) (). Indeed, the ECM is essential for biofilm development and maturation. The ECM is composed of extracellular polymeric substances secreted by cells within the biofilm, including a complex mixture of macromolecules comprising proteins (55%), carbohydrates (25%), lipids (15%) and nucleic acids (5%) (Zarnowski et al. Citation2014). The most common polysaccharide found in the ECM of C. albicans biofilms are mannans, which are often associated with β-1,6 glucans (Pierce et al. Citation2017). Proteomic analysis of C. albicans extracellular polymeric substances also reveals the presence of numerous host molecules, including those found in epithelial cells and neutrophils, suggesting a potential role for host components in biofilm development (Andes et al. Citation2004; Dongari-Bagtzoglou et al. Citation2009; Zarnowski et al. Citation2014; Nett et al. Citation2015).
The presence of nucleic acids in C. albicans biofilms contributes to the structural and protective properties of the ECM. Extracellular deoxyribonucleic acid (eDNA) released by C. albicans within the ECM may contribute to the maintenance and stability of mature biofilms, but is not required to establish a C. albicans biofilm (Al-Fattani and Douglas Citation2006; Martins et al. Citation2010; Rajendran et al. Citation2014), suggesting an important role for eDNA in ECM assembly. The release of eDNA confers heterogeneity during biofilm formation, with higher levels of eDNA observed in isolates with high biofilm-forming capacity compared to isolates with lower capacity (Rajendran et al. Citation2014). Moreover, eDNA may also play a role in modulating the host immune response by activating bone marrow-derived myeloid dendritic cells (Kasai et al. Citation2006; Miyazato et al. Citation2009; Martins et al. Citation2010). Treatment of C. albicans biofilm with deoxyribonuclease I (DNase) improved the efficacy of amphotericin B in vitro (Martins et al. Citation2012), particularly in the presence of the chitinase inhibitor acetazolamide (Rajendran et al. Citation2014), suggesting that eDNA also contributes to recalcitrance. Accordingly, targeting eDNA within the ECM may present a novel therapy to improve the activity of antifungal drugs and decrease biofilm formation (Martins et al. Citation2012; Sapaar et al. Citation2014).
The biofilm matrix has pleiotropic functionality; it contributes to biofilm structure, provides physical support (Nobile and Johnson Citation2015; Pierce et al. Citation2017) and is critical in enhancing tolerance to antifungal drugs (described below).
Biofilm dispersal
Dispersal is the final stage of the biofilm lifecycle and describes a process where cells are released from a mature biofilm and are thus free to disseminate and establish secondary sites of infection (). The majority of dispersal occurs from the uppermost layers of biofilm and is associated with yeast cells that have an elongated morphology (Uppuluri, Chaturvedi, et al. Citation2010). Several factors contribute to the process of dispersal. Candida albicans Nrg1p, Pes1p and Ume6p are known regulators of dispersal with opposing functions. Nrg1p is a negative regulator of the yeast-to-hypha morphological switch (Braun et al. Citation2001; Murad et al. Citation2001). Consistent with the release of yeast cells from biofilm during dispersal, overexpression of NRG1 promotes dispersal from biofilms in vitro (Uppuluri, Pierce, et al. Citation2010), while overexpression of the Pescadillo homologue PES1 also increases dispersal (Uppuluri, Chaturvedi, et al. Citation2010). In contrast, overexpression of the transcriptional regulator UME6 reduces dispersal (Uppuluri, Chaturvedi, et al. Citation2010), while transcriptional repression of the chaperone protein Hsp90p also reduces dispersal of C. albicans cells from biofilms in vitro (Robbins et al. Citation2011).
Candida albicans yeasts express Ywp1p, a cell-surface glycoprotein that is covalently linked to β-glucans in the fungal cell wall. Intriguingly, Ywp1p is reported to exhibit anti-adhesive properties consistent with a role in dispersal from biofilms. A C. albicans mutant unable to express YWP1 displays enhanced adhesion and biofilm formation (Granger et al. Citation2005). Members of the C. albicans histone deacetylase complex have also been implicated in biofilm dispersal, with C. albicans mutants unable to express HOS2, SET3, SIF2 or SNT1 exhibiting increased cohesiveness and impaired dispersal over a 60 h period (Nobile et al. Citation2014).
Notably, dispersed cells have a number of altered characteristics when compared to planktonic cells including an increased ability to filament and form biofilms, increased adherence to biotic and abiotic substrates, reduced susceptibility to antifungals and an increased ability to damage endothelial cells in vitro (Uppuluri, Chaturvedi, et al. Citation2010). Dispersed cells also upregulate the expression of genes whose products are involved in the acquisition of micronutrients, drug resistance, and hydrolysis of host substrates (Uppuluri et al. Citation2018). Moreover, dispersed cells display enhanced pathogenicity in vivo when compared with matched planktonic controls (Uppuluri, Chaturvedi, et al. Citation2010). Collectively, these observations suggest that dispersed cells are “pre-conditioned” for maximum virulence. More recent work suggests that the degree of fungal dispersal is influenced by the extracellular environment. Investigations with mixed-species biofilms reveal that the Gram-positive bacterium Rothia dentocariosa can inhibit the release of C. albicans cells in vitro (Uppuluri et al. Citation2017), thus the amount of dispersal that occurs in vivo is likely to be influenced by biofilm composition.
Transcriptional regulation of biofilm formation and maintenance
The formation and development of C. albicans biofilms are underpinned by complex changes in transcriptional circuitry. While 58 transcription factors have been identified to contribute to C. albicans biofilm formation and development (), an interdependent core network of transcriptional regulators has been identified (Bcr1p, Tec1p, Efg1p, Ndt80p, Rob1p and Brg1p) that is critical for biofilm formation (Nobile et al. Citation2012; Glazier et al. Citation2017; Figure 1). Candida albicans mutant strains unable to express BCR1, TEC1, EFG1, NDT8 or ROB1 fail to form biofilms in vivo, while ablation of BRG1 results in biofilms with abnormal structure (Ramage, Vande Walle, et al. Citation2002; Nobile and Mitchell Citation2005; Nobile et al. Citation2012; Yano et al. Citation2016; Panariello et al. Citation2017). Collectively, this core network of transcriptional regulators influences the expression of approximately 1000 genes involved in hypha formation, adhesion, metabolism, production of extracellular polymeric substances and drug resistance (Fox and Nobile Citation2012; Nobile and Johnson Citation2015).
Table 1. Transcription factors that contribute to C. albicans biofilm formation, development, and regulation.
More recent work has identified additional master regulators of biofilm formation including Flo8p, Gal4p and Rfx2p (Fox et al. Citation2015). Candida albicans mutant strains unable to express GAL4 or RFX2 form thicker biofilms (Fox et al. Citation2015), suggesting that these factors function as negative regulators. In contrast, strains unable to express FLO8 were markedly defective in biofilm formation in a rat central venous catheter model (Nobile et al. Citation2012; Fox et al. Citation2015). Bcr1p is critical for biofilm formation and influences the expression of several adhesins including Als3p, Hwp1p and Hyr1p (Nobile and Mitchell Citation2005; Nobile, Andes, et al. Citation2006; Dwivedi et al. Citation2011). A C. albicans bcr1ΔΔ mutant produces a defective biofilm in vitro and in vivo, but overexpression of ALS3 rescues the defective biofilm phenotype, highlighting the importance of Als3p (Nobile and Mitchell Citation2005; Nobile, Andes, et al. Citation2006; Nobile and Mitchell Citation2006; Liu and Filler Citation2011). Interestingly, Bcr1p also targets ECE1, which encodes the peptide toxin candidalysin (Nobile, Andes, et al. Citation2006; Moyes et al. Citation2016). RNA transcriptome analysis reveals that several genes (including ECE1) are upregulated in high biofilm-forming isolates of C. albicans (Rajendran, May, et al. Citation2016).
The production of biofilm ECM is predominantly controlled by two transcription factors: Zap1p and Rlm1p. Zap1p is a zinc regulator that negatively regulates the formation of β-1,3 glucan through its target genes GCA1 and GCA2 (Nobile et al. Citation2009; Mayer et al. Citation2013; Nobile and Johnson Citation2015). In contrast, Rlm1p is a positive regulator of ECM production (Nett et al. Citation2011; Nobile and Johnson Citation2015). Another transcription factor, Stp2p, is a key activator of amino acid permease genes that influences adherence and morphogenesis, adaptation to nutrients and biofilm longevity (Böttcher et al. Citation2020).
Candida albicans biofilms in vitro and in vivo: one morphology does not fit all
Most biofilm studies have been conducted in vitro. For the most part, data obtained from in vitro biofilm studies correlate well with data obtained from in vivo and ex vivo models. The structure of biofilms from rat denture stomatitis and indwelling urinary catheter models is similar to the architecture of biofilms produced in vitro (Andes et al. Citation2004; Johnson et al. Citation2012). The formation and structure of C. albicans biofilm is influenced by numerous factors including the nature of the contact surface, environmental factors, and C. albicans morphology. Maturation of C. albicans biofilms on polyvinyl chloride catheter discs occurs within 24–48 h (Hawser and Douglas Citation1994), whereas maturation on polymethylmethacrylate strips requires longer timeframes (38–72 h) (Chandra et al. Citation2001). Interestingly, Andes et al. reported differences in temporal developments between in vivo and in vitro studies. Utilising a rat central venous catheter model, the duration of the early phase and maturation of biofilm formation in vivo was shorter than that observed in vitro (Andes et al. Citation2004). While C. albicans forms biofilms readily on abiotic substrates and on the mucosal surfaces of the oral cavity, conflicting observations have been made regarding the formation of biofilms on the vaginal mucosa (Harriott et al. Citation2010; Paiva et al. Citation2012; Sherry et al. Citation2017; Noverr and Fidel Citation2019; Swidsinski et al. Citation2019; Xu et al. Citation2019), suggesting that tissue/anatomical location and surrounding microbiota are likely to influence biofilm development. These observations have led to some contention regarding the definition of clinical diseases that specifically involve biofilms as opposed to diseases that are associated with a diverse or altered microbiota (Harriott et al. Citation2010; Paiva et al. Citation2012; Sobel Citation2015; Sherry et al. Citation2017; Noverr and Fidel Citation2019; Swidsinski et al. Citation2019; Xu et al. Citation2019). Rather than being a simple binary event, biofilms comprise a spectrum of morphologies, the composition of which varies between diseases. Accordingly, the provision of an unambiguous definition of C. albicans biofilms in the clinical setting is not without significant challenge.
Advantages of the biofilm lifestyle
The environment within a biofilm differs markedly from that encountered during planktonic growth and offers numerous advantages to the microbes that reside within it. The morphological variation within biofilms (yeasts, pseudohyphae and hyphae) combined with the presence of other fungi and bacteria collectively confer an increased competitive fitness that is absent in planktonic cells (Lohse et al. Citation2018). Microbes encased within the ECM are more likely to withstand minor mechanical stress and are better protected from the external environment and hostile host immune responses (Mathé and Van Dijck Citation2013; Cavalheiro and Teixeira Citation2018; Kernien et al. Citation2018). Mixed-species biofilms can produce and utilise a greater variety of nutrients through the formation of metabolic consortiums in which the waste products of one species may be metabolised by another (Elias and Banin Citation2012). Such communities exhibit tremendous genomic diversity and, consequently, biofilms provide a suitable environment in which new genetic material can be acquired through the process of horizontal/lateral gene transfer, particularly between species of bacteria (Roberts and Kreth Citation2014).
Numerous differences have been reported between C. albicans biofilms and planktonic cells including alterations in transcriptional network activity (Yeater et al. Citation2007; Nobile et al. Citation2012; Fox et al. Citation2015), altered cell wall architecture (Nett et al. Citation2007), differences in ergosterol content of biofilm cell plasma membranes (Mukherjee et al. Citation2003) and, most notably, differences in sensitivities to antifungals. Candida albicans biofilms are substantially more difficult to eradicate with antifungals as compared to planktonic cells. Numerous mechanisms enable planktonic cells to withstand the toxicity of antifungals including constitutive expression and activation of drug efflux pumps, mutation of genes encoding drug targets, the acquisition of drug tolerance (Berman and Krysan Citation2020), alterations in the copy number of genes via aneuploidy (Yang et al. Citation2019) and adaptive mistranslation (Weil et al. Citation2017). Mature C. albicans biofilms are highly tolerant of fluconazole, amphotericin B, and caspofungin at concentrations that are lethal to planktonic cells (Vediyappan et al. Citation2010). Although numerous drug efflux pumps are expressed both transiently and permanently during the formation and maturation of C. albicans biofilm in vitro and in vivo (Mukherjee et al. Citation2003; Mateus et al. Citation2004; Yeater et al. Citation2007; Nett et al. Citation2009; Fox et al. Citation2015), the overall contribution of drug efflux to biofilm longevity appears to be relevant only during the early stages of biofilm formation (Mukherjee et al. Citation2003). Importantly, while C. albicans strains deleted for specific ATP-binding cassette transporters and major facilitator superfamily transporters (cdr1ΔΔ, cdr2ΔΔ, cdr1/cdr2ΔΔ, mdr1ΔΔ and mdr1/cdr1ΔΔ) were highly susceptible to fluconazole during planktonic growth, the same mutants exhibited resistance to fluconazole when grown as a biofilm (Ramage, Bachmann, et al. Citation2002), suggesting that factors distinct from drug efflux pumps contribute to drug tolerance within a biofilm.
Specific components of the biofilm ECM are instrumental in the orchestration of tolerance to antifungals. Candida albicans mutant strains unable to deliver β-1,3 glucan to the biofilm ECM are susceptible to fluconazole (Nett, Sanchez, et al. Citation2010; Nett et al. Citation2011; Taff et al. Citation2012) while reducing FKS1 (glucan synthase) expression renders cells susceptible to amphotericin B, anidulafungin, and flucytosine (Nett, Crawford, et al. Citation2010). Indeed, β-glucan is capable of binding fluconazole and amphotericin B (Nett et al. Citation2007; Vediyappan et al. Citation2010), while treatment of C. albicans biofilms with the enzyme β-1,3 glucanase increases sensitivity to fluconazole (Nett et al. Citation2007). Sequestration of fluconazole by β-glucan in the ECM combined with a reduction in antifungal penetrance into the biofilm often results in fungi receiving a sub-lethal dose of the drug, which can lead to the emergence of resistance.
Collectively, these observations are consistent with the notion that the ECM functions in part, as a “molecular trap” that impedes the diffusion of antifungals into the biofilm, causing a reduction in the local concentration of antifungal that can engage with target cells. Exposure of biofilms to diminished concentrations of an antifungal is likely to impose a positive selection pressure towards the acquisition of mutations that reduce susceptibility to subsequent therapy, an outcome that may have serious clinical implications should mutated cells be dispersed from a recalcitrant biofilm.
More recently, studies have highlighted an essential role for extracellular vesicles (EVs) in conferring the drug-resistant properties of C. albicans biofilms. Analyses of EVs released from C. albicans biofilms reveal a proteome that is distinct from that present in EVs released from planktonic cells, and which contain high levels of mannan and glucan (Zarnowski et al. Citation2018). Candida albicans phr1ΔΔ and sun41ΔΔ null mutants are devoid of specific EV cargo proteins and form biofilms that are sensitive to fluconazole. Strikingly, EVs that were harvested from wild type C. albicans biofilms and applied to phr1ΔΔ and sun41ΔΔ biofilms restored the ability to grow in the presence of the drug (Zarnowski et al. Citation2018), demonstrating a functional role for EVs in conferring drug resistance.
The effectiveness of the host immune response to biofilms is also diminished when compared to planktonic cells. The biofilm ECM protects underlying fungal cells from the innate immune system, particularly from killing by neutrophils and monocytes. Impaired recognition of biofilms by neutrophils and monocytes is evidenced by altered patterns of cytokine secretion when compared to those produced when encountering planktonic cells (Chandra et al. Citation2007). Monocytes can phagocytose planktonic C. albicans but not fungal cells associated with biofilms, while neutrophils fail to eradicate mature biofilms (Chandra et al. Citation2007; Katragkou et al. Citation2010; Xie et al. Citation2012; Pierce et al. Citation2017). Furthermore, neutrophils release extracellular traps in response to planktonic C. albicans yeast and short hyphae but fail to release extracellular traps in response to biofilm ECM (Nett Citation2016; Johnson et al. Citation2016, Citation2017; Pierce et al. Citation2017; Kernien et al. Citation2018).
Persister cells: a disputed presence in C. albicans biofilms
Persister cells are a contentious topic in the context of C. albicans biofilms. Defined as a subpopulation of metabolically dormant biofilm cells, these cells are only detected in the presence of antifungals. Persister cells express high levels of alkyl hydroperoxide reductase 1 (Ahp1p), which is positively correlated with prolonged survival following exposure to amphotericin B (Truong et al. Citation2016). When antifungal therapy is discontinued, persister cells proliferate to restore the biofilm. Patients with long-term oral carriage of C. albicans have a higher incidence of persister cells (LaFleur et al. Citation2010), suggesting that persister cells may be relevant in the context of continual colonisation or recurrent infections in vivo (Wuyts et al. Citation2018). However, despite observations which support the existence of persister cells they remain a controversial topic. Indeed, some studies suggest that persister cells are not a general characteristic of Candida biofilms (Al-Dhaheri and Douglas Citation2008; Denega et al. Citation2019).
Heteroresistance of C. albicans biofilms
As previously mentioned, C. albicans biofilms are difficult to eradicate. An issue that clinicians are facing is biofilm heterogeneity. Indeed, clinical isolates of C. albicans biofilms from the bloodstream or the vaginal mucosa seem to differ in biomass and therefore isolates can be defined as either high- or low-biofilm formers (Sherry et al. Citation2014, Citation2017). This inherent biological heterogeneity has led to patients responding differently to antifungals (Kean et al. Citation2018) and is thus called heteroresistance. Heteroresistance is defined as the presence of a subpopulation with lower susceptibility to specific antifungals within a larger microbial population. A recent study of C. glabrata clinical strains suggests that this heteroresistance might be linked to persistent infections (Ben-Ami et al. Citation2016).
Multispecies biofilms: C. albicans and polymicrobial interactions
Interactions between C. albicans and the microbes found on biotic and abiotic substrates are varied and complex, comprising synergistic, antagonistic, and neutral relationships (Delaney et al. Citation2018). These interactions involve direct cell to cell contact, chemical interactions via the secretion of small molecules, enhancement of colonisation, changes in the host environment, the use of metabolic by-products, changes in the host response or a combination of these. Unsurprisingly, these interactions are particularly relevant in the context of biofilms and are discussed below.
Adhesion in mixed species biofilms
The interactions of C. albicans with other microorganisms can occur via co-aggregation and co-adhesion. Candida albicans adhesins (e.g. Als1p, Als2p, Als3p, Hwp1p) facilitate interaction with bacterial species such as Streptococcus gordonii and Staphylococcus aureus (Peters et al. Citation2012; Hoyer et al. Citation2014; Schlecht et al. Citation2015; Allison et al. Citation2019; ). The adherence of C. albicans to pre-attached Streptococcus spp. in the oral cavity is mediated by polysaccharide receptors on the streptococcal cell surface (SspA and SspB) and C. albicans hypha-associated adhesins (Als1p and Als3p) (Holmes et al. Citation1996; Bamford et al. Citation2009; Silverman et al. Citation2010). Moreover, S. gordonii SspA and SspB mutants exhibit reduced expression of streptococcal adhesins for C. albicans, with less co-aggregation of C. albicans with these S. gordonii mutants (Klotz et al. Citation2007).
Microbial synergism and antagonism within mixed species biofilm
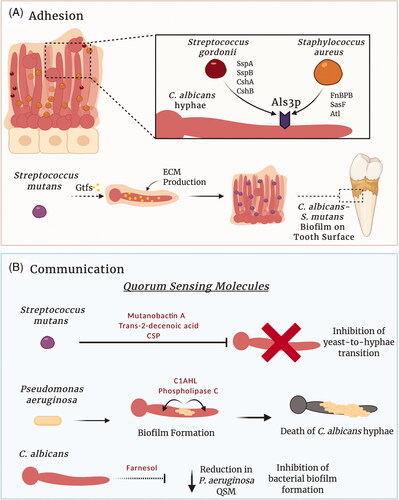
Interkingdom interactions between C. albicans and other microbes can be synergistic, mutually beneficial or antagonistic, and these interactions can dictate the composition of the microbiota. One of the most thoroughly characterised synergistic relationships is that of C. albicans and Streptococcus mutans. Within plaque biofilms, S. mutans glucosyltransferases bind directly to mannans on the surface of C. albicans yeast and hyphal cell walls, promoting the development of an extensive ECM and the formation of a mixed species biofilm (Hwang et al. Citation2015; Ellepola et al. Citation2017). Consistent with this, S. mutans mutants lacking glucosyltransferase genes demonstrate reduced ability to form mixed species biofilms with C. albicans (Falsetta et al. Citation2014; Hwang et al. Citation2017; Figure 2(A)). The presence of both S. mutans and C. albicans in the oral microbiota significantly increases bacterial colonisation and supports the development of dental caries, suggesting that this interaction may be a pivotal factor in increasing the risk of disease (de Carvalho et al. Citation2006; Khoury et al. Citation2020). In addition to physical associations, streptococci excrete lactate that acts as a carbon source for C. albicans yeast growth (Ene et al. Citation2012; Metwalli et al. Citation2013). The C. albicans quorum sensing (QS) molecule farnesol has known antibacterial effects, however, within a dual-species biofilm, farnesol enhances S. mutans cell growth and microcolony development (Kim et al. Citation2017). Conversely, the S. mutans QS molecules mutanobactin A, trans-2-decenoic acid and competence-stimulating peptide can inhibit C. albicans germ tube formation and the yeast-to-hypha transition (Jarosz et al. Citation2009; Joyner et al. Citation2010; Vílchez et al. Citation2010; ). Overall, this demonstrates a mutually beneficial relationship between C. albicans and S. mutans with a synergistic partnership that can alter the oral microbiota and influence disease.
Figure 2. Candida albicans interactions within a multispecies biofilm. Complex physical and chemical interactions govern the development of polymicrobial biofilms. (A) Several factors influence C. albicans–bacterial adhesion. Staphylococcus aureus and Streptococcus gordonii can utilise C. albicans adhesins to directly bind to hyphae. In contrast, glycosyltransferases (Gtfs) secreted by Streptococcus mutans within the oral cavity can bind to C. albicans mannans, increasing the production of glucans and ECM. Consequently, the glucan increases the ability of the bacterium to bind to C. albicans and forms a C. albicans–S. mutans biofilm on the tooth surface (dental plaque). (B) Signalling molecules produced by C. albicans and bacterial species enable inter-kingdom communication within multispecies biofilms. For example, S. mutans and Pseudomonas aeruginosa can secrete quorum sensing molecules that influence the behaviour of C. albicans within the biofilm. Likewise, the C. albicans quorum sensing molecule farnesol, can influence the behaviour of interacting bacteria. Figure created with BioRender.
Within a dual-species biofilm, C. albicans can protect anaerobic bacteria from high concentrations of oxygen by providing a hypoxic microenvironment that supports the growth of Bacteroides fragilis, B. vulgatus and Clostridium perfringens (Fox et al. Citation2014; Valentine et al. Citation2019). Accordingly, C. albicans affected the biofilm composition, suggesting a role for the fungus in maintaining homeostasis within the microbiota. Additionally, C. albicans β-1-3-glucan facilitates bacterial tolerance to ofloxacin (De Brucker et al. Citation2015) and vancomycin (Kong et al. Citation2016) in a dual-species biofilm with Escherichia coli and S. aureus respectively, demonstrating antimicrobial tolerance and microbial persistence within multispecies biofilms. However, interactions between C. albicans and bacteria are not always synergistic. Antagonistic effects have been reported between Lactobacillus spp. and C. albicans, with both hypha formation and biofilm development affected by the bacteria (Orsi et al. Citation2014). In the normal vaginal microbiota, Lactobacillus spp. compete with C. albicans for receptors present on the surface of genitourinary epithelium for adhesion; they also secrete lactic acid and hydrogen peroxide, lowering the pH, which in turn inhibits fungal attachment to the vaginal epithelium (Boris et al. Citation1998; Boris and Barbés Citation2000; Aroutcheva et al. Citation2001).
The antagonistic relationship between C. albicans and Pseudomonas aeruginosa is particularly well defined. Both P. aeruginosa and C. albicans are frequently co-isolated from contaminated catheters and chronic lung infections (El-Azizi et al. Citation2004; Williams and Cámara Citation2009). Numerous interactions occur between C. albicans and P. aeruginosa, the outcomes of which are greatly influenced by the environment. Pseudomonas aeruginosa can form dual-species biofilms with C. albicans in liquid medium through a physical association with C. albicans hyphae and germ tubes but not yeast cells. Once formed, C. albicans hyphae are killed by P. aeruginosa secreted virulence factors (phospholipase C and redox-active oxygen species) and quorum sensing molecules (3-oxo-C12-homoserine lactone (C12AHL)) (Hogan and Kolter Citation2002; Hogan et al. Citation2004; Gupta et al. Citation2005; Gibson et al. Citation2009; ). The destruction of hyphae but not C. albicans yeast cells suggests a potential survival mechanism developed by the fungus that allows yeast to evade destruction by P. aeruginosa.
Metabolic interactions play an important role in microbial adaptation within polymicrobial environments. Metabolic dependencies between microbes can be direct or indirect and involve the production of metabolites by one species and their subsequent consumption by another, or the localised depletion of a toxic metabolite by one species, enabling a second species to persist (Krom et al. Citation2014). Competition for nutrients is endemic within polymicrobial communities. In a mixed-species biofilm, P. aeruginosa sequesters iron from C. albicans, reducing the metabolic activity of the fungus (Purschke et al. Citation2012). Lactobacillus rhamnosus can protect oral epithelial cells from fungal damage by inducing metabolic reprogramming of C. albicans. This protective effect comprises a reduction in C. albicans adhesion, impaired hypha formation and glucose depletion (Mailänder-Sánchez et al. Citation2017). As the nature of fungal metabolism is adaptable, these metabolic interactions may be significant in maintaining homeostasis.
Intra-genus interactions within Candida biofilms
While the establishment of mixed-species biofilms involving C. albicans and bacteria has been extensively described, intra-genus interactions between Candida species are not as well characterised. Although C. albicans has been studied more extensively than any other Candida species, other species such as C. dubliniensis, C. glabrata, C. krusei, C. tropicalis and C. auris are becoming an increasing concern in the clinical setting. Though poorly studied, C. albicans, and other non-C. albicans species have been shown to form biofilms with each other (Pereira-Cenci et al. Citation2008; Pathak et al. Citation2012). Mixed species biofilms of C. albicans and non-C. albicans species have been identified on dental acrylic resin strips in which C. albicans was the predominant species (Pathak et al. Citation2012). Moreover, aggregates of C. albicans and C. dubliniensis have been isolated from the oral cavity of immunocompromised patients (Kirkpatrick et al. Citation2000), while C. albicans and C. glabrata were frequently co-isolated from patients with denture-associated stomatitis (Coco et al. Citation2008). In another study, the presence of C. albicans in a mixed biofilm facilitated the invasion of C. glabrata in an in vitro model of denture stomatitis and promoted significant tissue damage to the epithelium during vulvovaginal candidiasis (Silva et al. Citation2011; Alves et al. Citation2014). The increasing number of mixed Candida species biofilms poses a major problem in the clinical setting due to the resistance of Candida species to a variety of antifungal drugs (Al-Fattani and Douglas Citation2006; Bizerra et al. Citation2008). These interactions highlight the importance of further research into the mechanisms and consequences of C. albicans and non-C. albicans species biofilm formation.
Communication within mixed-species biofilms
Numerous chemical interactions occur between the microbes found within a mixed-species biofilm via the secretion of small molecules and the exchange of nutrients. Both bacterial and fungal populations within a biofilm can modulate their behaviour via quorum sensing; a mechanism of chemical interaction in which microbes communicate through the secretion of signalling molecules (Wongsuk et al. Citation2016; ). Quorum sensing induces changes in transcriptional activity, which in turn influences the behaviour, survival, or virulence of either microbe. Candida albicans secretes farnesol, a quorum-sensing molecule that inhibits the Ras1-cyclic AMP (cAMP)-protein kinase A (PKA) (Ras1-cAMP-PKA) signalling pathway to repress hyphal development (Hornby et al. Citation2001; Davis-Hanna et al. Citation2008), inhibit biofilm formation (Ramage, Saville, et al. Citation2002) and upregulate the expression of FCR1 and PDR16, which are involved in resistance to azole therapy (Cao et al. Citation2005). Notably, the addition of farnesol to cultures of P. aeruginosa decreases the production of Pseudomonas quinolone signalling (PQS) molecule, a known suppressor of C. albicans biofilm formation, and the PQS-controlled virulence factor pyocyanin and inhibits swarming motility (Cugini et al. Citation2007; McAlester et al. Citation2008). As these P. aeruginosa factors are toxic to C. albicans, the release of farnesol within this interaction is likely to promote C. albicans survival.
In the oral cavity, S. gordonii uses quorum sensing to suppress farnesol-mediated repression of C. albicans filamentation, enhancing biofilm formation (Bamford et al. Citation2009). Such behaviour is far from altruistic. Indeed, S. gordonii promotes fungal growth by providing a carbon source to C. albicans (Bamford et al. Citation2009) and, in return, C. albicans increases the likelihood that S. gordonii can survive and persist in the oral cavity by lowering oxygen tension and releasing bacterial growth factors derived from nutrient metabolism (Shirtliff et al. Citation2009).
Molecules secreted by microbes can have an inhibitory effect on C. albicans. In the gastrointestinal tract, Bacteroides thetaiotaomicron stimulates the host to express HIF-1α and LL-37, which inhibit gastrointestinal colonisation by C. albicans (Fan et al. Citation2015). Similarly, C. albicans is susceptible to type VI secretion system (T6SS)-mediated antifungal activity perpetrated by the Gram-negative opportunistic bacterium Serratia marcescens, which uses its T6SS to penetrate the fungal cell wall to deliver the toxic effector proteins Tfe1 and Tfe2 that disrupt nutrient uptake, amino acid metabolism and plasma membrane potential (Trunk et al. Citation2018). However, whether this occurs in the context of a multispecies biofilm has yet to be investigated.
Overall, polymicrobial interactions within multispecies biofilms are of great importance and further studies may aid better understanding of C. albicans co-infection and possible therapeutic strategies to combat inter and intra-kingdom biofilm infections. Moreover, our understanding from next-generation sequencing studies, accompanied by greater bioinformatics power, is likely to aid our discovery of key interactions between Candida spp. and bacteria from an array of clinical diseases.
Clinical impact of biofilms
Biofilms are intrinsically associated with highly virulent, persistent and drug-resistant infections. The dispersal of cells from an established biofilm facilitates whole-body dissemination and reduces the likelihood of a positive clinical outcome and compounds options for effective therapeutic intervention. As such, C. albicans biofilms are widely considered a major virulence factor and a key determinant to the high mortality rate attributed with candidiasis (Tumbarello et al. Citation2007; Tumbarello et al. Citation2012; Rajendran, Sherry, et al. Citation2016). Infections that arise from mixed-species biofilms are considerably more difficult to treat than their single-species counterparts, requiring complex multi-drug treatment strategies (Costa-Orlandi et al. Citation2017; Montelongo-Jauregui et al. Citation2019). Antimicrobials that target a specific group of species often enable other microbes within the biofilm to thrive and continue infection (Pammi et al. Citation2014; Kean et al. Citation2017; Allison et al. Citation2019); for instance within a S. aureus–C. albicans biofilm, the efficacy of miconazole is reduced, with a much higher concentration of the drug needed to induce an effect against the biofilm (Kean et al. Citation2017). This increases the challenge of finding effective treatments to tackle C. albicans multispecies biofilms and may exacerbate issues relating to drug resistance. While the treatment of single species C. albicans biofilms has been studied in vivo, in vitro and in clinical settings (Schinabeck et al. Citation2004; Shuford et al. Citation2006; Tsui et al. Citation2016), considerably less is known about targeted therapeutics to mixed-species biofilms. Understanding these polymicrobial interactions in mixed-species biofilms is necessary in order to improve current therapeutic strategies. Overall, we note a lack of anti-biofilm strategies, but an increasing need for efficacious therapies that specifically target C. albicans biofilms.
Therapeutic intervention
In recent years, attempts have been made to utilise natural or synthetic compounds with improved antimicrobial activity. The use of quaternary ammonium amphiphiles may be a useful application in preventing and treating C. albicans biofilms. Amphiphile screening has revealed putative antimycotic agents that target C. albicans membranes in biofilms (Gupta et al. Citation2020), and which prevent biofilm formation on acrylics and promote degradation. Natural compounds have served as a source of novel alternatives to C. albicans biofilm treatment (Manoharan et al. Citation2017). Shikonin, from the roots of the Lithospermum erythrorhizon plant, prevents biofilm formation and maintenance (Yan et al. Citation2019), while Thymus vulgaris, and its active compound thymol, has demonstrated efficacy against C. albicans biofilms in vitro when used alone or in combination with fluconazole, revealing disaggregation and deformity in C. albicans biofilms (Jafri and Ahmad Citation2020). Furthermore, thymol acts synergistically with standard antifungals (Sharifzadeh et al. Citation2018) and could be utilised in combination therapy. Recent studies have shown that miconazole combined with a chitosan-coated iron oxide nanoparticle could be used in anti-biofilm therapy against C. albicans (Arias, Pessan, et al. Citation2020), and that this was also efficacious within a range of different oral interkingdom biofilms (Arias, Brown, et al. Citation2020). These alternative methods of treatment may present a strategy to overcome tolerance in the treatment of C. albicans biofilms, but also in more complex interkingdom communities. The potential benefits of combination therapy include a broader spectrum of action, greater potency than agents used in monotherapy, reduction in the number of resistant organisms and lower tolerance to antifungal agents (Lewis and Kontoyiannis Citation2001; Mukherjee et al. Citation2005; De Cremer et al. Citation2015). More recently, drug combination therapy has been reported as an alternative to conventional treatment. The co-delivery of C12AHL (a quorum-sensing molecule) with fluconazole in a liposomal drug carrier has shown promise as a strategy to eliminate C. albicans biofilms (Bandara et al. Citation2020). In recent work, key components of the host innate immune response and fungal cell wall components have been a focus for the development of new antifungals. The antimicrobial peptides ToAP2 and NDBP-5.7, increased yeast cell permeability and generated morphological alterations in C. albicans cells in both the early and later stages of biofilm formation (do Nascimento Dias et al. Citation2020). Furthermore, the combination therapy utilised within this study increased the efficacy of fluconazole and amphotericin B, decreasing their active concentrations and potential toxicity (do Nascimento Dias et al. Citation2020). Finally, vaccination of mice with the N-terminus of recombinant Als3p (designated NDV-3A) markedly reduced the ability of C. albicans to colonise a medically implanted jugular vein catheter (Alqarihi et al. Citation2019), suggesting that targeting key adhesins may have a positive outcome in the clinical setting.
Conclusion
The ability of C. albicans to form biofilms is a complex process and complicates treatment of C. albicans biofilm-related infections. Biofilm development, antifungal resistance and polymicrobial interactions have been a continual focus of C. albicans biofilm research and while recent advances have begun to highlight the complex mechanisms that underpin the formation of C. albicans single and mixed-species biofilms, there is still much to be discovered. The prevention of biofilm formation and the eradication of mature biofilms in the clinical setting present formidable challenges that must be overcome if the mortality associated with biofilm infection is to be addressed. The question remains as to how we can utilise this knowledge to develop more efficacious strategies to treat C. albicans biofilms and biofilm-related infections.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Al-Dhaheri RS, Douglas LJ. 2008. Absence of amphotericin B-tolerant persister cells in biofilms of some Candida species. Antimicrob Agents Chemother. 52(5):1884–1887.
- Al-Fattani MA, Douglas LJ. 2006. Biofilm matrix of Candida albicans and Candida tropicalis: chemical composition and role in drug resistance. J Med Microbiol. 55(8):999–1008.
- Allison DL, Scheres N, Willems HME, Bode CS, Krom BP, Shirtliff ME. 2019. The host immune system facilitates disseminated Staphylococcus aureus disease due to phagocytic attraction to Candida albicans during coinfection: a case of bait and switch. Infect Immun. 87(11):e00137-19.
- Alqarihi A, Singh S, Edwards JE, Ibrahim AS, Uppuluri P. 2019. NDV-3A vaccination prevents C. albicans colonization of jugular vein catheters in mice. Sci Rep. 9(1):6194.
- Alsteens D, Garcia MC, Lipke PN, Dufrêne YF. 2010. Force-induced formation and propagation of adhesion nanodomains in living fungal cells. Proc Natl Acad Sci USA. 107(48):20744–20749.
- Alves CT, Wei XQ, Silva S, Azeredo J, Henriques M, Williams DW. 2014. Candida albicans promotes invasion and colonisation of Candida glabrata in a reconstituted human vaginal epithelium. J Infect. 69(4):396–407.
- Andes D, Nett J, Oschel P, Albrecht R, Marchillo K, Pitula A. 2004. Development and characterization of an in vivo central venous catheter Candida albicans biofilm model. Infect Immun. 72(10):6023–6031.
- Arias LS, Brown JL, Butcher MC, Delaney C, Monteiro DR, Ramage G. 2020. A nanocarrier system that potentiates the effect of miconazole within different interkingdom biofilms. J Oral Microbiol. 12(1):1771071.
- Arias LS, Pessan JP, de Souza Neto FN, Lima BHR, de Camargo ER, Ramage G, Delbem ACB, Monteiro DR. 2020. Novel nanocarrier of miconazole based on chitosan-coated iron oxide nanoparticles as a nanotherapy to fight Candida biofilms. Colloids Surfaces B Biointerfaces. 192:111080.
- Aroutcheva A, Gariti D, Simon M, Shott S, Faro J, Simoes JA, Gurguis A, Faro S. 2001. Defense factors of vaginal lactobacilli. Am J Obstet Gynecol. 185(2):375–379.
- Askew C, Sellam A, Epp E, Mallick J, Hogues H, Mullick A, Nantel A, Whiteway M. 2011. The zinc cluster transcription factor Ahr1p directs Mcm1p regulation of Candida albicans adhesion. Mol Microbiol. 79(4):940–953.
- Awad A, El Khoury P, Wex B, Khalaf RA. 2018. Proteomic analysis of a Candida albicans pga1 Null Strain. EuPA Open Proteom. 18:1–6.
- Bailey DA, Feldmann PJF, Bovey M, Gow NAR, Brown AJP. 1996. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 178(18):5353–5360.
- Baillie GS, Douglas LJ. 1999. Role of dimorphism in the development of Candida albicans biofilms. J Med Microbiol. 48(7):671–679.
- Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. 2009. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infect Immun. 77(9):3696–3704.
- Bandara HMHN, Hewavitharana AK, Shaw PN, Smyth HDC, Samaranayake LP. 2020. A novel, quorum sensor-infused liposomal drug delivery system suppresses Candida albicans biofilms. Int J Pharm. 578:119096.
- Ben-Ami R, Zimmerman O, Finn T, Amit S, Novikov A, Wertheimer N, Lurie-Weinberger M, Berman J. 2016. Heteroresistance to fluconazole is a continuously distributed phenotype among Candida glabrata clinical strains associated with in vivo persistence. MBio. 7(4):e00655–16.
- Berman J, Krysan DJ. 2020. Drug resistance and tolerance in fungi. Nat Rev Microbiol. 18(6):319–331.
- Bizerra FC, Nakamura CV, de Poersch C, Estivalet Svidzinski TI, Borsato Quesada RM, Goldenberg S, Krieger MA, Yamada-Ogatta SF. 2008. Characteristics of biofilm formation by Candida tropicalis and antifungal resistance. FEMS Yeast Res. 8(3):442–450.
- Bongomin F, Gago S, Oladele RO, Denning DW. 2017. Global and multi-national prevalence of fungal diseases—estimate precision. J Fungi. 3(4):57.
- Bonhomme J, Chauvel M, Goyard S, Roux P, Rossignol T, D’Enfert C. 2011. Contribution of the glycolytic flux and hypoxia adaptation to efficient biofilm formation by Candida albicans. Mol Microbiol. 80(4):995–1013.
- Boris S, Barbés C. 2000. Role played by lactobacilli in controlling the population of vaginal pathogens. Microbes Infect. 2(5):543–546.
- Boris S, Suárez JE, Vázquez F, Barbés C. 1998. Adherence of human vaginal lactobacilli to vaginal epithelial cells and interaction with uropathogens. Infect Immun. 66(5):1985–1989.
- Böttcher B, Hoffmann B, Garbe E, Weise T, Cseresnyés Z, Brandt P, Dietrich S, Driesch D, Figge MT, Vylkova S, et al. 2020. The transcription factor Stp2 is important for Candida albicans biofilm establishment and sustainability. Front Microbiol. 11:794.
- Braun BR, Kadosh D, Johnson AD. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. Embo J. 20(17):4753–4761.
- Cao YY, Cao YB, Xu Z, Ying K, Li Y, Xie Y, Zhu Z-Y, Chen W-S, Jiang Y-Y. 2005. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob Agents Chemother. 49(2):584–589.
- Cavalheiro M, Teixeira MC. 2018. Candida biofilms: threats, challenges, and promising strategies. Front Med. 5:28.
- Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. 2001. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 183(18):5385–5394.
- Chandra J, McCormick TS, Imamura Y, Mukherjee PK, Ghannoum MA. 2007. Interaction of Candida albicans with adherent human peripheral blood mononuclear cells increases C. albicans biofilm formation and results in differential expression of pro- and anti-inflammatory cytokines. Infect Immun. 75(5):2612–2620.
- Chen H-F, Lan C-Y. 2015. Role of SFP1 in the regulation of Candida albicans biofilm formation. PLoS One. 10(6):e0129903.
- Chong PP, Chin VK, Wong WF, Madhavan P, Yong VC, Looi CY. 2018. Transcriptomic and genomic approaches for unravelling Candida albicans biofilm formation and drug resistance—an update. Genes. 9(11):540.
- Coco BJ, Bagg J, Cross LJ, Jose A, Cross J, Ramage G. 2008. Mixed Candida albicans and Candida glabrata populations associated with the pathogenesis of denture stomatitis. Oral Microbiol Immunol. 23(5):377–383.
- Costa-Orlandi CB, Sardi JCO, Pitangui NS, de Oliveira HC, Scorzoni L, Galeane MC, Medina-Alarcón KP, Melo WCMA, Marcelino MY, Braz JD, et al. 2017. Fungal biofilms and polymicrobial diseases. J Fungi. 3(2):22.
- Crump JA, Collignon PJ. 2000. Intravascular catheter-associated infections. Eur J Clin Microbiol Infect Dis. 19(1):1–8.
- Cugini C, Calfee MW, Farrow JM, Morales DK, Pesci EC, Hogan DA. 2007. Farnesol, a common sesquiterpene, inhibits PQS production in Pseudomonas aeruginosa. Mol Microbiol. 65(4):896–906.
- Daniels KJ, Srikantha T, Pujol C, Park YN, Soll DR. 2015. Role of Tec1 in the development, architecture, and integrity of sexual biofilms of Candida albicans. Eukaryot Cell. 14(3):228–240.
- Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. 2008. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol. 67(1):47–62.
- De Brucker K, Tan Y, Vints K, De Cremer K, Braem A, Verstraeten N, Michiels J, Vleugels J, Cammue BPA, Thevissen K, et al. 2015. Fungal β-1,3-Glucan Increases Ofloxacin Tolerance of Escherichia coli in a Polymicrobial E. coli/Candida albicans Biofilm. Antimicrob Agents Chemother. 59(6):3052–3058.
- de Carvalho FG, Silva DS, Hebling J, Spolidorio LC, Spolidorio DMP. 2006. Presence of mutans streptococci and Candida spp. in dental plaque/dentine of carious teeth and early childhood caries. Arch Oral Biol. 51(11):1024–1028.
- De Cremer K, Staes I, Delattin N, Cammue BPA, Thevissen K, De Brucker K. 2015. Combinatorial drug approaches to tackle Candida albicans biofilms. Expert Rev anti Infect Ther. 13(8):973–984.
- de Groot PWJ, Bader O, de Boer AD, Weig M, Chauhan N. 2013. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell. 12(4):470–481.
- de Groot PWJ, Hellingwerf KJ, Klis FM. 2003. Genome-wide identification of fungal GPI proteins. Yeast. 20(9):781–796.
- Delaney C, Kean R, Short B, Tumelty M, McLean W, Nile CJ, Ramage G. 2018. Fungi at the scene of the crime: innocent bystanders or accomplices in oral infections? Curr Clin Micro Rpt. 5(3):190–200.
- Denega I, D’Enfert C, Bachellier-Bassi S. 2019. Candida albicans biofilms are generally devoid of persister cells. Antimicrob Agents Chemother. 63(5):e01979–18.
- Desai JV, Mitchell AP. 2015. Candida albicans biofilm development and its genetic control. Microbiol Spectr. 3(3)
- do Nascimento Dias J, de Souza Silva C, de Araújo AR, Souza JMT, de Holanda Veloso Júnior PH, Cabral WF, da Glória da Silva M, Eaton P, de Souza de Almeida Leite JR, Nicola AM, et al. 2020. Mechanisms of action of antimicrobial peptides ToAP2 and NDBP-5.7 against Candida albicans planktonic and biofilm cells. Sci Rep. 10(1):1–14.
- Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. 2009. Characterization of mucosal Candida albicans biofilms. PLoS One. 4(11):e7967.
- Douglas LJ. 1987. Adhesion of Candida species to epithelial surfaces. Crit Rev Microbiol. 15(1):27–43.
- Dwivedi P, Thompson A, Xie Z, Kashleva H, Ganguly S, Mitchell AP, Dongari-Bagtzoglou A. 2011. Role of Bcr1-activated genes Hwp1 and Hyr1 in Candida albicans oral mucosal biofilms and neutrophil evasion. PLoS One. 6(1):e16218.
- El-Azizi MA, Starks SE, Khardori N. 2004. Interactions of Candida albicans with other Candida spp. and bacteria in the biofilms. J Appl Microbiol. 96(5):1067–1073.
- Elias S, Banin E. 2012. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 36(5):990–1004.
- Ellepola K, Liu Y, Cao T, Koo H, Seneviratne CJ. 2017. Bacterial GtfB augments Candida albicans accumulation in cross-kingdom biofilms. J Dent Res. 96(10):1129–1135.
- Ene IV, Bennett RJ. 2009. Hwp1 and related adhesins contribute to both mating and biofilm formation in Candida albicans. Eukaryot Cell. 8(12):1909–1913.
- Ene IV, Heilmann CJ, Sorgo AG, Walker LA, de Koster CG, Munro CA, Klis FM, Brown AJP. 2012. Carbon source-induced reprogramming of the cell wall proteome and secretome modulates the adherence and drug resistance of the fungal pathogen Candida albicans. Proteomics. 12(21):3164–3179.
- Falsetta ML, Klein MI, Colonne PM, Scott-Anne K, Gregoire S, Pai C-H, Gonzalez-Begne M, Watson G, Krysan DJ, Bowen WH, et al. 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 82(5):1968–1981.
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, Koh AY, et al. 2015. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 21(7):808–814.
- Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, et al. 2012. Portrait of Candida albicans adherence regulators. PLoS Pathog. 8(2):e1002525.
- Fox EP, Bui CK, Nett JE, Hartooni N, Mui MC, Andes DR, Nobile CJ, Johnson AD. 2015. An expanded regulatory network temporally controls Candida albicans biofilm formation. Mol Microbiol. 96(6):1226–1239.
- Fox EP, Cowley ES, Nobile CJ, Hartooni N, Newman DK, Johnson AD. 2014. Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr Biol. 24(20):2411–2416.
- Fox EP, Nobile CJ. 2012. A sticky situation: untangling the transcriptional network controlling biofilm development in Candida albicans. Transcription. 3(6):315–322.
- Ganguly S, Bishop AC, Xu W, Ghosh S, Nikerson KW, Lanni F, Patton-Vogt J, Mitchell AP. 2011. Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot Cell. 10(11):1448–1454.
- Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrêne YF, Lipke PN. 2011. A role for amyloid in cell aggregation and biofilm formation. PLoS One. 6(3):e17632.
- García-Sánchez S, Iraqui AS, Janbon I, Ghigo G, D’Enfert JM. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell. 3(2):536–545.
- Ghosh AK, Wangsanut T, Fonzi WA, Rolfes RJ. 2015. The GRF10 homeobox gene regulates filamentous growth in the human fungal pathogen Candida albicans. FEMS Yeast Res. 15. DOI:10.1093/femsyr/fov093
- Gibson J, Sood A, Hogan DA. 2009. Pseudomonas aeruginosa-Candida albicans interactions: localization and fungal toxicity of a phenazine derivative. Appl Environ Microbiol. 75(2):504–513.
- Glazier VE, Murante T, Murante D, Koselny K, Liu Y, Kim D, Koo H, Krysan DJ. 2017. Genetic analysis of the Candida albicans biofilm transcription factor network using simple and complex haploinsufficiency. PLoS Genet. 13(8):e1006948.
- Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. 2005. Yeast wall protein 1 of Candida albicans. Microbiology. 151(5):1631–1644.
- Gupta N, Haque A, Mukhopadhyay G, Narayan RP, Prasad R. 2005. Interactions between bacteria and Candida in the burn wound. Burns. 31(3):375–378.
- Gupta R, Thakur J, Pal S, Mishra D, Rani P, Kumar S, Saini A, Singh A, Yadav K, Srivastava A, et al. 2020. Cholic-acid-derived amphiphiles can prevent and degrade fungal biofilms. ACS Appl Bio Mater.
- Hajjeh RA, Sofair AN, Harrison LH, Lyon GM, Arthington-Skaggs BA, Mirza SA, Phelan M, Morgan J, Lee-Yang W, Ciblak MA, et al. 2004. Incidence of bloodstream infections due to Candida species and in vitro susceptibilities of isolates collected from 1998 to 2000 in a population-based active surveillance program. J Clin Microbiol. 42(4):1519–1527.
- Harriott MM, Lilly EA, Rodriguez TE, Fidel PL, Noverr MC. 2010. Candida albicans forms biofilms on the vaginal mucosa. Microbiology. 156(12):3635–3644.
- Harriott MM, Noverr MC. 2009. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob Agents Chemother. 53(9):3914–3922.
- Hashash R, Younes S, Bahnan W, El Koussa J, Maalouf K, Dimassi HI, Khalaf RA. 2011. Characterisation of Pga1, a putative Candida albicans cell wall protein necessary for proper adhesion and biofilm formation. Mycoses. 54(6):491–500.
- Hawser SP, Douglas LJ. 1994. Biofilm formation by Candida species on the surface of catheter materials in vitro. Infect Immun. 62(3):915–921.
- Hazen KC, Lay JG, Hazen BW, Fu RC, Murthy S. 1990. Partial biochemical characterization of cell surface hydrophobicity and hydrophilicity of Candida albicans. Infect Immun. 58(11):3469–3476.
- Hazen KC. 1989. Participation of yeast cell surface hydrophobicity in adherence of Candida albicans to human epithelial cells. Infect Immun. 57(7):1894–1900.
- Hobden C, Teevan C, Jones L, O’Shea P. 1995. Hydrophobic properties of the cell surface of Candida albicans: a role in aggregation. Microbiology. 141(8):1875–1881.
- Hogan DA, Kolter R. 2002. Pseudomonas-Candida interactions: an ecological role for virulence factors. Science. 296(5576):2229–2232.
- Hogan DA, Vik Å, Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol Microbiol. 54(5):1212–1223.
- Holmes AR, McNab R, Jenkinson HF. 1996. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infect Immun. 64(11):4680–4685.
- Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl Environ Microbiol. 67(7):2982–2992.
- Hoyer LL, Cota E. 2016. Candida albicans agglutinin-like sequence (Als) family vignettes: a review of als protein structure and function. Front Microbiol. 7:280.
- Hoyer LL, Oh SH, Jones R, Cota E. 2014. A proposed mechanism for the interaction between the Candida albicans Als3 adhesin and streptococcal cell wall proteins. Front Microbiol. 5:564
- Hwang G, Liu Y, Kim D, Li Y, Krysan DJ, Koo H. 2017. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLoS Pathog. 13(6):e1006407.
- Hwang G, Marsh G, Gao L, Waugh R, Koo H. 2015. Binding force dynamics of Streptococcus mutans -glucosyltransferase B to Candida albicans. J Dent Res. 94(9):1310–1317.
- Jafri H, Ahmad I. 2020. Thymus vulgaris essential oil and thymol inhibit biofilms and interact synergistically with antifungal drugs against drug resistant strains of Candida albicans and Candida tropicalis. J Mycol Med. 30(1):100911.
- Jarosz LM, Deng DM, Van Der Mei HC, Crielaard W, Krom BP. 2009. Streptococcus mutans competence-stimulating peptide inhibits Candida albicans hypha formation. Eukaryot Cell. 8(11):1658–1664.
- Johnson CC, Yu A, Lee H, Fidel PL, Noverr MC. 2012. Development of a contemporary animal model of Candida albicans-associated denture stomatitis using a novel intraoral denture system. Infect Immun. 80(5):1736–1743.
- Johnson CJ, Cabezas-Olcoz J, Kernien JF, Wang SX, Beebe DJ, Huttenlocher A, Ansari H, Nett JE. 2016. The extracellular matrix of Candida albicans biofilms impairs formation of neutrophil extracellular traps. PLoS Pathog. 12(9):e1005884.
- Johnson CJ, Kernien JF, Hoyer AR, Nett JE. 2017. Mechanisms involved in the triggering of neutrophil extracellular traps (NETs) by Candida glabrata during planktonic and biofilm growth. Sci Rep. 7(1):1–13.
- Jones DS, Schep LJ, Shepherd MG. 1995. The effect of cetylpyridinium chloride (CPC) on the cell surface hydrophobicity and adherence of Candida albicans to human buccal epithelial cells in vitro. Pharm Res an off J Am Assoc Pharm Sci. 12(12):1896–1900.
- Joyner PM, Liu J, Zhang Z, Merritt J, Qi F, Cichewicz RH. 2010. Mutanobactin A from the human oral pathogen Streptococcus mutans is a cross-kingdom regulator of the yeast-mycelium transition. Org Biomol Chem. 8(24):5486–5489.
- Kakade P, Mahadik K, Balaji KN, Sanyal K, Nagaraja V. 2019. Two negative regulators of biofilm development exhibit functional divergence in conferring virulence potential to Candida albicans. FEMS Yeast Res. 19. DOI:10.1093/femsyr/foy078
- Kakade P, Sadhale P, Sanyal K, Nagaraja V. 2016. ZCF32, a fungus specific Zn(II)2 Cys6 transcription factor, is a repressor of the biofilm development in the human pathogen Candida albicans. Sci Rep. 6(1). DOI:10.1038/srep31124
- Kasai M, Francesconi A, Petraitiene R, Petraitis V, Kelaher AM, Kim H-s, Meletiadis J, Sein T, Bacher J, Walsh TJ, et al. 2006. Use of quantitative real-time PCR to study the kinetics of extracellular DNA released from Candida albicans, with implications for diagnosis of invasive candidiasis. J Clin Microbiol. 44(1):143–150.
- Katragkou A, Kruhlak MJ, Simitsopoulou M, Chatzimoschou A, Taparkou A, Cotten CJ, Paliogianni F, Diza-Mataftsi E, Tsantali C, Walsh TJ, et al. 2010. Interactions between human phagocytes and Candida albicans biofilms alone and in combination with antifungal agents. J Infect Dis. 201(12):1941–1949.
- Kean R, Delaney C, Rajendran R, Sherry L, Thomas R, Williams C, Ramage G, Metcalfe R, McLean W. 2018. Gaining insights from Candida biofilm heterogeneity: one size does not fit all. J Fungi. 4(1):12.
- Kean R, Rajendran R, Haggarty J, Townsend EM, Short B, Burgess KE, Lang S, Millington O, Mackay WG, Williams C, et al. 2017. Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front Microbiol. 8:258.
- Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJP, Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 53(3):969–983.
- Kernien JF, Snarr BD, Sheppard DC, Nett JE. 2018. The interface between fungal biofilms and innate immunity. Front Immunol. 8:1968.
- Khoury ZH, Vila T, Puthran TR, Sultan AS, Montelongo-Jauregui D, Melo MAS, Jabra-Rizk MA. 2020. The Role of Candida albicans Secreted Polysaccharides in Augmenting Streptococcus mutans Adherence and Mixed Biofilm Formation: In vitro and in vivo Studies. Front Microbiol. 11: 307.
- Kim D, Sengupta A, Niepa THR, Lee B-H, Weljie A, Freitas-Blanco VS, Murata RM, Stebe KJ, Lee D, Koo H, et al. 2017. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci Rep. 7:41332.
- Kirkpatrick WR, Lopez-Ribot JL, Mcatee RK, Patterson TF. 2000. Growth competition between Candida dubliniensis and Candida albicans under broth and biofilm growing conditions. J Clin Microbiol. 38(2):902–904.
- Klotz SA, Gaur NK, De Armond R, Sheppard D, Khardori N, Edwards JE, Lipke PN, El-Azizi M. 2007. Candida albicans Als proteins mediate aggregation with bacteria and yeasts. Med Mycol. 45(4):363–370.
- Kojic EM, Darouiche RO. 2004. Candida infections of medical devices. Clin Microbiol Rev. 17(2):255–267.
- Kong EF, Tsui C, Kucharíková S, Andes D, Van Dijck P, Jabra-Rizk MA. 2016. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. MBio. 7(5):e01365–16.
- Krom BP, Kidwai S, ten Cate JM. 2014. Candida and other fungal species: forgotten players of healthy oral microbiota. J Dent Res. 93(5):445–451.
- LaFleur MD, Qi Q, Lewis K. 2010. Patients with long-term oral carriage harbor high-persister mutants of Candida albicans. Antimicrob Agents Chemother. 54(1):39–44.
- Lagree K, Woolford CA, Huang MY, May G, McManus CJ, Solis NV, Filler SG, Mitchell AP. 2020. Roles of Candida albicans Mig1 and Mig2 in glucose repression, pathogenicity traits, and SNF1 essentiality. PLoS Genet. 16(1):e1008582.
- Lee JA, Robbins N, Xie JL, Ketela T, Cowen LE. 2016. Functional genomic analysis of Candida albicans adherence reveals a key role for the Arp2/3 complex in cell wall remodelling and biofilm formation. PLoS Genet. 12(11):e1006452.
- Lewis RE, Kontoyiannis DP. 2001. Rationale for combination antifungal therapy. Pharmacotherapy. 21(8):149S–164S.
- Li F, Svarovsky MJ, Karlsson AJ, Wagner JP, Marchillo K, Oshel P, Andes D, Palecek SP. 2007. Eap1p, an adhesin that mediates Candida albicans biofilm formation in vitro and in vivo. Eukaryot Cell. 6(6):931–939.
- Li WS, Chen YC, Kuo SF, Chen FJ, Lee CH. 2018. The impact of biofilm formation on the persistence of candidemia. Front Microbiol. 9:1196.
- Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrêne YF. 2012. Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 20(2):59–65.
- Lipke PN, Ramsook C, Garcia-Sherman MC, Jackson DN, Chan CXJ, Bois M, Klotz SA. 2014. Between amyloids and aggregation lies a connection with strength and adhesion. New J Sci. 2014:815102.
- Liu Y, Filler SG. 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell. 10(2):168–173.
- Lohse MB, Gulati M, Johnson AD, Nobile CJ. 2018. Development and regulation of single- and multi-species Candida albicans biofilms. Nat Rev Microbiol. 16(1):19–31.
- Mailänder-Sánchez D, Braunsdorf C, Grumaz C, Müller C, Lorenz S, Stevens P, Wagener J, Hebecker B, Hube B, Bracher F, et al. 2017. Antifungal defense of probiotic Lactobacillus rhamnosus GG is mediated by blocking adhesion and nutrient depletion. PLoS One. 12(10):e0184438.
- Manoharan RK, Lee JH, Kim YG, Kim SI, Lee J. 2017. Inhibitory effects of the essential oils α-longipinene and linalool on biofilm formation and hyphal growth of Candida albicans. Biofouling. 33(2):143–155.
- Martins M, Henriques M, Lopez-Ribot JL, Oliveira R. 2012. Addition of DNase improves the in vitro activity of antifungal drugs against Candida albicans biofilms. Mycoses. 55(1):80–85.
- Martins M, Uppuluri P, Thomas DP, Cleary IA, Henriques M, Lopez-Ribot JL, Oliveira R. 2010. Presence of extracellular DNA in the Candida albicans biofilm matrix and its contribution to biofilms. Mycopathologia. 169(5):323–331.
- Mateus C, Crow SA, Ahearn DG. 2004. Adherence of Candida albicans to silicone induces immediate enhanced tolerance to fluconazole. Antimicrob Agents Chemother. 48(9):3358–3366.
- Mathé L, Van Dijck P. 2013. Recent insights into Candida albicans biofilm resistance mechanisms. Curr Genet. 59(4):251–264.
- Mayer FL, Wilson D, Hube B. 2013. Candida albicans pathogenicity mechanisms. Virulence. 4(2):119–128.
- McAlester G, O’Gara F, Morrissey JP. 2008. Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J Med Microbiol. 57(5):563–569.
- McCall AD, Pathirana RU, Prabhakar A, Cullen PJ, Edgerton M. 2019. Candida albicans biofilm development is governed by cooperative attachment and adhesion maintenance proteins. Npj Biofilms Microbiomes. 5(1):1–12.
- Metwalli KH, Khan SA, Krom BP, Jabra-Rizk MA. 2013. Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 9(10):e1003616.
- Miyazato A, Nakamura K, Yamamoto N, Mora-Montes HM, Tanaka M, Abe Y, Tanno D, Inden K, Gang X, Ishii K, et al. 2009. Toll-like receptor 9-dependent activation of myeloid dendritic cells by deoxynucleic acids from Candida albicans. Infect Immun. 77(7):3056–3064.
- Monniot C, Boisramé A, Da Costa G, Chauvel M, Sautour M, Bougnoux M-E, Bellon-Fontaine M-N, Dalle F, d’Enfert C, Richard ML, et al. 2013. Rbt1 protein domains analysis in Candida albicans brings insights into hyphal surface modifications and Rbt1 potential role during adhesion and biofilm formation. PLoS One. 8(12):e82395.
- Montelongo-Jauregui D, Saville SP, Lopez-Ribot JL. 2019. Contributions of Candida albicans dimorphism, adhesive interactions, and extracellular matrix to the formation of dual-species biofilms with Streptococcus gordonii. MBio. 10(3):e01179–19.
- Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, et al. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 532(7597):64–68.
- Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. 2003. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. IAI. 71(8):4333–4340.
- Mukherjee PK, Sheehan DJ, Hitchcock CA, Ghannoum MA. 2005. Combination treatment of invasive fungal infections. Clin Microbiol Rev. 18(1):163–194.
- Mulhern SM, Logue ME, Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot Cell. 5(12):2001–2013.
- Murad AMA, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, et al. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20(17):4742–4752.
- Nett J, Lincoln L, Marchillo K, Massey R, Holoyda K, Hoff B, VanHandel M, Andes D. 2007. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother. 51(2):510–520.
- Nett JE, Andes DR. 2020. Contributions of the biofilm matrix to Candida pathogenesis. J Fungi. 6(1):21.
- Nett JE, Crawford K, Marchillo K, Andes DR. 2010. Role of Fks1p and matrix glucan in Candida albicans biofilm resistance to an echinocandin, pyrimidine, and polyene. Antimicrob Agents Chemother. 54(8):3505–3508.
- Nett JE, Lepak AJ, Marchillo K, Andes DR. 2009. Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis. 200(2):307–313.
- Nett JE, Sanchez H, Cain MT, Andes DR. 2010. Genetic basis of Candida Biofilm resistance due to drug-sequestering matrix glucan. J Infect Dis. 202(1):171–175.
- Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. 2011. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell. 10(12):1660–1669.
- Nett JE, Zarnowski R, Cabezas-Olcoz J, Brooks EG, Bernhardt J, Marchillo K, Mosher DF, Andes DR. 2015. Host contributions to construction of three device-associated Candida albicans biofilms. Infect Immun. 83(12):4630–4638.
- Nett JE. 2016. The host’s reply to Candida biofilm. Pathogens. 5(1):33.
- Nobile CJ, Andes DR, Nett JE, Smith FJ, Yue F, Phan Q-T, Edwards JE, Filler SG, Mitchell AP. 2006. Critical role of Bcr1-dependent adhesins in C. albicans biofilm formation in vitro and in vivo. PLoS Pathog. 2(7):e63.
- Nobile CJ, Fox EP, Hartooni N, Mitchell KF, Hnisz D, Andes DR, Kuchler K, Johnson AD. 2014. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. MBio. 5(3):e01201–14.
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 148(1–2):126–138.
- Nobile CJ, Johnson AD. 2015. Candida albicans biofilms and human disease. Annu Rev Microbiol. 69(1):71–92.
- Nobile CJ, Mitchell AP. 2005. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr Biol. 15(12):1150–1155.
- Nobile CJ, Mitchell AP. 2006. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 8(9):1382–1391.
- Nobile CJ, Nett JE, Andes DR, Mitchell AP. 2006. Function of Candida albicans adhesin hwp1 in biofilm formation. Eukaryot Cell. 5(10):1604–1610.
- Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault J-S, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol. 7(6):e1000133.
- Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr Biol. 18(14):1017–1024.
- Noverr MC, Fidel PL. 2019. Questions remain regarding the presence of fungal species biofilm in women with vulvovaginal candidiasis. Am J Obstet Gynecol. 221(2):169.
- Orsi CF, Sabia C, Ardizzoni A, Colombari B, Neglia RG, Peppoloni S, Morace G, Blasi E. 2014. Inhibitory effects of different lactobacilli on Candida albicans hyphal formation and biofilm development. J Biol Regul Homeost Agents. 28(4):743–752.
- Orsi GB, Stefano LD, Noah N. 2002. Hospital-acquired, laboratory-confirmed bloodstream infection: increased hospital stay and direct costs. Infect Control Hosp Epidemiol. 23(4):190–197.
- Pérez A, Pedros B, Murgui A, Casanova M, Lopez-Ribot JÃL, Martinez JÃP. 2006. Biofilm formation by Candida albicans mutants for genes coding fungal proteins exhibiting the eight-cysteine-containing CFEM domain. FEMS Yeast Res. 6(7):1074–1084.
- Paiva LCF, Vidigal PG, Donatti L, Svidzinski TIE, Consolaro MEL. 2012. Assessment of in vitro biofilm formation by Candida species isolates from vulvovaginal candidiasis and ultrastructural characteristics. Micron. 43(2–3):497–502.
- Pammi M, Zhong D, Johnson Y, Revell P, Versalovic J. 2014. Polymicrobial bloodstream infections in the neonatal intensive care unit are associated with increased mortality: a case-control study. BMC Infect Dis. 14(1):390.
- Panariello BHD, Klein MI, Pavarina AC, Duarte S. 2017. Inactivation of genes TEC1 and EFG1 in Candida albicans influences extracellular matrix composition and biofilm morphology. J Oral Microbiol. 9(1):1385372.
- Pappas PG, Kauffman CA, Andes DR, Clancy CJ, Marr KA, Ostrosky-Zeichner L, Reboli AC, Schuster MG, Vazquez JA, Walsh TJ, et al. 2016. Clinical practice guideline for the management of candidiasis: 2016 update by the infectious diseases society of America. Clin Infect Dis. 62(4):e1–e50.
- Park SE, Blissett R, Susarla SM, Weber HP. 2008. Candida albicans adherence to surface-modified denture resin surfaces. J Prosthodont. 17(5):365–369.
- Pathak AK, Sharma S, Shrivastva P. 2012. Multi-species biofilm of Candida albicans and non-Candida albicans Candida species on acrylic substrate. J Appl Oral Sci. 20(1):70–75.
- Pereira-Cenci T, Deng DM, Kraneveld EA, Manders EMM, Del Bel Cury AA, ten Cate JM, Crielaard W. 2008. The effect of Streptococcus mutans and Candida glabrata on Candida albicans biofilms formed on different surfaces. Arch Oral Biol. 53(8):755–764.
- Peters BM, Ovchinnikova ES, Krom BP, Schlecht LM, Zhou H, Hoyer LL, Busscher HJ, van der Mei HC, Jabra-Rizk MA, Shirtliff ME, et al. 2012. Staphylococcus aureus adherence to Candida albicans hyphae is mediated by the hyphal adhesin Als3p. Microbiology. 158(12):2975–2986.
- Pierce CG, Vila T, Romo JA, Montelongo-Jauregui D, Wall G, Ramasubramanian A, Lopez-Ribot L. 2017. The Candida albicans biofilm matrix: composition, structure and function. J Fungi. 3(1).
- Pittet D. 1994. Nosocomial bloodstream infection in critically ill patients. Excess length of stay, extra costs, and attributable mortality. JAMA. 271(20):1598.
- Puri S, Kumar R, Chadha S, Tati S, Conti HR, Hube B, Cullen PJ, Edgerton M. 2012. Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One. 7(11):e46020.
- Purschke FG, Hiller E, Trick I, Rupp S. 2012. Flexible survival strategies of Pseudomonas aeruginosa in biofilms result in increased fitness compared with Candida albicans. Mol Cell Proteomics. 11(12):1652–1669.
- Rajendran R, May A, Sherry L, Kean R, Williams C, Jones BL, Burgess KV, Heringa J, Abeln S, Brandt BW, et al. 2016. Integrating Candida albicans metabolism with biofilm heterogeneity by transcriptome mapping. Sci Rep. 6:35436.
- Rajendran R, Sherry L, Lappin DF, Nile CJ, Smith K, Williams C, Munro CA, Ramage G. 2014. Extracellular DNA release confers heterogeneity in Candida albicans biofilm formation. BMC Microbiol. 14(1):303.
- Rajendran R, Sherry L, Nile CJ, Sherriff A, Johnson EM, Hanson MF, Williams C, Munro CA, Jones BJ, Ramage G, et al. 2016. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012-2013. Clin Microbiol Infect. 22(1):87–93.
- Ramage G, Bachmann S, Patterson TF, Wickes BL, López-Ribot JL. 2002. Investigation of multidrug efflux pumps in relation to fluconazole resistance in Candida albicans biofilms. J Antimicrob Chemother. 49(6):973–980.
- Ramage G, Saville SP, Wickes BL, López-Ribot JL. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol. 68(11):5459–5463.
- Ramage G, VandeWalle K, López-Ribot JL, Wickes BL. 2002. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol Lett. 214(1):95–100.
- Ramsook CB, Tan C, Garcia MC. 2010. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot Cell. 9(3):393–404.
- Rauceo JM, Gaur NK, Lee KG, Edwards JE, Klotz SA, Lipke PN. 2004. Global cell surface conformational shift mediated by a Candida albicans adhesin. Infect Immun. 72(9):4948–4955.
- Richard ML, Nobile CJ, Bruno VM, Mitchell AP. 2005. Candida albicans biofilm-defective mutants. Eukaryot Cell. 4(8):1493–1502.
- Robbins N, Uppuluri P, Nett J, Rajendran R, Ramage G, Lopez-Ribot JL, Andes D, Cowen LE. 2011. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 7(9):e1002257.
- Roberts AP, Kreth J. 2014. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front Cell Infect Microbiol. 4:124.
- Rouabhia M, Semlali A, Chandra J, Mukherjee P, Chmielewski W, Ghannoum MA. 2012. Disruption of the ECM33 gene in Candida albicans prevents biofilm formation, engineered human oral mucosa tissue damage and gingival cell necrosis/apoptosis. Mediators Inflamm. 2012:398207–398209.
- Sahni N, Yi S, Daniels KJ, Srikantha T, Pujol C, Soll DR. 2009. Genes selectively up-regulated by pheromone in white cells are involved in biofilm formation in Candida albicans. PLoS Pathog. 5(10):e1000601.
- Sandini S, Stringaro A, Arancia S, Colone M, Mondello F, Murtas S, Girolamo A, Mastrangelo N, De Bernardis F. 2011. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol. 11:106.
- Sapaar B, Nur A, Hirota K, Yumoto H, Murakami K, Amoh T, Matsuo T, Ichikawa T, Miyake Y. 2014. Effects of extracellular DNA from Candida albicans and pneumonia-related pathogens on Candida biofilm formation and hyphal transformation. J Appl Microbiol. 116(6):1531–1542.
- Schierholz JM, Beuth J. 2001. Implant infections: a haven for opportunistic bacteria. J Hosp Infect. 49(2):87–93.
- Schinabeck MK, Long LA, Hossain MA, Chandra J, Mukherjee PK, Mohamed S, Ghannoum MA. 2004. Rabbit model of Candida albicans biofilm infection: liposomal amphotericin B antifungal lock therapy. AAC. 48(5):1727–1732.
- Schlecht LM, Peters BM, Krom BP, Freiberg JA, Hänsch GM, Filler SG, Jabra-Rizk MA, Shirtliff ME. 2015. Systemic Staphylococcus aureus infection mediated by Candida albicans hyphal invasion of mucosal tissue. Microbiology. 161(1):168–181.
- Sellam A, Askew C, Epp E, Tebbji F, Mullick A, Whiteway M, Nantel A. 2010. Role of transcription factor CaNdt80p in cell separation, hyphal growth, and virulence in Candida albicans. Eukaryot Cell. 9(4):634–644.
- Sharifzadeh A, Khosravi AR, Shokri H, Shirzadi H. 2018. Potential effect of 2-isopropyl-5-methylphenol (thymol) alone and in combination with fluconazole against clinical isolates of Candida albicans, C. glabrata and C. krusei. J Mycol Med. 28(2):294–299.
- Shen J, Cowen LE, Griffin AM, Chan L, Köhler JR. 2008. The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc Natl Acad Sci USA. 105(52):20918–20923.
- Sherry L, Kean R, McKloud E, O’Donnell LE, Metcalfe R, Jones BL, Ramage G. 2017. Biofilms formed by isolates from recurrent vulvovaginal candidiasis patients are heterogeneous and insensitive to fluconazole. Antimicrob Agents Chemother. 61(9):e01065–17.
- Sherry L, Rajendran R, Lappin DF, Borghi E, Perdoni F, Falleni M, Tosi D, Smith K, Williams C, Jones B, et al. 2014. Biofilms formed by Candida albicans bloodstream isolates display phenotypic and transcriptional heterogeneity that are associated with resistance and pathogenicity. BMC Microbiol. 14(1):182.
- Shirtliff ME, Peters BM, Jabra-Rizk MA. 2009. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 299(1):1–8.
- Shuford JA, Rouse MS, Piper KE, Steckelberg JM, Patel R. 2006. Evaluation of caspofungin and amphotericin B deoxycholate against Candida albicans biofilms in an experimental intravascular catheter infection model. J Infect Dis. 194(5):710–713.
- Silva S, Henriques MC, Hayes A, Oliveira R, Azeredo J, Williams DW. 2011. Candida glabrata and Candida albicans co-infection of an in vitro oral epithelium. J Oral Pathol Med. 40(5):421–427.
- Silva-Dias A, Miranda IM, Branco J, Monteiro-Soares M, Pina-Vaz C, Rodrigues AG. 2015. Adhesion, biofilm formation, cell surface hydrophobicity, and antifungal planktonic susceptibility: relationship among Candida spp. Front Microbiol. 6: 205.
- Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. 2010. Interaction of Candida albicans cell wall Als3 Protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 78(11):4644–4652.
- Sobel JD. 2015. Editorial commentary: vaginal biofilm: much ado about nothing, or a new therapeutic challenge? Clin Infect Dis. 61(4):607–608.
- Staab JF. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 283(5407):1535–1538.
- Swidsinski A, Guschin A, Tang Q, Dorffel Y, Verstraelen H, Tertychnyy A, Khayrullina G, Luo Z, Sobel JD, Jiang X. 2019. Vulvovaginal candidiasis: histologic lesions are primarily polymicrobial and invasive and do not contain biofilms. Am J Obstet Gynecol. 220(1):91.e1–91.e8.
- Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. 2012. A candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 8(8):e1002848.
- Trunk K, Peltier J, Liu Y-C, Dill BD, Walker L, Gow NAR, Stark MJR, Quinn J, Strahl H, Trost M, et al. 2018. The type VI secretion system deploys antifungal effectors against microbial competitors. Nat Microbiol. 3(8):920–931.
- Truong T, Zeng G, Qingsong L, Kwang LT, Tong C, Chan FY, Wang Y, Seneviratne CJ. 2016. Comparative ploidy proteomics of Candida albicans biofilms unraveled the role of the AHP1 gene in the biofilm persistence against amphotericin B. Mol Cell Proteomics. 15(11):3488–3500.
- Tsai P-W, Chen Y-T, Yang C-Y, Chen H-F, Tan T-S, Lin T-W, Hsieh W-P, Lan C-Y. 2014. The role of Mss11 in Candida albicans biofilm formation. Mol Genet Genomics. 289(5):807–819.
- Tsui C, Kong EF, Jabra-Rizk MA. 2016. Pathogenesis of Candida albicans biofilm. Pathog Dis. 74(4):ftw018.
- Tumbarello M, Fiori B, Trecarichi EM, Posteraro P, Losito AR, De Luca A, Sanguinetti M, Fadda G, Cauda R, Posteraro B, et al. 2012. Risk factors and outcomes of candidemia caused by biofilm-forming isolates in a tertiary care hospital. PLoS One. 7(3):e33705.
- Tumbarello M, Posteraro B, Trecarichi EM, Fiori B, Rossi M, Porta R, de Gaetano Donati K, La Sorda M, Spanu T, Fadda G, et al. 2007. Biofilm production by Candida species and inadequate antifungal therapy as predictors of mortality for patients with candidemia. J Clin Microbiol. 45(6):1843–1850.
- Uppuluri P, Acosta Zaldívar M, Anderson MZ, Dunn MJ, Berman J, Lopez Ribot JL, Köhler JR. 2018. Candida albicans dispersed cells are developmentally distinct from biofilm and planktonic cells. MBio. 9(4):e01338–18.
- Uppuluri P, Busscher HJ, Chakladar J, van der Mei HC, Chaffin L. 2017. Transcriptional profiling of C. albicans in a two species biofilm with Rothia dentocariosa. Front Cell Infect Microbiol. 7: 311.
- Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Köhler JR, Kadosh D, Lopez-Ribot JL. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 6(3):e1000828.
- Uppuluri P, Pierce CG, Thomas DP, Bubeck SS, Saville SP, Lopez-Ribot JL. 2010. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot Cell. 9(10).
- Valentine M, Benadé E, Mouton M, Khan W, Botha A. 2019. Binary interactions between the yeast Candida albicans and two gut-associated Bacteroides species. Microb Pathog. 135:103619.
- Vediyappan G, Rossignol T, d’Enfert C. 2010. Interaction of Candida albicans biofilms with antifungals: transcriptional response and binding of antifungals to beta-glucans. Antimicrob Agents Chemother. 54(5):2096–2111.
- Vílchez R, Lemme A, Ballhausen B, Thiel V, Schulz S, Jansen R, Sztajer H, Wagner-Döbler I. 2010. Streptococcus mutans inhibits Candida albicans hyphal formation by the fatty acid signaling molecule trans-2-decenoic acid (SDSF). Chem Eur J of Chem Bio. 11(11):1552–1562.
- Villa S, Hamideh M, Weinstock A, Qasim MN, Hazbun TR, Sellam A, Hernday AD, Thangamani S. 2020. Transcriptional control of hyphal morphogenesis in Candida albicans. FEMS Yeast Res. 20(1):foaa005.
- Weil T, Santamaría R, Lee W, Rung J, Tocci N, Abbey D, Bezerra AR, Carreto L, Moura GR, Bayés M, et al. 2017. Adaptive mistranslation accelerates the evolution of fluconazole resistance and induces major genomic and gene expression alterations in Candida albicans. mSphere. 2(4):e00167–17.
- Williams DW, Jordan RPC, Wei X-Q, Alves CT, Wise MP, Wilson MJ, Lewis MAO. 2013. Interactions of Candida albicans with host epithelial surfaces. J Oral Microbiol. 5(1):22434.
- Williams P, Cámara M. 2009. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 12(2):182–191.
- Wongsuk T, Pumeesat P, Luplertlop N. 2016. Fungal quorum sensing molecules: role in fungal morphogenesis and pathogenicity. J Basic Microbiol. 56(5):440–447.
- Wuyts J, Van Dijck P, Holtappels M. 2018. Fungal persister cells: the basis for recalcitrant infections? PLoS Pathog. 14(10):e1007301.
- Xie Z, Thompson A, Sobue T, Kashleva H, Xu H, Vasilakos J, Dongari-Bagtzoglou A. 2012. Candida albicans biofilms do not trigger reactive oxygen species and evade neutrophil killing. J Infect Dis. 206(12):1936–1945.
- Xu B, Qu Y, Deighton M. 2019. Should we absolutely reject the hypothesis that epithelium-based Candida biofilms contribute to the pathogenesis of human vulvovaginal candidiasis? Am J Obstet Gynecol. 221(4):372–373.
- Yan Y, Tan F, Miao H, Wang H, Cao YY. 2019. Effect of shikonin against Candida albicans biofilms. Front Microbiol. 10:1085.
- Yang F, Teoh F, Tan ASM, Cao Y, Pavelka N, Berman J. 2019. Aneuploidy enables cross-adaptation to unrelated drugs. Mol Biol Evol. 36(8):1768–1782.
- Yano J, Yu A, Fidel PL, Noverr MC. 2016. Transcription factors Efg1 and Bcr1 regulate biofilm formation and virulence during Candida albicans-associated denture stomatitis. PLoS One. 11(7):e0159692.
- Yeater KM, Chandra J, Cheng G, Mukherjee PK, Zhao X, Rodriguez-Zas SL, Kwast KE, Ghannoum MA, Hoyer LL. 2007. Temporal analysis of Candida albicans gene expression during biofilm development. Microbiology. 153(8):2373–2385.
- Younes S, Bahnan W, Dimassi HI, Khalaf RA. 2011. The Candida albicans Hwp2 is necessary for proper adhesion, biofilm formation and oxidative stress tolerance. Microbiol Res. 166(5):430–436.
- Yousif A, Jamal MA, Raad I. 2015. Biofilm-based central line-associated bloodstream infections. Adv Exp Med Biol. 830:157–79.
- Zarnowski R, Sanchez H, Covelli AS, Dominguez E, Jaromin A, Bernhardt J, Mitchell KF, Heiss C, Azadi P, Mitchell A, et al. 2018. Candida albicans biofilm–induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 16(10):e2006872.
- Zarnowski R, Westler WM, Lacmbouh GA, Marita JM, Bothe JR, Bernhardt J, Lounes-Hadj Sahraoui A, Fontaine J, Sanchez H, Hatfield RD, et al. 2014. Novel entries in a fungal biofilm matrix encyclopedia. MBio. 5(4):1–13.
- Zhao X, Daniels KJ, Oh SH, Greeen CB, Yeater KM, Soll DR, Hoyer LL. 2006. Candida albicans Als3p is required for wild-type biofilm formation on silicone elastomer surfaces. Microbiology. 152(8):2287–2299.
- Zhao X, Oh SH, Yeater KM, Hoyer LL. 2005. Analysis of the Candida albicans Als2p and Als4p adhesins suggests the potential for compensatory function within Als family. Microbiology. 151(5):1619–1630.

