Abstract
Microbial natural products from microbes in extreme environments, including haloarchaea, and halophilic bacteria, possess a huge capacity to produce novel antibiotics. Additionally, enhanced isolation techniques and improved tools for genomic mining have expanded the efficiencies in the antibiotic discovery process. This review article provides a detailed overview of known antimicrobial compounds produced by halophiles from all three domains of life. We summarize that while halophilic bacteria, in particular actinomycetes, contribute the vast majority of these compounds the importance of understudied halophiles from other domains of life requires additional consideration. Finally, we conclude by discussing upcoming technologies- enhanced isolation and metagenomic screening, as tools that will be required to overcome the barriers to antimicrobial drug discovery. This review highlights the potential of these microbes from extreme environments, and their importance to the wider scientific community, with the hope of provoking discussion and collaborations within halophile biodiscovery. Importantly, we emphasize the importance of bioprospecting from communities of lesser-studied halophilic and halotolerant microorganisms as sources of novel therapeutically relevant chemical diversity to combat the high rediscovery rates. The complexity of halophiles will necessitate a multitude of scientific disciplines to unravel their potential and therefore this review reflects these research communities.
Introduction
Life exists in almost all environmental niches on the earth. Our planet plays host to various unique environments, and the diversity of these ecosystems is reflected by the diversity of microbial communities that exists within them (Rothschild and Mancinelli Citation2001; Maheshwari and Saraf Citation2015). Extremophiles are organisms that survive and even thrive in bizarre and extreme ecosystems – both physical (temperature, radiation, or pressure) and geochemical extremes (desiccation, salinity, pH, oxygen species, or redox potential) (Rothschild and Mancinelli Citation2001). Extremophiles are some of the oldest life forms on earth. Their existence is thought to be an evolutionary adaptive response to the extreme environments that were present in the earth’s primordial atmosphere (Rothschild and Mancinelli Citation2001). The selective nature of these environments resulted in the evolutionary development of the unique metabolism thought to be fundamental to their survival (Maheshwari and Saraf Citation2015). Today, extremophiles live in habitats that will support no other form of life.
Halophiles are extremophiles that can exist in a range of salt concentrations, from freshwater habitats to the hypersaline lakes of the Dead Sea, and even the salt flats of Salar de Atacama (Rothschild and Mancinelli Citation2001). The world of halophilic microorganisms is highly diverse and contains taxonomic representatives from all three domains of life – Archaea, Bacteria, and Eucarya (Oren Citation2002).
Originally thought to be bacterial, their distinctive 16S ribosomal RNA sequences and lipid compositions revealed that archaea were a separate and distinct third domain of life (Woese and Fox Citation1977). Archaea are comprised of three phylogenetically distinct groups: The Crenarchaeota (mainly consisting of hyperthermophilic sulphur-dependent organisms), the Euryarchaeota (the methanogens), and the extreme halophiles. The classification of archaea is a rapidly evolving one, most recently, a superphylum Asgard was proposed whose members include Lokiarchaeota and Thorarchaeota (Zaremba-Niedzwiedzka et al. Citation2017), with the eventual cultivation of an Asgard archaeon Candidatus Prometheoarchaeum syntrophicum after 12 years of growth (Imachi et al. Citation2020). Numerous genera of archaea live and thrive in extreme niches, and therefore a significant portion of halophilic microorganisms belongs to the Archaea domain (Kessel and Klink Citation1980).
Among the bacteria, halophiles exist within the phyla Cyanobacteria, α-Proteobacteria, Firmicutes, Actinobacteria, Spirochaetes, and Bacteroidetes (Sorokin et al. Citation2006; Oren Citation2008). Halophilic bacteria offer a high potential for biotechnological applications due to their adaptations to extreme natural environments particularly concerning their contribution to biogeochemical processes, the formation, and dissolution of carbonates, the immobilization of phosphate, and the production of growth factors and nutrients (Rodriguez-Valera Citation1993).
Eukaryotic organisms contribute to a smaller percentage of life in hypersaline environments; representatives include the unicellular green algae Dunaliella (Wegmann et al. Citation1980), the brine shrimps Artemia salina and Artemia franciscana (Gajardo and Beardmore Citation2012), and the fungus Trimmatostroma salinum and Hortaea werneckii (Gunde-Cimermana et al. Citation2000).
Through evolution, these salt-loving organisms have adapted to the high osmotic pressures that exist within their environments and their unique biochemical and genetic characteristics have resulted in the production of a multitude of biotechnologically relevant bioactive compounds including antimicrobials (Maheshwari and Saraf Citation2015).
Once considered the “wonder drugs” of the twentieth century (Bud Citation2007), the disproportionate and indiscriminate usage of antibiotics has accelerated the emergence of clinically significant multiple drug-resistant (MDR) pathogenic strains (Ciofu et al. Citation1994). The dynamic nature of pathogenic microorganisms to evolve sophisticated mechanisms that inactivate antibiotics (Butler et al. Citation2017), and the continued global recognition that the emergence of MDR pathogens pose a serious threat to human health, have substantially emphasized the urgent need for new antibiotics that would target this emerging crisis (Butler et al. Citation2017). Pathogenic strains that are resistant to most, or all, available antibiotics are now routinely isolated (Brown and Wright Citation2016).
Microbial natural products
The beginning of the modern “antibiotic era” is often associated with Paul Ehrlich and Alexander Fleming, replacing the antibacterial sulphonamide, and the toxic arsphenamine/606. Ehrlich with his “magic bullet” argued that chemical compounds could be synthesized and would “exert their full action exclusively on the parasite harbored within the organism” (Aminov Citation2010). While Sir Alexander Fleming’s serendipitous discovery of penicillin in 1928 (Fleming Citation1929), would revolutionize medical practice, dramatically decreasing fatality rates from bacterial infections (Kovac and Sneader Citation2006) and in the midst of World War Two they saved millions of lives because “Thanks to Penicillin…He Will Come Home!” (Levy Citation2001).
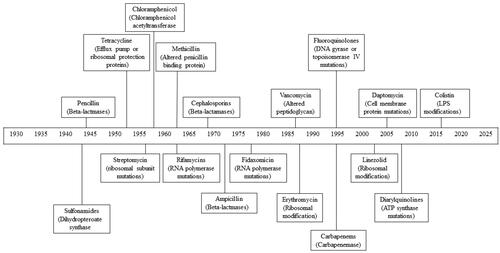
Fleming’s discovery kick-started a concerted search for other antimicrobial agents. Over the next 30 years, the “Golden age” (1940–1962) of antibiotic discovery, resulted in the development of half of the clinically used antibiotics today (). Throughout the remainder of the twentieth century, two parallel lines of drug discovery were employed; the isolation of antibiotics from natural sources and the chemical synthesis of antibiotics (Davies Citation2006).
Figure 1. Timeline depicting the year in which antibiotic resistance in each major class of antibiotics was first observed. The main mechanism of resistance for each antibiotic class is also included.
Microbial natural products have made an incredible contribution to the antibiotic drug discovery and development process over the last seven decades, with 70 out of 90 antibiotics marketed in the years 1982–2002 originating from natural products (Newman et al. Citation2003). Between 1981 and 2010, 34% of all medicines approved by the FDA were natural products or their derivatives (Newman and Cragg Citation2016). Synthetic antibiotics followed James Black’s famous observation that “the most fruitful basis for the discovery of a new drug is to start with an old drug” (Raju Citation2000).
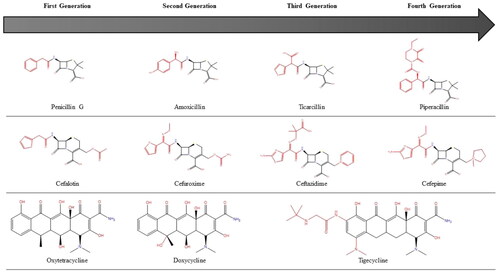
Almost all of the antibiotics approved between the early 1960s and 2000 were iterative, semisynthetic tailored derivatives of previous natural scaffolds () (Von Nussbaum et al. Citation2006; Fischbach and Walsh Citation2009). The beginning of a discovery drought, with a void in antibiotic discovery since lipopeptides in 1987 (Friedman and Alper Citation2014), marked the end of the “Golden Age” of antibiotic drug discovery, the law of diminishing returns had forced the hands of the drug discovery sector to increasingly turn their efforts towards high-throughput screens of synthetic compounds. However, the chemical diversity offered by synthetic and combinatorial chemical libraries was not sufficient to provide the required number of quality hits for hit-to-lead programs in drug discovery and efforts were largely unsuccessful (Demain Citation2014).
Figure 2. Successive generations of major antibiotic classes over time. Black bonds represent the original skeletal structure while red bonds are structural changes.
The milestone of the failed synthetic approach was noted in 2007 when Payne et al. detailed the unsuccessful hits of a huge screening program by GSK (Payne et al. Citation2007). This effect was amplified at the turn of the century with further disengagement and withdrawal by pharmaceutical companies in antibiotic research and development, which had previously played a significant role in bringing many compounds to market (Koehn and Carter Citation2005).
Numerous factors prompted this withdrawal, including high rediscovery rates of known natural products and the challenges associated with the purification and structure elucidation of natural products (Clardy et al. Citation2006). Economically, antibiotics have a poor return on investment due to their short-term curative nature compared with chronic drug therapies (Projan and Shlaes Citation2004). Additionally, regulatory hurdles within clinical trials (Projan Citation2003; Brown Citation2013; Lewis Citation2013), increased market competition from more affordable generic drugs, and marketplace and industry consolidation (Projan and Shlaes Citation2004), has led to the virtual collapse of antimicrobial research and development by the pharmaceutical industry (Simpkin et al. Citation2017). Even within research companies, compound libraries employing inappropriate filters such as Lipinski’s rules (to attain desirable physicochemical properties for improved oral bioavailability of drugs (Lipinski et al. Citation1997)), ignore the principles required for targeting prokaryotes (Lewis Citation2013). For the companies that remain in the area, cautionary tales with antibiotics such as solithromycin (Owens Citation2017) demonstrate the risk of antibiotic research, and recent insolvency in smaller start-up companies such as Achaogen (Dall Citation2019) pose a warning that stifles antimicrobial discovery.
The emergence of AMR and the lack of novel antibiotics in the pharmaceutical pipeline has prompted a resurgence to screen natural products, which have provided and continue to provide a significant number of clinically approved drugs (Newman and Cragg Citation2012; Thaker et al. Citation2013). There is a necessity to improve the efficiency of the discovery process, and bioprospecting of antibiotics from underexplored habitats represents a largely untapped repository of compounds with unique chemical structures (Clardy et al. Citation2006; Mohammadipanah and Wink Citation2016).
Within extreme environments, natural forces already eradicate a large percentage of the competition by creating a habitat in which it is difficult to thrive. Therefore there would be a reduced necessity for the remaining microbes to utilize secondary metabolites for competition (Pettit Citation2011). However, recent studies have demonstrated that extremophiles are capable of producing a diverse range of secondary metabolites (Wilson and Brimble Citation2009). The hypersaline environment is one such niche, and the following sections explore the antimicrobial potential of each halophilic domain – the Archaea, the Bacteria, and the Eukarya.
Halophiles as a source of antibiotic chemical diversity
Halophilic and halotolerant bacteria
When compared to their terrestrial and marine counterparts, bacteria from hypersaline environments are largely under-explored as a source of antimicrobial agents. Previous studies have described the antimicrobial activity of halophilic bacteria from marine invertebrates, solar salterns, soil, and salt lakes. A predominant genus of antimicrobial-producing strains are Bacillus (Pinchuk et al. Citation2002), but other species including, Halomonas salifodinae (Velmurugan et al. Citation2013), halophilic cyanobacteria (Pramanik et al. Citation2011) have also been described.
Kamat et al. (Citation2011) demonstrated the antimicrobial potential of a range of marine Bacillus spp. from the salt pans of Goa, India. In a study by Velho-Pereira, (Velho-Pereira and Furtado Citation2012) 100 halophilic bacteria from marine invertebrates were screened for their antibacterial activity. Forty-six of the isolates with the most potent activity belonged to the genera Chromohalobacter, Bacillus, and Corynebacterium. A further study confirmed their abundance, which revealed that amongst the 200 halophilic bacteria isolated, the dominant genus was Bacillus compromising 24% of the total isolates (Irshad et al. Citation2014). Sawale et al. (Citation2014) reported similar findings with two halophilic bacterial strains, Bacillus pumilus NKCM 8905 and Bacillus pumilus AB211228 displaying antimicrobial production against Staphylococcus aureus, Bacillus subtilis, Aspergillus niger, and Aspergillus flavus.
Li et al. cultured 152 halophilic bacterial isolates from Yuncheng Salt Lake, China; 34 of the isolates were related to the phylum Firmicutes with representatives in three families Bacillaceae, Clostridiaceae, and Staphylococcaceae, while other strains belonged to the phylum γ-Proteobacteria and were identified as members of Halomonadaceae and Idiomarinaceae. Six of the isolates were capable of inhibiting both bacterial and fungal pathogens, the majority of which belonged to the Bacillaceae family (Li and Yu Citation2015).
However, Chen et al. (Citation2010) reported not only the bioactivity of halophilic Bacillus, but also the potential of other moderately halophilic bacterial strains, with 23 of the 45 strains inhibiting the growth of Bacillus subtilis, one strain inhibiting Escherichia coli, and two strains inhibiting Pseudomonas fluorescens. Additionally, almost all the halophilic strains had inhibitory effects on one or several indicator fungal strains. Phylogenetic analysis revealed that only one of the strains was related to the genus of Bacillus, and 12 strains were related to Halobacillus.
Halophilic microorganisms have also been shown to possess bioactivity beyond antibacterial and antifungal activities as reported by Velmurugan et al. (Citation2013). The organic extracts of Halomonas salifodinae MPM-TC displayed not only antibacterial activity against several aquatic bacterial pathogens (Vibrio harveyi, Vibrio parahaemolyticus, Pseudomonas aeruginosa, and Aeromonas hydrophila) but also possessed in vitro antiviral activity against white spot syndrome virus and immunostimulatory activity in prawns.
The saline ecosystem of Sundarbans’ mangrove forests serves as an example of a unique environment with the potential for antimicrobial discovery. Pramanik et al. (Citation2011) reported the first study of antimicrobial production from obligate halophilic cyanobacteria against several human pathogens. Marine cyanobacteria are known for their ability to produce a diverse array of biologically active secondary metabolites, which have attracted significant research interest due to their potent biological activities. Some of these compounds such as cryptophycin 52 (LY355703) advanced to Phase II clinical trial, but failed due to its high neurotoxicity (Weiss et al. Citation2017), and curacin A was evaluated for its potential therapeutic as an antimitotic agent (Simmons et al. Citation2005). In addition to the antimicrobial potential of halophilic cyanobacteria, a further study isolated 54 halophilic actinomycetes with nine of the isolates showing antimicrobial activity against both bacterial and fungal test organisms (Sengupta et al. Citation2015).
Halophilic and halotolerant Actinobacteria
This bacterial phylum, the Actinobacteria, has proven to be a vibrant source of microbial natural products. Since the discovery of streptomycin from Streptomyces griseus by Selman Waksman’s group in 1942 (Schatz et al. Citation1944), the greatest number of clinically relevant antibiotics discovered have been from the phylum Actinobacteria (Baltz Citation2007). Actinomycetes account for 45% of all known microbial secondary metabolites (Bérdy Citation2005) and over two-thirds of the clinically useful antibiotics (Lam Citation2006; Papagianni Citation2012) including gentamicin, vancomycin, clindamycin, erythromycin, amphotericin, rifampicin, and tetracycline (Baltz Citation2007). Actinobacteria are recognized as being the richest microbial source of natural products, their unique chemical scaffolds (Bérdy Citation2015) has meant that they have traditionally been viewed as the most prolific group of antibiotic producers amongst the prokaryotes (Bérdy Citation2005). As such, this phylum is routinely screened for new bioactive structures (Baltz Citation2007). Actinobacteria have been the focus of many antimicrobial discovery programs, and in recent decades, isolation and exploitation of Actinobacteria for novel compounds from conventional environments have led to the perennial rediscovery of known compounds (Tulp and Bohlin Citation2005). It has been argued that Actinobacteria have been so extensively studied that the chances of finding novel chemistries are too low to make them viable research leads and despite years of success, this over-mining of soil bacteria has resulted in diminishing returns (Lewis Citation2012). Watve et al. (Citation2001) have disputed this maintaining that only a minor percentile of all the species of Streptomyces existing in nature have been grown in culture and their mathematical models suggest that the number of antibiotics still to be discovered from Streptomyces spp. could well be above 100,000. However, this high rate of compound rediscovery in culture-based screening programs has meant that it is now crucial that new groups of Actinobacteria be pursued as sources of novel therapeutically relevant compounds (Bull et al. Citation2000). Actinobacteria have a ubiquitous distribution in the biosphere, including the extremobiosphere. In the same regard, there is evidence that suggests Actinobacteria isolated from the extremobiosphere will be a rich source of novel natural products (Bull Citation2011). These unique environments will undoubtedly harbor novel strains of actinomycetes, which are likely to yield novel metabolites. It has been nearly 50 years since the occurrence of actinomycetes in saline environments and their tolerance to high salt concentrations was first described (Tresner et al. Citation1968; Gottlieb Citation1973). Since this time, Hamedi et al. estimated 70 species of halotolerant and halophilic actinomycetes from at least 24 genera have been successfully described (Hamedi et al. Citation2013). summarises currently known antimicrobial compounds from halophilic and halotolerant actinomycete.
Table 1. Antimicrobial compounds from halophilic and halotolerant actinomycetes.
Despite this, halophilic (and more generally extremophilic) actinomycetes remain largely underexplored for the discovery of novel bioactive secondary metabolites (Hamedi et al. Citation2013). The unique metabolic diversity and biotechnological potential found in halophilic and halotolerant microorganisms demonstrate that this phylum of Actinobacteria are an invaluable source to be exploited (Cragg and Newman Citation2013; Hamedi et al. Citation2013). Jose et al. have reviewed the biodiscovery potential of novel hypersaline actinomycetes and conclude that halophilic and halotolerant actinomycetes have an enormous capacity to produce a range of bioactive secondary metabolites with diverse activities (Jose and Jebakumar Citation2014). The discovery of taxonomically unique strains of extremophilic Actinobacteria has led to the anticipation that mining these groups could add an alternative dimension to the line of natural product drug discovery (Thumar et al. Citation2010) that promises to raise the prospect of discovering novel bioactive compounds (Jose et al. Citation2011; Hamedi et al. Citation2013). In this regard, isolation programs have been overhauled towards largely unexplored, unusual, and extreme environments like hypersaline environments (Hamedi et al. Citation2013; Jose and Jebakumar Citation2013). The determination towards understanding these species has led to the creation of online databases such as actinobase, which compiles information on the diversity, molecular phylogeny and biotechnological potential of actinomycetes (Sharma et al. Citation2012).
The presence of such activity is widely described in the literature, and numerous authors have reported the occurrence of antimicrobial-producing halophilic actinomycetes (Kokare et al. Citation2004; Li et al. Citation2005; Manam et al. Citation2005; Vasavada et al. Citation2006; Sarkar et al. Citation2008; Suthindhiran and Kannabiran Citation2009; Thumar et al. Citation2010). Similarly, their halotolerant counterparts have also been reported for their bioactivity (Maskey, Helmke, et al. Citation2003; Maskey, Li, et al. Citation2003; Maskey et al. Citation2004; Mahyudin et al. Citation2012).
Bioactives from halophilic actinomycetes have been previously described since the early 1990s, with the isolation of the first moderately halophilic actinomycete, Actinopolyspora mortivallis JCM7550, capable of producing nucleoside antibiotics isolated from saline soils, sampled from Death Valley, California, USA (Yoshida et al. Citation1991). To date, several studies have observed actinomycetes-like-halophiles in many diverse hypersaline environments from salt lakes (Swan et al. Citation2010; Guan et al. Citation2011), solar salterns (Baati et al. Citation2008), salt mines (Chen et al. Citation2007) to brine wells (Xiang et al. Citation2008), and particularly from salt pans (Jensen et al. Citation1991; Dhanasekaran et al. Citation2004; Roshan et al. Citation2013). Following the isolation of Actinopolyspora, another halophilic Actinopolyspora was isolated, on this occasion, from marine saltpans. This isolate demonstrated antibacterial and antifungal activity that was dependent on the growth medium (Kokare et al. Citation2004), the significance of this finding highlights the importance of utilising an array of media designed to promote the maximum production of bioactives.
Irshad et al. (Citation2013) demonstrated that the foreshore soil of Daecheon Beach and Saemangeum Sea (Korea), was a rich source of halophilic Actinobacteria. Out of the 63 strains examined, 12 isolates exhibited pathogenic bacteria, pathogenic fungi, and yeast. Two isolates, Microbacterium oxydans, and Streptomyces fradiae displayed antibacterial activity against all pathogens tested.
Ballav et al. assessed the capacity of halotolerant and halophilic Actinomycetales from the crystallizer pond sediments of the Ribandar saltern, Goa, for their capacity to produce antibacterial secondary metabolites (Ballav et al. Citation2015). These isolates demonstrated antimicrobial activity against S. aureus, Staphylococcus citreus, and Vibrio cholerae.
The importance of a biologically competitive environment was emphasized by Imada et al. the study described an actinomycete with high similarity to Micromonospora globosa, whose antibacterial production was dependent on seawater (Imada et al. Citation2007). A previous study reported that a similar strain, M. globosa JCM 3126, did not produce any antibiotics in the presence of seawater (Shiomi et al. Citation1990), speculating that environmental factors that influence antibiotic production could exert strain-specific, rather than universal, effects. Many Streptomyces species are reported to secrete antibiotics against bacteria, fungi, and yeasts at higher salinity and alkaline pH (Basilio et al. Citation2003). For example, the alkaliphilic Streptomyces sp. AK409 secretes the antimicrobial compound pyrocoll, but only under alkaline conditions (Dietera et al. Citation2003). Another species, Streptomyces sannanensis RJT-1, a halophile isolated from alkaline soil, was also reported to be a potent antibiotic producer of compounds active against Gram-positive bacteria (Singh et al. Citation2006). Gohel et al. highlighted again the role of biologically competitive environments, with unique conditions such as pH, temperature, pressure, oxygen, light, nutrients, and salinity, in the production of novel antibiotic compounds from actinomycetes isolated from saline and alkaline habitats (Gohel et al. Citation2015). Another example provided by Okami et al. is the isolate Streptomyces griseus, cultivated from marine sediment. This isolate exhibited no streptomycin production, however when grown in a medium containing low concentrations of organic nutrients and a high concentration of NaCl, it produced a new ionophoric polyether antibiotic (Okami et al. Citation1976). While it is unclear if this production was an epigenetic trigger or an adaptive response to the saline environment, it does signify the importance of utilizing a growth medium that produces maximum bioactivity.
Amongst the most known and most extensively characterized antimicrobial compounds produced by Actinobacteria are polyketides. In 2011, a moderately halophilic Streptomyces strain JAJ06 isolated from saltpan soil in Tuticorin (India) produced a seawater-dependent antimicrobial activity against a panel of bacteria and yeast strains with high potency, e.g. MIC of 1–2 μg/mL against MRSA. Subsequent sequential analysis revealed that this compound was a polyketide (Jose et al. Citation2011). Zhao et al. discovered further linear polyketides, actinopolysporins A, B, and C, as well as the known antineoplastic antibiotic tubercidin from the halophilic actinomycete Actinopolyspora erythraea YIM 90600 (Zhao et al. Citation2011). Later Jang et al. described the noteworthy discovery of the polyketide antibiotic, anthracimycin, produced by the marine-derived Actinobacteria, Streptomyces sp. strain CNH365, which had significant activity against Bacillus anthracis (Jang et al. Citation2013). In 2014 Chen et al. isolated the halophilic Actinobacterium, Actinopolyspora erythraea YIM90600, from the Baicheng salt field, the only known erythromycin-producing extremophile. Additionally, the new erythromycin biosynthetic gene cluster had a high identity and similarity to Saccharopolyspora erythraea, a soil actinomycete known to produce erythromycins (McGuire et al. Citation1952; Chen et al. Citation2014).
Other antibiotic chemical classes were isolated in 2014, from the soil-associated halophile Nocardiopsis terrae strain YIM 90022. This strain produced five types of secondary metabolites, two of which, a quinoline alkaloid and N-acetyl-anthranilic acid, displayed antimicrobial activities. The quinoline alkaloid displayed antibacterial activity against S. aureus, B. subtilis, and E. coli and antifungal activity Pyricularia oryzae. N-acetyl-anthranilic acid inhibited the growth of S. aureus and E. coli and antifungal activity against Fusarium avenaceum, Fusarium graminearum, and Fusarium culmorum.
Tian et al. reported the isolation of another halophilic species Nocardiopsis gilva YIM 90087. The Actinobacteria produced three p-terphenyl derivatives with antifungal activity against some pathogenic fungi (including F. avenaceum, F. graminearum, F. culmorum, and C. albicans) and antibacterial activity against B. subtilis (Tian et al. Citation2013).
The high-altitude deserts of Chile, such as the Atacama, are amongst the oldest and driest deserts on the earth, long considered to be too extreme to support any form of life given their harsh conditions, high levels of UV radiation, and lack of liquid water (Gómez-Silva et al. Citation2008). However, despite these seemingly inhabitable conditions, many authors have reported the isolation of actinomycetes from soils and halites collected from these arid deserts (Okoro et al. Citation2009; Bull and Asenjo Citation2013). The Atacama has also yielded interesting organisms with unique biosynthetic potential such as Streptomyces leeuwenhoekii, which produces the novel antimicrobials the chaxalactins and chaxamycins (Busarakam et al. Citation2014).
While halophilic actinomycetes are a demonstrable resource of antimicrobials, strains such as Saccharopolyspora salina VITSDK4 from salt pan soil of the Marakkanam coast of the Bay of Bengal, displayed not only antagonistic properties to various bacterial and fungal pathogens but additionally, extracellular bioactives were found to inhibit proliferation of HeLa cells indicating potential cytotoxic activity (Suthindhiran and Kannabiran Citation2009).
The scaffolds of about 80% of antibiotics approved for clinical use over the past decade are derived from microbial natural products (Thaker et al. Citation2013). However, traditional screens of natural product extracts for compounds with antibiotic activity are faced with low hit rates and high rates of rediscovery, a characteristic of conventional discovery approaches (Thaker et al. Citation2013). Modern approaches should be utilised to exploit halophilic actinomycetes, given their enormous capacity to produce a range of bioactive secondary metabolites with diverse activities.
Halophilic archaea
Halotolerant and halophilic bacteria actively pump ions out of cells and produce organic osmotic solutes to maintain a low intracellular ionic concentration and prevent the “salting-out” of enzymes. Archaea, however, adopt a different strategy to achieve osmotic balance and their cytoplasm contains molar concentrations of salts, mainly KCl, which provides the necessary osmotic equilibrium (Oren Citation2006). Difficulties in the isolation and cultivation of archaea have meant that they have been vastly under-investigated as a source of novel bioactive compounds.
Traditionally, natural product chemistry has focussed primarily on bacteria, fungi, plants, and the marine environment as sources of bioactive compounds. By comparison, research into the extreme biosphere of archaea has largely been overlooked, since their first description and proposal as a distinct branch, a third domain, of the tree of life by Woese in the late 1970s (Woese and Fox Citation1977). With the unbounded rise of AMR, archaea could be an important source of antimicrobial compounds. Given that archaea survive and even thrive in unique extreme environments, the compounds they produce could also be both unique in terms of structure and pharmacology, and distinct from other clinically used antibiotics.
While traditional antibiotics production have been demonstrated from the bacterial and eukaryotic domains of life, the archaeal third domain is yet to yield such compounds. Given the similarity of archaea both to bacterial and eukarya and the co-colonization by archaea of the dynamic and competitive environmental niches inhabited by the other domains of life where antibiotic production has proven advantageous, we propose that the archaea are a potentially untapped source of novel, clinically useful antibiotic compounds.
The most widely studied antimicrobial compounds isolated from archaea are the archaeocins. Thought to endow archaea with a competitive advantage for nutrients and other resources, archaeocins are ribosomally synthesized bacteriocin-like antimicrobial peptides (AMPs) or proteins with antagonistic activity against other microorganisms (Rodriguez-Valera et al. Citation1982). Archaeocins fall into two phylogenetic groups: halocins produced by haloarchaea (Shand and Leyva Citation2007) and sulfolobicins produced by the genus Sulfolobus (Prangishvili et al. Citation2000). Halocins have been shown to possess activity against haloarchaea, thermophilic crenarchaea (Haseltine et al. Citation2001) and some bacterial strains. In comparison with bacteriocins and eucaryocins, there is much to be learnt from archaea-derived AMPs (O’Connor and Shand Citation2002).
The first halocin was described almost four decades ago by Rodriguez-Valera et al. (Citation1982), – secreted by Haloferax mediterranei (Rodriguez-Valera et al. Citation1982). In 1994 Torreblanca et al. proposed that halocins were a near-universal feature in all haloarchaea (Torreblanca et al. Citation1994). However, despite this apparent abundance, only 12 archaeocins (11 halocins and one sulfolobicin) have been partially or fully characterized to date, with hundreds of archaeocins thought to exist within the haloarchaea (Shand and Leyva Citation2008). Only halocins H4, H6, and RI have been characterized to a protein level (Meseguer and Rodriguez-Valera Citation1985; Rdest and Sturm Citation1987; Torreblanca et al. Citation1989), while halocins S8, H4, and C8 are the most characterized halocins, at both a genetic and mRNA transcription level. shows the known primer sequences for these halocins (Sun et al. Citation2005).
Table 2. Forward and Reverse primer sequences for Halocins S8, H4 and C8.
Most halocin genes follow the pattern of induction during the transition into the stationary phase. Halocins, such as H4, H6, and S8, have low detectable levels during exponential growth, but as the media becomes depleted during the stationary phase, these levels increase drastically (Cheung et al. Citation1997). An exception to this is halocin H1, which possesses activity first detectable earlier during the mid-exponential phase (Platas et al. Citation1996; O’Connor and Shand Citation2002). Furthermore, while the activity of most of these halocins decreases again during the stationary phase, others such as HalR1 maintain a high level of activity for a prolonged time (Shand et al. Citation1998).
Archaeal proteins undergo secretion in either an unfolded (via the Sec pathway) or a folded conformation (via the Twin arginine transport (Tat) pathway). Halocins contain a predicted Tat signal peptide synthesized at the N-terminal that targets them for secretion from the cell, and this would suggest that they are generally secreted by the Tat pathway (Price and Shand Citation2000; O’Connor and Shand Citation2002). Halocin genes are plasmid-encoded, with typical archaeal TATA boxes found within promoter regions, although for halocin C8 the TATA box is closer to the transcription site compared to the other halocins, 18 bp rather than 22–25 bp (Sun et al. Citation2005). Their transcription is described as leaderless given their transcription site either coincidences with or is only a few bps upstream from the translational start codon ATG (Shand and Leyva Citation2007).
Halocins can be divided into two classes based on size: the smaller microhalocin peptides (<10 kDa but as small as 3.6 kDa) and the larger halocin proteins (>10 kDa up to 35 kDa) Microhalocins are cleaved from a larger precursor protein, the preproprotein (O’Connor and Shand Citation2002).
The hydrophobic microhalocins are much more robust than larger halocin proteins. They can be desalted without losing activity, are insensitive to organic solvents (such as acetone or acetonitrile), and are relatively insensitive to heat (O’Connor and Shand Citation2002). Microhalocins such as R1 can also be stored at 4 °C for long periods, retaining activity after seven years (O’Connor and Shand Citation2002). The larger protein halocins are much more fragile, they are generally more heat-labile, and lose their activity when desalted below 5% w/v NaCl. The addition of one or more serine proteases, typically proteinase K and trypsin, confirms the presence of mature halocins. summarises many of the known features of several well-described halocins.
Table 3. Characterisation of known halocins, with thermal stability, salt dependency, the spectrum of activity, mechanism, and known sequences.
Halocin H4
HalH4 was the first reported halocin, and was isolated from Haloferax mediterranei. It has characterized at both the protein and genomic levels. HalH4 is cleaved from a preprotein (39.6 kDa) to yield the mature HalH4 protein (34.9 kDa), and its activity against other haloarchaea has been described as archeaolytic. Halocin H4 appears to disrupt the permeability of the membrane on sensitive Hbt. salinarum cells, leading to an ionic imbalance and eventual cell lysis. Halocin H4 can be desalted to ∼0.06% w/v NaCl (10 mM) and with only a twofold loss in activity, against Halobacterium sp. (Rodriguez-Valera et al. Citation1982; Meseguer and Rodriguez-Valera Citation1986; Cheung et al. Citation1997).
Halocin S8
The halS8 gene has an open reading frame of 933 base pairs long, which when translated yields a 311 amino acid in length pre-pro-protein. Catalytic cleavage of the pre-pro-protein generates separate peptides, a 230 amino acid N-terminal protein, a 45 amino acid C-terminal peptide, and the 36 amino acids mature microhalocin S8. The mature halocin S8 has a molecular mass of 3.58 kDa with four cysteine residues and can form potentially two disulfide bridges. Whether the release of this active halocin from the pre-pro-protein is due to autocatalysis or the activity of a protease is unknown (Price and Shand Citation2000).
Halocin R1
R1 is a microhalocin produced by Halobacterium salinarum GN101 (O’Connor Citation2002). While initial studies suggested a molecular mass of 6.2 kDa(Rdest and Sturm Citation1987), it was later revealed that R1 was part of a larger preproprotein that had a mass of 29 kDa (Shand et al. Citation1998; O’Connor and Shand Citation2002). The mature HalR1 is made up of 38 amino acids and shares 71% similarity to S8 (Price and Shand Citation2000; O’Connor and Shand Citation2002), this difference is proposed to account for the differences in the spectral activity and thermostability, between the two peptides as previously reported (Shand and Leyva Citation2007). The sequences of these two halocins are:
–R1: lqsNINiNTAAaVILiFNQVqvgALCaPTpVsGGgPpP
–S8: sdcNINsNTAAdVILcFNQVgscALCsPTIV–GG–PvP
Halocin H7
HalH7 (formerly known as HalH6) is produced by Haloferax gibbonsii Ma2.39 (Torreblanca et al. Citation1989). Halocin H7 is a stable halocin that retains its activity when desalted and is heat resistant. Initially determined to have a molecular mass of 32 kDa (Torreblanca et al. Citation1989), as with halocin R1, this halocin is released from a larger preproprotein, yielding a smaller 3 kDa active peptide, thus classifying halocin H7 as a microhalocin. This halocin is lytic, as sensitive cells were exposed to the protein, they would swell and eventually lyse. Further studies revealed that this mechanism was due to the action of the halocin on the Na+/H+ antiporter system on the cell membrane of both haloarchaea and mammalian cells (HEK293, NIH3T3, Jurkat, and HL-1) (Meseguer et al. Citation1995).
Halocin A4
Halocin A4, another microhalocin was produced by an uncharacterized haloarchaeon isolated from a Tunisian saltern. It has a molecular mass of 7.435 kDa, is hydrophobic and acidic. It demonstrates a broad spectrum of activity, compared to other halocins with activity against haloarchaea and the crenarchaeal hyperthermophile Sulfolobus solfataricus (Shand Citation2006). This indicates that there may be a conserved biological target between euryarchaeal halophiles and crenarchaeal hyperthermophiles, where the halocin acts (Haseltine et al. Citation2001).
Halocin C8
Halocin C8 is produced by Halobacterium strain AS7092 (Li et al. Citation2003; Sun et al. Citation2005) and contains ten cysteine residues and is the largest microhalocin at 7.44 kDa. Like S8, it is cleaved from a preproprotein, at the C-terminal end, by an unknown mechanism to produce the active mature peptide HalC8 and an immunity protein HalI. HalI functions to protect the producer organism against its native halocin, by binding, to and inactivating the halocin (Sun et al. Citation2005).
Halocin H1
H1 is a larger halocin peptide produced by Haloferax mediterranei M2a. Halocin H1 has a molecular mass of 31 kDa and is heat-labile. It loses activity if desalted and requires a minimum of 5% w/v NaCl to maintain activity. Additionally, the compounds within the media influence the production of this halocin with optimal antimicrobial activity being obtained when 0.5% of N-Z-amine E was used as a nutrient (Platas et al. Citation1996). This halocin displayed activity against Haloferax gibbonsii ATCC 33959, Halobacterium salinarum NRC 34002 and Haloferax volcanii NCMB 2012.
The spectrum of activity of the halocins varies for each compound, halocins H4 and H6 exhibit a narrow range of activity while others such as halocin H1 possess a wider range of activity towards numerous haloarchaea as mentioned previously (Blum Citation2008; Clark et al. Citation2008). Initially, halocins were thought to display activity confined to closely related Halobacteria sp. such as C8 (Li et al. Citation2003). However, the microhalocins A4 and S8 demonstrate a broader activity with cross-phylum inhibition of Sulfolobus solfataricus (Haseltine et al. Citation2001) and halocin R1 inhibiting the growth of Methanosarcina thermophile (Shand et al. Citation1999).
A relatively restricted number of studies have demonstrated the potential of archaea to interact with and inhibit the growth of bacteria. Atanasova et al. demonstrated this interaction, they screened 68 haloarchaea of different genera (Halorubrum, Haloferax, Haloplanus, Haloarcula, Halogranum, Halobacterium, Halosarcina, and Halogeometricum), and found they produced halocins that had antagonistic interactions on 22 halophilic bacteria (Halomonas, Rhodovibrio, Salisaeta, and Pontibacillus). Additionally, this study also found that halophilic bacteria from various genera could be identified as halocin producers (Atanasova et al. Citation2013). More recently Ghanmi et al. (Ghanmi et al. Citation2016), screened 35 halophilic bacterial and archaeal strains for antagonism, and three cases of cross-domain inhibition (archaea/bacteria or bacteria/archaea) were observed. Four archaeal strains inhibited the growth of several other strains (both bacterial and archaeal). Three archaeal strains were further investigated, and these strains were shown to encode and express either halocins S8 or H4, demonstrating that these halocins have both anti archaeal and antibacterial properties. Another study by Shand and Leyva (Citation2008), screened 75 strains isolated from the Wilson Hot Springs (48 archaeal and 27 bacterial strains) for growth inhibition and found that 77% inhibited the growth of at least one other isolate. Inter-domain interactions were also present with 43 haloarchaea inhibiting halophilic members of the domain Bacteria, while seven bacteria were capable of antagonising haloarchaea. The 75 isolates possessed no bioactivity against Gram-negative bacteria, but inhibited, the growth of several Gram-positive bacteria was inhibited (Shand and Leyva Citation2008).
Microhalocins like KPS1 produced by Haloferax volcanii KPS1 exhibited inhibitory activity towards Streptococcus mutans, S. aureus, E. coli, and P. aeruginosa (Kavitha et al. Citation2011). Another microhalocin, C8 or C8-like, produced by Natrinema gari QI1, was shown to have antimicrobial activity towards both archaea and the bacterium, Micrococcus luteus, but not against any other pathogens tested (S. aureus, B. subtilis, B. cereus, and Sarcina lutea) (Quadri et al. Citation2016). The body of evidence demonstrating the potential of halocins as anti-bacterial agents is growing. While halocins display anti-archaeal activity, further investigation to assess their potential against Gram-positive and Gram-negative bacteria requires investigation. Microhalocins remain active after desalting and based on previous studies are likely to be a stronger candidate for antibacterial activity. Additionally in order to fully correctly assess the potential of haloarchaea supernatants extraction of microhalocins from culture supernatant requires the use of non-polar solvent extractions instead of traditional liquid-liquid extraction solvents such as ethyl acetate,
To date, few studies have addressed the mechanism of action of halocins. While it has been shown that some halocins such as A4 are cytocidal, and others like S8 and R1 are cytostatic, only the mechanism of one halocin (H7/6) has been elucidated and involved inhibition of the Na+/H+ antiporter (Lequerica et al. Citation2006). Microhalocin peptides have either little or no net charge and therefore are unable to interact with the negatively charged bacterial membrane in the same way many bacteriocins and eucaryocins do, suggesting that their mechanism is distinct from these other classes.
The most recently identified halocins have been HA3 (Kumar and Tiwari Citation2017a), H17 (Mazguene et al. Citation2018), and HA1(Kumar and Tiwari Citation2017b) however, a deeper understanding of the mechanism of action of halocins along with structural elucidation, will undoubtedly prove highly beneficial with regards to their future applications. Equally, investigation at both a protein and gene expression level will allow for the screening and discovery of novel AMPs with genomic mining.
Halophilic eukaryotes
Fungi from hypersaline environments do not require salt for viability but can tolerate salt to very high concentrations from 0 to 30% NaCl (Plemenitaš et al. Citation2008), a much broader range than bacteria or archaea (Plemenitaš et al. Citation2014). They have developed survival strategies to adapt to low water activity (aw) such as the accumulation of compatible solutes such as glycerol, which do not interfere with the vital functions of their cellular proteins while counteracting changes in turgor pressure (Gunde-Cimerman and Zalar Citation2014). Only a few fungal species display true halophilic behaviour (Zalar et al. Citation2005), such as Debaryomyces hansenii, Hortaea werneckii, and Wallemia ichthyophaga, these fungi have been isolated from natural hypersaline environments (Gunde-Cimerman et al. Citation2009).
While halotolerant fungi such as Aspergillus variecolor B-17 have already proven to be useful sources of cytotoxic agents (Wang et al. Citation2007), extremophilic fungi have been largely overlooked as a reservoir of potential antibiotic compounds.
A previous study demonstrated that stress conditions (high NaCl concentration and low aw) could induce the production of compounds with antibacterial activity. Methanol and acetone extracts from different fungal strains demonstrated antimicrobial activity against B. subtilis. The study revealed that low aw induces the production of active metabolites in halophilic and halotolerant fungi from hypersaline waters (Sepcic et al. Citation2010).
However, other eukaryotes such as the halophilic yeasts isolated from cocoa bean pulp displayed antimicrobial activity against Salmonella spp. (Indah et al. Citation2015). The halotolerant microalgae Dunaliella, and its close relatives have been reported for their potential to produce a multitude of biologically relevant compounds. Of clinical relevance, Ohta et al. reported the ability of methanol extracts of D. bioculata and D. primolecta to inhibit the cytopathic effect of herpes simplex virus type 1 (Ohta et al. Citation1998).
Quorum sensing inhibition as an anti-virulent strategy
Within bacterial infections, an estimated 65–80% (Potera Citation1999; NIH Citation2003) are attributable to the occurrence of specialized surface-adhered bacterial communities known as biofilms (Lewis Citation2007). Bacterial biofilms are characterized by their highly elevated tolerance to antimicrobial challenges, with biofilms requiring up to 1000 times higher concentrations of antibiotics compared to their planktonic equivalents to obtain complete eradication (Koch and Høiby Citation2000). Bacterial behaviors within a biofilm are regulated, in part, by the phenomenon of quorum sensing (QS), a population density-dependent gene regulation system whereby bacteria release chemical signaling molecules and, when a threshold concentration has been achieved, express genes in a cell density-dependent manner (Bhargava et al. Citation2010). Bacterial pathogens employ QS to regulate virulence and QS has emerged as a promising target for anti-virulence therapeutic approaches in the past decade, and several quorum sensing inhibitors (QSI) from halophiles have been identified – as summarised in .
Table 4. Examples of QSI compounds and their corresponding activity produced from halophilic and halotolerant bacteria.
Teasdale et al. (Citation2009) isolated two phenethylamide-based metabolites from the marine bacterium, Halobacillus salinus C42, which were capable of inhibiting a range of QS-controlled phenotypes in bioreporter strains including; violacein production by Chromobacterium violaceum; bioluminescence in Vibrio harveyi; and QS controlled Green Fluorescent Protein (GFP) production in E. coli JB525. These compounds act as antagonists in Gram-negative bacterial QS by competing with N-acyl homoserine lactones (AHL) for receptor binding. Further investigation into the widespread production of these QS inhibitory molecules, identified five isolates, either Bacillus or Halobacillus, that were capable of interfering with QS-controlled bioluminescence in V. harveyi and inhibiting violacein production by C. violaceum, with the elucidation of the QSI dipeptide cyclo-L-proline-L-tyrosine produced by Bacillus cereus D28 (Teasdale et al. Citation2011).
Hypersaline mats have previously been shown to be a source of QS-inhibitory compounds, following the total ethyl acetate extraction of the mat, extracts demonstrated QS-inhibitory properties against bioreporter strains Agrobacterium tumefaciens NTL4 (pZLR4) and Salmonella enterica S235, but not C. violaceum CV017 (Abed et al. Citation2011). Abed et al. (Citation2013) subsequently screened the halophilic and moderately thermophilic strains from a hypersaline microbial mat for QSI molecules, resulting in the structural elucidation of four related diketopiperazines (DKPs) from Marinobacter sp. SK-3. The compounds capable of inhibiting violacein production by C. violaceum and reduced QS-dependent luminescence of the bioreporter E. coli pSB401. DKPs have previously been shown to have antimicrobial, antifungal, antiviral, and antitumor properties (Herbert Citation1992; Martins and Carvalho Citation2007) and their production has been observed across many genera, including Bacillus (Wang et al. Citation2010), Streptomyces (Li et al. Citation2006), and Pseuoalteromonas (Qi et al. Citation2009), and from a wide range of environments. In addition, DKPs are capable of activating QS-regulated pathways as well as inhibiting them, as observed by the haloarchaea Haloterrigena hispanica (Tommonaro et al. Citation2012). In Abed et al. (Abed et al. Citation2013), the supernatant and water extract of the cell pellet from the isolate, Haloterrigena saccharevitan SK-6, was found to possess QSI compounds capable of inhibiting violacein production in the bioreporter strain CV017.
Numerous studies have indicated the existence of QSI compounds from the marine environment, particularly from marine cyanobacteria. For example, the marine cyanobacterium, Blennothrix cantharidosmum, was found to produce several QSI tumonoic acids with tumonoic acid F being the most potent (Clark et al. Citation2008). Dobretsov et al. revealed that another marine cyanobacterium, Lyngbya majuscule, secreted the δ-lactone QSI compound, malyngolide. This compound inhibits the violacein production in C. violaceum CV017 and las QS-dependent production of elastase by P. aeruginosa PAO1 (Dobretsov et al. Citation2010).
More recently Choi et al. isolated, Leptolyngbya crossbyana, from corals on the Hawaiian coast, which produced the AHL mimetic compounds, honaucins A–C, which inhibited QS-regulated bioluminescence in V. harevyi BB120 (Choi et al. Citation2012).
The discovery of compounds and enzymes which interrupt QS via inhibition of signal generation, degradation of generated signals, or suppression of QS receptors, have important clinical applications (Hong et al. Citation2012; Tang and Zhang Citation2014). For pathogens like Burkholderia species, P. aeruginosa, Erwinia carotovora, and Vibrio spp., biofilm formation, toxin production, and virulence factor production are regulated by QS (Tang and Zhang Citation2014). QSI can be considered as an alternative antimicrobial strategy, that must be considered to control bacterial disease, by targeting the virulence factors essential for an infection rather than the viability of pathogens (Clatworthy et al. Citation2007). Additionally, the delayed emergence of resistance which results from imposing less selective pressure on pathogens (Clatworthy et al. Citation2007) makes QSI an ideal candidate as an adjunct therapy to improve antibiotic drug treatment (Busetti et al. Citation2014). QSI may be used as antifouling agents capable of inhibiting biofilm formation in medicine, industry, and environmental settings (Abed et al. Citation2013).
Important tools and techniques for antimicrobial drug discovery from halophiles
As the prevalence of AMR increases, there is a critical and urgent need to develop new technologies and strategies to combat this major global public health concern (Bakour et al. Citation2016). Particular focus has surrounded the development of methods that can extract novel antimicrobials from microbes uncultivable under standard laboratory conditions. “The Great Plate Count Anomaly” accounts for the 99% of microorganisms in the natural environment that cannot be isolated using conventional culture techniques (Amann et al. Citation1995). The development of methods that either enhance current culturing or utilise a culture-independent approach such as genome mining are indispensable if these “hidden” antimicrobial metabolites are to be investigated (Challis and Ravel Citation2000; Hug et al. Citation2018).
Genomic mining
It is now widely accepted that most of the investigated culturable microorganisms have a broader genetic capacity to produce a greater number of natural products than the number of compounds that they produce under conditions of laboratory culture (Bussi and Gutierrez Citation2019). Following the full genome sequencing of Streptomyces coelicolor (Bentley et al. Citation2002), it was clear that there is underexplored potential within bacterial genomes for drug discovery. S. coelicolor could produce 20 secondary metabolites when only three secondary metabolites were identified (Bentley et al. Citation2002). Within the Streptomyces genus, conservative estimates suggest that this genus alone can produce over 150,000 natural products, with less than 5% of these compounds characterized (Watve et al. Citation2001).
This classical approach of genome mining involves the prediction of cryptic metabolic pathways and genes that encode putatively in the biosynthesis of secondary metabolites of interest, such as biosynthetic gene clusters (BGC) (León et al. Citation2014; Blin et al. Citation2019; Zheng et al. Citation2019). The most typical examples of BGCs, with regards to antimicrobials, are two multifunctional mega-synthases, non-ribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) (Fischbach and Walsh Citation2006; Gross Citation2007). NRPS and PKS have long been utilised by bacteria for cellular defence and offense against competitors (Czaran et al. Citation2002). They are prolific and generate many of the antibiotics used clinically, such as penicillin, erythromycin, vancomycin, and daptomycin among numerous others (Fischbach and Walsh Citation2006).
The continued enhancement of computational biology has resulted in many of these approaches carried out in silico. In silico mining has significantly advanced the analyses of microbial genome sequences, allowing the determination of BGC involved in the production of new antibiotics from different organisms (Challis Citation2008; Scheffler et al. Citation2013), with the use of sequence-based comparison software such as DIAMOND (Buchfink et al. Citation2015) or BLAST (Altschul et al. Citation1990). One of the more popular and straightforward tools used in genome mining is the antiSMASH- a web-based bioinformatics server. It allows users to identify antiSMASH-annotated BGCs from its database with the most recent version containing 6200 bacterial genomes (Blin et al. Citation2019).
Genome mining has led to the discovery of many antimicrobial compounds (Nikolouli and Mossialos Citation2012; Scheffler et al. Citation2013) including orfamide A (Gross Citation2007), salinilactam (Udwary et al. Citation2007), holomycin (Li and Walsh Citation2010), and emericellamide A, C–E (Chiang et al. Citation2008) (summarised in ). Within halophiles, Schäberle et al. reported three strains of Enhygromyxa salina, that were producers of novel secondary metabolites (Schäberle et al. Citation2010). Furthermore, Meklat et al. revealed that halophilic actinomycetes, such as Nocardiopsis had a high frequency of NRPS genes (Meklat et al. Citation2011). Genome mining of the marine halophile, Haliangium ochraceum, identified the compound haliamide, the product of a gene cluster containing one NRPS module followed by four PKS modules. Five more PKS-NRPS gene clusters were identified in the genome of H. ochraceum, indicating that there is further antimicrobial potential (Dávila-Céspedes et al. Citation2016; Sun et al. Citation2016). Despite the success of genome mining in the discovery of many natural products, it does pose several limitations (Gross Citation2007). The prediction algorithms of the bioinformatic tools are based on the identification of known biosynthetic activities, and therefore truly novel biosynthetic pathways are not identified (Challis Citation2008; Weber Citation2014). In addition to unreliable predictions, if the genes are “silent”, it becomes much more difficult to produce, identify, and test the biological activity of the product. In such cases, growing the organism under many different conditions may activate the bioactive (Okami et al. Citation1976; Scherlach and Hertweck Citation2006). Genome mining methods include the genomisotopic approach, gene inactivation method, heterologous expression approach, the use of transcription activators, inactivation of a silent gene inhibitor (Scheffler et al. Citation2013), and the resistance-guided approach (Thaker et al. Citation2013).
Table 5. Structures of antimicrobials identified through genome mining from five species of bacteria.
The resistance-based mining technique is a genome-mining approach that based on the identification of self-protection mechanisms that antibiotic producers need to avoid autotoxicity. This self-resistant mechanism acts as a filter with the potential to identify antibiotic-producing bacteria and therefore could be exploited for the discovery of novel bioactive (Thaker et al. Citation2013). This resistance-guided approach is directly relatable to many halophilic organisms and studies by Khelaifia et al. have reported AMR in the haloarchaea Halalkalicoccus tibetensis and Natronococcus amylolyticus (Khelaifia and Drancourt Citation2012). A similar approach Culp et al. recently described an evolution-guided antibiotic discovery approach that combines phylogenetic divergence of BGCs along with the absence of self-resistance determinants to predict the novel glycopeptide corbomycin (Culp et al. Citation2020).
Another widely used method for antibiotic discovery involves metagenomic screening, which circumvents the difficulties associated with cultivation. Metagenomic screening relays on prior examination for BGCs using in silico bioinformatics tools, then a heterologous expression of selected pathways originating from uncultured microorganisms. The current problem with this approach is the discovery of novel antimicrobials requires a large reference dataset of “meta-omics” derived compounds (Hamm et al. Citation2019; Wang et al. Citation2019).
There are two main strategies utilized in the metagenomic approach – a classical functional metagenomic screening approach, and a targeted sequence-based metagenomic screening approach- summarises the main steps for both methods. In both cases, environmental samples are collected and environmental DNA (eDNA) is extracted and purified. In the classical approach, metagenomic libraries are created by extracting, cloning, and ligating the eDNA into shuttle vectors and transforming them into appropriate heterologous hosts, e.g. E. coli. The metagenomic library is screened for bioactivity using standard antimicrobial tests such as disc diffusions, MTT, or MIC assays. Positive clones are recovered from the metagenomic library, and the eDNA insert is sequenced. The antibacterial violacein was isolated from a soil metagenome using this approach (Brady et al. Citation2001).
Figure 3. Summary describing the two main approaches that used for antimicrobial discovery from metagenomes. Adapted from Hug et al. (Citation2018).
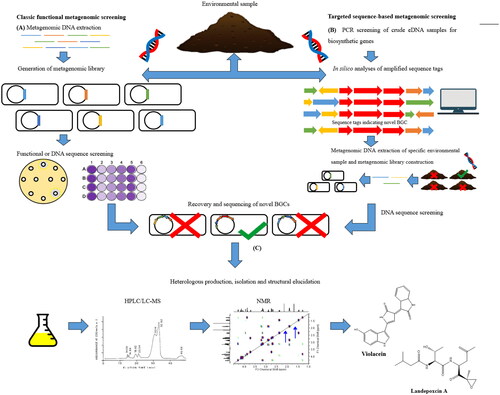
In the targeted approach, eDNA is screened by PCR amplicons for specific sequences within BGCs. The sequence tags are evaluated for their biosynthetic origin, compared to a reference database, and biosynthetic gene clusters are reassembled in silico. A metagenomic library is generated with samples harboring the in silico reassembled BGC of interest. The proteasome inhibitor landepoxin A was identified using this targeted approach from metagenomic samples (Lok Citation2015). For both approaches, the novel BGCs are assembled, modified for heterologous expression, and then the bioactive natural product is isolated and structurally elucidated. Charlop-Powers et al. used a metagenomic approach and revealed that there is a rich biosynthetic diversity of clinically relevant BGCs in eDNA from New York City park soils (Charlop-Powers et al. Citation2016).
Enhanced culture techniques
In 2007 Coates and others, argued that the genomics route has proven to be target-rich, but has not led to the introduction of a marketed antibiotic as yet and suggested that non-culturable bacteria may be an alternative source of new antibiotics (Coates and Hu Citation2007). Moreover, while researchers involved in genome mining would debate this lack of value in their approach, the prospect of tapping into these non-culturable organisms offers the unquestionable opportunity to tap into a rich vein of drug discovery. Strategies used to increase bacterial diversity have ranged from soil-extract agar (Hamaki et al. Citation2005) to simulated environments with diffusion chambers (Kaeberlein et al. Citation2002) and the iChip (Ling et al. Citation2015), to variation of culturing environments (co-cultures or mixed cultures) (Tanaka et al. Citation2004) and to the role of host-associated environments (Vartoukian et al. Citation2010). An extensive review of these technologies is provided by Stewart et al. (Stewart Citation2012).
The discovery of teixobactin is probably the most significant example of a compound discovered as a direct result of using advanced isolation technologies (Ling et al. Citation2015). More recently, a screening of symbionts from the microbiome of entomopathogenic nematodes revealed a Gram-negative specific antibiotic termed darobactin (Imai et al. Citation2019). These studies demonstrate that the cultivation of microbes from unexplored territories (and not genomic mining) continues to provide the mainstay of novel classes of antimicrobials with unique natural product chemistry. Concerning extremophiles, access to their bioactives is already limited with difficulty in their cultivation using traditional laboratory conditions (Hug et al. Citation2018). Technologies such as that of the iChip device are likely to improve isolation efficiency and fermentation of previously uncultured bacteria and archaea from these largely unexplored habitats. Therefore, extreme environments possess an under-explored microbiome whose metabolites require interrogation to assess their antimicrobial potential.
Conclusion
We must rejuvenate the antibiotic arsenal with novel classes of antibiotics that possess unique modes of action that can overcome traditional mechanisms of drug resistance. To discover these therapeutically unique classes, we must shift our emphasis towards organisms from an equally unique environment. This review demonstrates that halophiles from all domains are a promising source of antimicrobials. Among the halophilic microorganisms, Actinobacteria, are of noteworthy interest for their unique metabolic diversity, biological activity, and biotechnological potential. The fastidious nature of extremophiles, especially haloarchaea, requires that biotechnologies such as enhanced isolation e.g. the iChip, genomic mining e.g. antiSMASH, and metagenomic discovery be further exploited to investigate the antimicrobial potential of these underexplored ecological niches. The future of natural product discovery will be the application of a combination of multi-omics approaches to improve the access of these non-culturable microbes, investigate the potential of their silent operons and reduce the high rediscovery rate of known compounds. Both genomic mining and enhanced culture techniques will prove invaluable tools for unlocking new avenues for antimicrobial natural products.
Authors contributions
TT and BG conceived the idea. TT collected literature data, created the tables and figures, and wrote the first draft of the manuscript. BG reviewed and edited the manuscript. Both authors approved the final version of the manuscript.
Acknowledgments
We acknowledge our colleagues at Queen’s University Belfast for the great discussions and feedback. Also, Simon Gibbons from the University of East Anglia for the thoughtful advices.
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Funding
References
- Abed RMM, Dobrestov S, Al-Kharusi S, Schramm A, Jupp B, Golubic S. 2011. Cyanobacterial diversity and bioactivity of inland hypersaline microbial mats from a desert stream in the Sultanate of Oman. Fottea. 11(1):215–224.
- Abed RMM, Dobretsov S, Al-Fori M, Gunasekera SP, Sudesh K, Paul VJ. 2013. Quorum-sensing inhibitory compounds from extremophilic microorganisms isolated from a hypersaline cyanobacterial mat. J Ind Microbiol Biotechnol. 40(7):759–772.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410.
- Amann RI, Ludwig W, Schleifer KH. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 59(1):143–169.
- Aminov RI. 2010. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 1:134.
- Atanasova NS, Pietila MK, Oksanen HM. 2013. Diverse antimicrobial interactions of halophilic archaea and bacteria extend over geographical distances and cross the domain barrier. Microbiologyopen. 2(5):811–825.
- Baati H, Guermazi S, Amdouni R, Gharsallah N, Sghir A, Ammar E. 2008. Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles. 12(4):505–518.
- Bakour S, Sankar SA, Rathored J, Biagini P, Raoult D, Fournier PE. 2016. Identification of virulence factors and antibiotic resistance markers using bacterial genomics. Future Microbiol. 11(3):455–466.
- Ballav S, Kerkar S, Thomas S, Augustine N. 2015. Halophilic and halotolerant actinomycetes from a marine saltern of Goa, India producing anti-bacterial metabolites. J Biosci Bioeng. 119(3):323–330. [doi].
- Baltz RH. 2007. Antimicrobials from Actinomycetes: back to the future. Microbe. 2:125–131.
- Basilio A, González I, Vicente MF, Gorrochategui J, Cabello A, González A, Genilloud O. 2003. Patterns of antimicrobial activities from soil actinomycetes isolated under different conditions of pH and salinity. J Appl Microbiol. 95(4):814–823.
- Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature. 417(6885):141–147.
- Bérdy J. 2005. Bioactive microbial metabolites. J Antibiot. 58(1):1–26. .
- Bérdy J. 2015. Microorganisms producing antibiotics. In: Sanchez S, Demain AL, editors. Antibiot Curr Innov Futur trends. Norfolk (UK): Caister Academic Press; p. 49–64.
- Bhargava N, Sharma P, Capalash N. 2010. Quorum sensing in Acinetobacter: an emerging pathogen. Crit Rev Microbiol. 36(4):349–360.
- Blin K, Kim HU, Medema MH, Weber T. 2019. Recent development of antiSMASH and other computational approaches to mine secondary metabolite biosynthetic gene clusters. Brief Bioinform. 20(4):1103–1113.
- Blin K, Pascal Andreu V, de Los Santos ELC, Del Carratore F, Lee SY, Medema MH, Weber T. 2019. The antiSMASH database version 2: a comprehensive resource on secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 47(D1):D625–D630.
- Blum P. 2008. Archaea: new models for prokaryotic biology. Norwich: Horizon Scientific Press.
- Boubetra D, Sabaou N, Zitouni A, Bijani C, Lebrihi A, Mathieu F. 2013. Taxonomy and chemical characterization of new antibiotics produced by Saccharothrix SA198 isolated from a Saharan soil. Microbiol Res. 168(4):223–230.
- Brady SF, Chao CJ, Handelsman J, Clardy J. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org Lett. 3(13):1981–1984.
- Brown ED. 2013. Is the GAIN Act a turning point in new antibiotic discovery? Can J Microbiol. 59(3):153–156.
- Brown ED, Wright GD. 2016. Antibacterial drug discovery in the resistance era. Nature. 529(7586):336–343.
- Buchanan GO, Williams PG, Feling RH, Kauffman CA, Jensen PR, Fenical W. 2005. Sporolides A and B: structurally unprecedented halogenated macrolides from the marine actinomycete Salinispora tropica. Org Lett. 7(13):2731–2734.
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60.
- Bud R. 2007. Antibiotics: the epitome of a wonder drug. BMJ. 334(Suppl 1):s6–s6.
- Bull AT. 2011. Actinobacteria of the extremobiosphere. In: Horikoshi K, editor. Extremophiles handbook. Tokyo: Springer; p. 1203–1240.
- Bull AT, Asenjo JA. 2013. Microbiology of hyper-arid environments: recent insights from the Atacama Desert, Chile. Antonie Van Leeuwenhoek. 103(6):1173–1179.
- Bull AT, Ward AC, Goodfellow M. 2000. Search and discovery strategies for biotechnology: the paradigm shift. Microbiol Mol Biol Rev. 64(3):573–606.
- Busarakam K, Bull AT, Girard G, Labeda DP, Van Wezel GP, Goodfellow M. 2014. Streptomyces leeuwenhoekii sp. nov., the producer of chaxalactins and chaxamycins, forms a distinct branch in Streptomyces gene trees. Antonie Van Leeuwenhoek. 105(5):849–861.
- Busetti A, Shaw G, Megaw J, Gorman SP, Maggs CA, Gilmore BF. 2014. Marine-derived quorum-sensing inhibitory activities enhance the antibacterial efficacy of tobramycin against Pseudomonas aeruginosa. Mar Drugs. 13(1):1–28.
- Bussi C, Gutierrez MG. 2019. Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol Rev. 43(4):341–361.
- Butler MS, Blaskovich MA, Cooper MA. 2017. Antibiotics in the clinical pipeline at the end of 2015. J Antibiot. 70(1):3–24.
- Challis GL. 2008. Mining microbial genomes for new natural products and biosynthetic pathways. Microbiology. 154(Pt 6):1555–1569.
- Challis GL, Ravel J. 2000. Coelichelin, a new peptide siderophore encoded by the Streptomyces coelicolor genome: structure prediction from the sequence of its non-ribosomal peptide synthetase. FEMS Microbiol Lett. 187(2):111–114.
- Charan RD, Schlingmann G, Janso J, Bernan V, Feng X, Carter GT. 2004. Diazepinomicin, a new antimicrobial alkaloid from a marine Micromonospora sp. J Nat Prod. 67(8):1431–1433.
- Charlop-Powers Z, Pregitzer CC, Lemetre C, Ternei MA, Maniko J, Hover BM, Calle PY, McGuire KL, Garbarino J, Forgione HM, et al. 2016. Urban park soil microbiomes are a rich reservoir of natural product biosynthetic diversity. Proc Natl Acad Sci U S A. 113(51):14811–14816.
- Chen D, Feng J, Huang L, Zhang Q, Wu J, Zhu X, Duan Y, Xu Z. 2014. Identification and characterization of a new erythromycin biosynthetic gene cluster in Actinopolyspora erythraea YIM90600, a novel erythronolide-producing halophilic actinomycete isolated from salt field. PLOS One. 9(9):e108129.
- Chen L, Wang G, Bu T, Zhang Y, Wang Y, Liu M, Lin X. 2010. Phylogenetic analysis and screening of antimicrobial and cytotoxic activities of moderately halophilic bacteria isolated from the Weihai Solar Saltern (China). World J Microbiol Biotechnol. 26(5):879–888.
- Chen Y-G, Cui X-L, Pukall R, Li H-M, Yang Y-L, Xu L-H, Wen M-L, Peng Q, Jiang C-L. 2007. Salinicoccus kunmingensis sp. nov., a moderately halophilic bacterium isolated from a salt mine in Yunnan, south-west China. Int J Syst Evol Microbiol. 57(Pt 10):2327–2332.
- Cheung J, Danna KJ, O'Connor EM, Price LB, Shand RF. 1997. Isolation, sequence, and expression of the gene encoding halocin H4, a bacteriocin from the halophilic archaeon Haloferax mediterranei R4. J Bacteriol. 179(2):548–551.
- Chiang Y-M, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo H-C, Ho W-Y, Simityan H, Kuo E, Praseuth A, et al. 2008. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol. 15(6):527–532.
- Choi H, Mascuch SJ, Villa FA, Byrum T, Teasdale ME, Smith JE, Preskitt LB, Rowley DC, Gerwick L, Gerwick WH, et al. 2012. Honaucins A-C, potent inhibitors of inflammation and bacterial quorum sensing: synthetic derivatives and structure-activity relationships. Chem Biol. 19(5):589–598.
- Ciofu O, Giwercman B, Høiby N, Pedersen SS. 1994. Development of antibiotic resistance in Pseudomonas aeruginosa during two decades of antipseudomonal treatment at the Danish CF Center. APMIS. 102(9):674–680.
- Clardy J, Fischbach MA, Walsh CT. 2006. New antibiotics from bacterial natural products. Nat Biotechnol. 24(12):1541–1550.
- Clark BR, Engene N, Teasdale ME, Rowley DC, Matainaho T, Valeriote FA, Gerwick WH. 2008. Natural products chemistry and taxonomy of the marine cyanobacterium Blennothrix cantharidosmum. J Nat Prod. 71(9):1530–1537.
- Clatworthy AE, Pierson E, Hung DT. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 3(9):541–548.
- Coates ARM, Hu Y. 2007. Novel approaches to developing new antibiotics for bacterial infections. Br. J. Pharmacol. 152(8):1147–1154.
- Cragg GM, Newman DJ. 2013. Natural products: a continuing source of novel drug leads. Biochim Biophys Acta. 1830(6):3670–3695.
- Culp EJ, Waglechner N, Wang W, Fiebig-Comyn AA, Hsu Y-P, Koteva K, Sychantha D, Coombes BK, Van Nieuwenhze MS, Brun YV, et al. 2020. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature. 578(7796):582–587.
- Czaran TL, Hoekstra RF, Pagie L. 2002. Chemical warfare between microbes promotes biodiversity. Proc Natl Acad Sci U S A. 99(2):786–790.
- Dall C. 2019. Achaogen bankruptcy raises worry over antibiotic pipeline. CIDRAP News. [cited 2020 Feb 7]. http://www.cidrap.umn.edu/news-perspective/2019/04/achaogen-bankruptcy-raises-worry-over-antibiotic-pipeline
- Davies J. 2006. Where have all the antibiotics gone? Can J Infect Dis Med Microbiol. 17(5):287–290.
- Dávila-Céspedes A, Hufendiek P, Crüsemann M, Schäberle TF, König GM. 2016. Marine-derived myxobacteria of the suborder Nannocystineae: an underexplored source of structurally intriguing and biologically active metabolites. Beilstein J Org Chem. 12:969–984.
- Demain AL. 2014. Importance of microbial natural products and the need to revitalize their discovery. J Ind Microbiol Biotechnol. 41(2):185–201.
- Dhanasekaran D, Rajakumar G, Sivamani P, Selvamani S, Panneerselvam A. 2004. Screening of salt pans actinomycetes for antibacterial agents. Internet J Microbiol. 1:2–5.
- Dietera A, Hamm A, Fiedler H-P, Goodfellow M, Müller WEG, Brun R, Beil W, Bringmann G. 2003. Pyrocoll, an antibiotic, antiparasitic and antitumor compound produced by a novel alkaliphilic Streptomyces strain. J Antibiot (Tokyo). 56(7):639–646.
- Dobretsov S, Teplitski M, Alagely A, Gunasekera SP, Paul VJ. 2010. Malyngolide from the cyanobacterium Lyngbya majuscula interferes with quorum sensing circuitry. Environ Microbiol Rep. 2(6):739–744.
- Fischbach MA, Walsh CT. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 106(8):3468–3496.
- Fischbach MA, Walsh CT. 2009. Antibiotics for emerging pathogens. Science. 325(5944):1089–1093.
- Fleming A. 1929. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br J Exp Pathol. 10(3):226–236.
- Friedman D, Alper J. 2014. Challenges in overcoming antibiotic resistance. In: Technological challenges in antibiotic discovery and development. Washington: National Academies Press; p. 7–18. [cited 2020 Feb 7]. www.nap.edu.
- Gajardo GM, Beardmore JA. 2012. The brine shrimp Artemia: adapted to critical life conditions. Front Physiol. 3:185.
- Ghanmi F, Carré-Mlouka A, Vandervennet M, Boujelben I, Frikha D, Ayadi H, Peduzzi J, Rebuffat S, Maalej S. 2016. Antagonistic interactions and production of halocin antimicrobial peptides among extremely halophilic prokaryotes isolated from the solar saltern of Sfax, Tunisia. Extremophiles. 20(3):363–374.
- Gohel SD, Sharma AK, Dangar KG, Thakrar FJ, Singh SP. 2015. Antimicrobial and biocatalytic potential of haloalkaliphilic actinobacteria. In: Halophiles. Cham: Springer; p. 29–55.
- Gómez-Silva B, Rainey FA, Warren-Rhodes KA, McKay CP, Navarro-González R. 2008. Atacama desert soil microbiology. In: Dion P, editor. Nautiyal CS microbiology of extreme soils. Berlin Heidelberg: Springer; p. 117–132.
- Gottlieb D. 1973. General consideration and implications of the Actinomycetales. Symp Ser Soc Appl Bacteriol. 2:1–10.
- Gross H. 2007. Strategies to unravel the function of orphan biosynthesis pathways: recent examples and future prospects. Appl Microbiol Biotechnol. 75(2):267–277.
- Guan T-W, Wu N, Xia Z-F, Ruan J-S, Zhang X-P, Huang Y, Zhang L-L. 2011. Saccharopolyspora lacisalsi sp. nov., a novel halophilic actinomycete isolated from a salt lake in Xinjiang, China. Extremophiles. 15(3):373–378.
- Gunde-Cimerman N, Ramos J, Plemenitaš A. 2009. Halotolerant and halophilic fungi. Mycol Res. 113(Pt 11):1231–1241.
- Gunde-Cimerman N, Zalar P. 2014. Extremely halotolerant and halophilic fungi inhabit brine in solar salterns around the globe. Food Technol Biotechnol. 52:170–179.
- Gunde-Cimermana N, Zalarb P, de Hoogc S, Plemenitasd A. 2000. Hypersaline waters in salterns – natural ecological niches for halophilic black yeasts. FEMS Microbiol Ecol. 32(3):235–240.
- Hamaki T, Suzuki M, Fudou R, Jojima Y, Kajiura T, Tabuchi A, Sen K, Shibai H. 2005. Isolation of novel bacteria and actinomycetes using soil-extract agar medium. J Biosci Bioeng. 99(5):485–492.
- Hamedi J, Mohammadipanah F, Ventosa A. 2013. Systematic and biotechnological aspects of halophilic and halotolerant actinomycetes. Extremophiles. 17(1):1–13.
- Hamm JN, Erdmann S, Eloe-Fadrosh EA, Angeloni A, Zhong L, Brownlee C, Williams TJ, Barton K, Carswell S, Smith MA, et al. 2019. Unexpected host dependency of Antarctic nanohaloarchaeota. Proc Natl Acad Sci U S A. 116(29):14661–14670.
- Haseltine C, Hill T, Montalvo-Rodriguez R, Kemper SK, Shand RF, Blum P. 2001. Secreted euryarchaeal microhalocins kill hyperthermophilic crenarchaea. J Bacteriol. 183(1):287–291.
- Herbert RA. 1992. A perspective on the biotechnological potential of extremophiles. Trends Biotechnol. 10(11):395–402.
- Hong K-W, Koh C-L, Sam C-K, Yin W-F, Chan K-G. 2012. Quorum quenching revisited—from signal decays to signalling confusion. Sensors. 12(4):4661–4696.
- Hug JJ, Bader CD, Remškar M, Cirnski K, Müller R. 2018. Concepts and methods to access novel antibiotics from actinomycetes. Antibiotics. 7(2):44.
- Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, et al. 2020. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature. 577(7791):519–525.
- Imada C, Koseki N, Kamata M, Kobayashi T, Hamada-Sato N. 2007. Isolation and characterization of antibacterial substances produced by marine actinomycetes in the presence of seawater. Actinomycetologica. 21(1):27–31.
- Imai Y, Meyer KJ, Iinishi A, Favre-Godal Q, Green R, Manuse S, Caboni M, Mori M, Niles S, Ghiglieri M, et al. 2019. A new antibiotic selectively kills gram-negative pathogens. Nature. 576(7787):459–464.
- Indah H, Putri F, Utama GL. 2015. Preliminary studies of halophilic yeasts antimicrobial activities isolated from cocoa bean pulp towards E. coli and Salmonella spp. Int J Adv Sci Eng Inf Technol. 5(2):107–109.
- Irshad A, Ahmad I, Kim SB. 2013. Isolation, characterization and antimicrobial activity of halophilic bacteria in foreshore soils. African J Microbiol Res. 7:164–173.
- Irshad A, Kim SB, Ahmad I. 2014. Culturable diversity of halophilic bacteria in foreshore soils. Braz J Microbiol. 45(2):563–571.
- Jang KH, Nam S-J, Locke JB, Kauffman CA, Beatty DS, Paul LA, Fenical W. 2013. Anthracimycin, a potent anthrax antibiotic from a marine-derived actinomycete. Angew Chem Int Ed Engl. 52(30):7822–7824.
- Jensen PR, Dwight R, Fenical W. 1991. Distribution of actinomycetes in near-shore tropical marine sediments. Appl Environ Microbiol. 57(4):1102–1108.
- Jose PA, Jebakumar SR. 2014. Unexplored hypersaline habitats are sources of novel actinomycetes. Front Microbiol. 5:242.
- Jose PA, Jebakumar SRD. 2013. Diverse actinomycetes from Indian coastal solar salterns-a resource for antimicrobial screening. J.Pure Appl.Microbiol. 7:2569–2575.
- Jose PA, Santhi VS, Jebakumar SR. 2011. Phylogenetic-affiliation, antimicrobial potential and PKS gene sequence analysis of moderately halophilic Streptomyces sp. inhabiting an Indian saltpan. J Basic Microbiol. 51(4):348–356.
- Kaeberlein T, Lewis K, Epstein SS. 2002. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 296(5570):1127–1129.
- Kamat TK, Kiran S, Kerkar S. 2011. Antimicrobial potential of Bacillus marismortal, a salt pan isolate of Cavellosim, Goa-India. Int J Adv Biotechnol Res. 2:321–328.
- Kavitha A, Prabhakar P, Vijayalakshmi M, Venkateswarlu Y. 2010. Purification and biological evaluation of the metabolites produced by Streptomyces sp. TK-VL_333. Res Microbiol. 161(5):335–345.
- Kavitha P, Lipton AP, Sarika AR, Aishwarya MS. 2011. Growth characteristics and halocin production by a new isolate, Haloferax volcanii KPS1 from Kovalam solar saltern (India). Res J Biol Sci. 6:257–262.
- Kessel M, Klink F. 1980. Archaebacterial elongation factor is ADP-ribosylated by diphtheria toxin. Nature. 287(5779):250–251.
- Khelaifia S, Drancourt M. 2012. Susceptibility of archaea to antimicrobial agents: applications to clinical microbiology. Clin Microbiol Infect. 18(9):841–848. .
- Koch C, Høiby N. 2000. Diagnosis and treatment of cystic fibrosis. Respiration. 67(3):239–247.
- Koehn FE, Carter GT. 2005. The evolving role of natural products in drug discovery. Nat Rev Drug Discov. 4(3):206–220.
- Kokare CR, Mahadik KR, Kadam SS, Chopade BA. 2004. Isolation, characterization and antimicrobial activity of marine halophilic Actinopolyspora species AH1 from the west coast of India. Curr Sci. 86:593–597.
- Kovac J, Sneader W. 2006. Chemical education today book & media reviews drug discovery: a history. J Chem Educ. 83(8):1139.
- Kumar V, Tiwari SK. 2017a. Activity-guided separation and characterization of new halocin HA3 from fermented broth of Haloferax larsenii HA3. Extremophiles. 21(3):609–621.
- Kumar V, Tiwari SK. 2017b. Halocin HA1: an archaeocin produced by the haloarchaeon Haloferax larsenii HA1. Process Biochem. 61:202–208.
- Lam KS. 2006. Discovery of novel metabolites from marine actinomycetes. Curr Opin Microbiol. 9(3):245–251.
- León MJ, Fernández AB, Ghai R, Sánchez-Porro C, Rodriguez-Valera F, Ventosa A. 2014. From metagenomics to pure culture: isolation and characterization of the moderately halophilic bacterium Spiribacter salinus gen. nov., sp. nov. Appl Environ Microbiol. 80(13):3850–3857.
- Lequerica JL, O'Connor JE, Such L, Alberola A, Meseguer I, Dolz M, Torreblanca M, Moya A, Colom F, Soria B, et al. 2006. A halocin acting on Na+/H + exchanger of haloarchaea as a new type of inhibitor in NHE of mammals. J Physiol Biochem. 62(4):253–262.
- Levy S. 2001. Thanks to Penicillin…He Will Come Home, The Antibiotic Paradox. New York: The National WWII Museum.
- Lewis K. 2007. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 5(1):48–56.
- Lewis K. 2012. Antibiotics: recover the lost art of drug discovery. Nature. 485(7399):439–440.
- Lewis K. 2013. Platforms for antibiotic discovery. Nat Rev Drug Discov. 12(5):371–387.
- Li B, Walsh CT. 2010. Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc Natl Acad Sci U S A. 107(46):19731–19735.
- Li F, Maskey RP, Qin S, Sattler I, Fiebig HH, Maier A, Zeeck A, Laatsch H. 2005. Chinikomycins A and B: isolation, structure elucidation, and biological activity of novel antibiotics from a marine Streptomyces sp. isolate M045#, 1. J Nat Prod. 68(3):349–353.
- Li X, Dobretsov S, Xu Y, Xiao X, Hung O, Qian PY. 2006. Antifouling diketopiperazines produced by a deep-sea bacterium, Streptomyces fungicidicus. Biofouling. 22(3):187–194.
- Li X, Yu YH. 2015. Biodiversity and screening of halophilic bacteria with hydrolytic and antimicrobial activities from Yuncheng Salt Lake, China. Biologia. 70(2):151–156.
- Li Y, Xiang H, Liu J, Zhou M, Tan H. 2003. Purification and biological characterization of halocin C8, a novel peptide antibiotic from Halobacterium strain AS7092. Extremophiles. 7(5):401–407.
- Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S, et al. 2015. A new antibiotic kills pathogens without detectable resistance. Nature. 517(7535):455–459.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. 1997. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 23(1–3):3–25.
- Lok C. 2015. Mining the microbial dark matter. Nature. 522(7556):270–273.
- Maheshwari DK, Saraf M. 2015. Halophiles: biodiversity and sustainable exploitation. Switzerland: Springer.
- Mahyudin NA, Blunt JW, Cole AL, Munro MH. 2012. The isolation of a new S-methyl benzothioate compound from a marine-derived Streptomyces sp. J Biomed Biotechnol. 2012:894708.
- Manam RR, Teisan S, White DJ, Nicholson B, Grodberg J, Neuteboom STC, Lam KS, Mosca DA, Lloyd GK, Potts BCM, et al. 2005. Lajollamycin, a nitro-tetraene spiro-beta-lactone-gamma-lactam antibiotic from the marine actinomycete Streptomyces nodosus. J Nat Prod. 68(2):240–243.
- Martins MB, Carvalho I. 2007. Diketopiperazines: biological activity and synthesis. Tetrahedron. 63(40):9923–9932.
- Maskey RP, Helmke E, Laatsch H. 2003. Himalomycin A and B: isolation and structure elucidation of new fridamycin type antibiotics from a marine Streptomyces isolate. J Antibiot. 56(11):942–949.
- Maskey RP, Li F, Qin S, Fiebig HH, Laatsch H. 2003. Chandrananimycins A approximately C: production of novel anticancer antibiotics from a marine Actinomadura sp. isolate M048 by variation of medium composition and growth conditions. J Antibiot. 56(7):622–629.
- Maskey RP, Sevvana M, Usón I, Helmke E, Laatsch H. 2004. Gutingimycin: a highly complex metabolite from a marine streptomycete. Angew Chem Int Ed Engl. 43(10):1281–1283.
- Mazguene S, Rossi M, Gogliettino M, Palmieri G, Cocca E, Mirino S, Imadalou-Idres N, Benallaoua S. 2018. Isolation and characterization from solar salterns of North Algeria of a haloarchaeon producing a new halocin. Extremophiles. 22(2):259–270.
- McGuire JM, Bunch RL, Anderson RC, Boaz HE, Flynn EH, Powell HM, et al. 1952. Ilotycin, a new antibiotic. Antibiot Chemother. 2:281–283.
- Meklat A, Sabaou N, Zitouni A, Mathieu F, Lebrihi A. 2011. Isolation, taxonomy, and antagonistic properties of halophilic actinomycetes in Saharan soils of Algeria. Appl Environ Microbiol. 77(18):6710–6714.
- Meknaci R, Lopes P, Servy C, Le Caer J-P, Andrieu J-P, Hacène H, Ouazzani J. 2014. Agar-supported cultivation of Halorubrum sp. SSR, and production of halocin C8 on the scale-up prototype Platotex. Extremophiles. 18(6):1049–1055.
- Meseguer I, Rodriguez-Valera F. 1985. Production and purification of halocin H4. FEMS Microbiol. Lett. 28(2):177–182.
- Meseguer I, Rodriguez-Valera F. 1986. Effect of halocin H4 on cells of Halobacterium halobium. J Gen Microbiol. 132(11):3061–3068.
- Meseguer I, Torreblanca M, Konishi T. 1995. Specific inhibition of the halobacterial Na+/H + antiporter by halocin H6. J Biol Chem. 270(12):6450–6455.
- Mohammadipanah F, Wink J. 2016. Actinobacteria from arid and desert habitats: diversity and biological activity. Front Microbiol. 6:1541.
- Newman DJ, Cragg GM. 2012. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 75(3):311–335.
- Newman DJ, Cragg GM. 2016. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 79(3):629–661.
- Newman DJ, Cragg GM, Snader KM. 2003. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. 66(7):1022–1037. [doi].
- NIH. 2003. Research on microbial biofilms: PA-03-047. J Investig Med. 51:162–163.
- Nikolouli K, Mossialos D. 2012. Bioactive compounds synthesized by non-ribosomal peptide synthetases and type-I polyketide synthases discovered through genome-mining and metagenomics. Biotechnol Lett. 34(8):1393–1403.
- O’Connor E. 2002. Purification and characterization of microhalocin R1 from Halobacterium salinarum GN101 [Dosctoral dissertation]. Flagstaff (AZ): Northern Arizona University.
- O’Connor EM, Shand RF. 2002. Halocins and sulfolobicins: the emerging story of archaeal protein and peptide antibiotics. J Ind Microbiol Biotech. 28(1):23–31.
- Oh DC, Williams PG, Kauffman CA, Jensen PR, Fenical W. 2006. Cyanosporasides A and B, chloro- and cyano-cyclopenta[a]indene glycosides from the marine actinomycete “Salinispora pacifica”. Org Lett. 8(6):1021–1024.
- Ohta S, Ono F, Shiomi Y, Nakao T, Aozasa O, Nagate T, Kitamura K, Yamaguchi S, Nishi M, Miyata H. 1998. Anti-herpes simplex virus substances produced by the marine green alga, Dunaliella primolecta. J Appl Phycol. 10(4):349–356.
- Okami Y, Okazaki T, Kitahara T, Umezawa H. 1976. Studies on marine microorganisms. V. A new antibiotic, aplasmomycin, produced by a streptomycete isolated from shallow sea mud. J Antibiot. 29(10):1019–1025.
- Okoro CK, Brown R, Jones AL, Andrews BA, Asenjo JA, Goodfellow M, Bull AT. 2009. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek. 95(2):121–133.
- Oren A. 2002. Diversity of halophilic microorganisms: environments, phylogeny, physiology, and applications. J Ind Microbiol Biotechnol. 28(1):56–63.
- Oren A. 2006. Adaptation of halophilic archaea to life at high salt concentrations. In: Läuchli A, Lüttge U, editors. Salinity: environment – plants – molecules. Dordrecht: Kluwer Academic Publishers; p. 81–96.
- Oren A. 2008. Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst. 4:2.
- Owens B. 2017. Solithromycin rejection chills antibiotic sector. Nat Biotechnol. 35(3):187–188.
- Papagianni M. 2012. Recent advances in engineering the central carbon metabolism of industrially important bacteria. Microb Cell Fact. 11:50.
- Pašić L, Velikonja BH, Ulrih NP. 2008. Optimization of the culture conditions for the production of a bacteriocin from halophilic archaeon Sech7a. Prep Biochem Biotechnol. 38(3):229–245.
- Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 6(1):29–40.
- Pettit RK. 2011. Culturability and secondary metabolite diversity of extreme microbes: expanding contribution of deep sea and deep-sea vent microbes to natural product discovery. Mar Biotechnol. 13(1):1–11.
- Pinchuk IV, Bressollier P, Sorokulova IB, Verneuil B, Urdaci MC. 2002. Amicoumacin antibiotic production and genetic diversity of Bacillus subtilis strains isolated from different habitats. Res Microbiol. 153(5):269–276.
- Platas G, Meseguer I, Amils R. 1996. Optimization of the production of a bacteriocin from Haloferax mediterranei Xia3. Microbiologia. 12(1):75–84.
- Plemenitaš A, Lenassi M, Konte T, Kejžar A, Zajc J, Gostinčar C, Gunde-Cimerman N. 2014. Adaptation to high salt concentrations in halotolerant/halophilic fungi: a molecular perspective. Front Microbiol. 5:199.
- Plemenitaš A, Vaupotič T, Lenassi M, Kogej T, Gunde-Cimerman N. 2008. Adaptation of extremely halotolerant black yeast Hortaea werneckii to increased osmolarity: a molecular perspective at a glance. Stud Mycol. 61:67–75.
- Potera C. 1999. Forging a link between biofilms and disease. Science. 283(5409):1837–1839.
- Pramanik A, Sundararaman M, Das S, Ghosh U, Mukherjee J. 2011. Isolation and characterization of cyanobacteria possessing antimicrobial activity from the Sundarbans, the World’s largest tidal mangrove forest. J Phycol. 47(4):731–743.
- Prangishvili D, Holz I, Stieger E, Nickell S, Kristjansson JK, Zillig W. 2000. Sulfolobicins, specific proteinaceous toxins produced by strains of the extremely thermophilic archaeal genus Sulfolobus. J Bacteriol. 182(10):2985–2988.
- Price LB, Shand RF. 2000. Halocin S8: a 36-amino-acid microhalocin from the haloarchaeal strain S8a. J Bacteriol. 182(17):4951–4958.
- Projan SJ. 2003. Why is big pharma getting out of antibacterial drug discovery? Curr Opin Microbiol. 6(5):427–430.
- Projan SJ, Shlaes DM. 2004. Antibacterial drug discovery: is it all downhill from here? Clin Microbiol Infect. 10(Suppl 4):18–22.
- Qi SH, Xu Y, Gao J, Qian PY, Zhang S. 2009. Antibacterial and antilarval compounds from marine bacterium Pseudomonas rhizosphaerae. Ann Microbiol. 59(2):229–233.
- Quadri I, Hassani II, l‘Haridon S, Chalopin M, Hacène H, Jebbar M. 2016. Characterization and antimicrobial potential of extremely halophilic archaea isolated from hypersaline environments of the Algerian Sahara. Microbiol Res. 186-187:119–131.
- Raju TN. 2000. The Nobel chronicles. 1988: James Whyte Black, (b 1924), Gertrude Elion (1918–99), and George H Hitchings (1905–98). Lancet. 355(9208):1022.
- Rateb ME, Houssen WE, Arnold M, Abdelrahman MH, Deng H, Harrison WTA, Okoro CK, Asenjo JA, Andrews BA, Ferguson G, et al. 2011. Chaxamycins A–D, bioactive ansamycins from a hyper-arid desert Streptomyces sp. J Nat Prod. 74(6):1491–1499.
- Rateb ME, Houssen WE, Harrison WTA, Deng H, Okoro CK, Asenjo JA, Andrews BA, Bull AT, Goodfellow M, Ebel R, et al. 2011. Diverse metabolic profiles of a Streptomyces strain isolated from a hyper-arid environment. J Nat Prod. 74(9):1965–1971.
- Rdest U, Sturm M. 1987. Bacteriocins from halobacteria. In: Burgess R, editor. Protein purification: micro to macro. New York (NY): Alan R. Liss Inc; p. 271–278.
- Rodriguez-Valera F. 1993. Introduction to saline environments. In: Vreeland RH, Hochstein LI, editors. The Biology Halophilic Bacteria. Boca Raton (FL): CRC Press; p. 1–23.
- Rodriguez-Valera F, Juez G, Kushner DJ. 1982. Halocins: salt-dependent bacteriocins produced by extremely halophilic rods. Can J Microbiol. 28(1):151–154.
- Roshan K, Tarafdar A, Saurav K, Ali S, Lone SA, Pattnaik S, et al. 2013. Isolation and screening of bioactive compound from actinomycetes isolated from salt pan of Marakanam district of the state Tamil Nadu, India. Elixir Int J:61:16826–16831.
- Rothschild LJ, Mancinelli RL. 2001. Life in extreme environments. Nature. 409(6823):1092–1101.
- Sarkar S, Saha M, Roy D, Jaisankar P, Das S, Gauri Roy L, Gachhui R, Sen T, Mukherjee J. 2008. Enhanced production of antimicrobial compounds by three salt-tolerant actinobacterial strains isolated from the Sundarbans in a niche-mimic bioreactor. Mar Biotechnol. 10(5):518–526.
- Sawale A, Kadam TA, Karale MA, Kadam OA. 2014. Antimicrobial Activity of Secondary Metabolites from Halophilic Bacillus pumilus sp. Int J Curr Microbiol Applied Sci. 3:506–512.
- Schäberle TF, Goralski E, Neu E, Erol O, Hölzl G, Dörmann P, Bierbaum G, König GM. 2010. Marine myxobacteria as a source of antibiotics—comparison of physiology, polyketide-type genes and antibiotic production of three new isolates of Enhygromyxa salina. Mar Drugs. 8(9):2466–2479.,.
- Schatz A, Bugle E, Waksman SA. 1944. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Proc Soc Exp Biol Med. 55(1):66–69.
- Scheffler RJ, Colmer S, Tynan H, Demain AL, Gullo VP. 2013. Antimicrobials, drug discovery, and genome mining. Appl Microbiol Biotechnol. 97(3):969–978.
- Scherlach K, Hertweck C. 2006. Discovery of aspoquinolones A–D, prenylated quinoline-2-one alkaloids from Aspergillus nidulans, motivated by genome mining. Org Biomol Chem. 4(18):3517–3520.
- Schulz D, Beese P, Ohlendorf B, Erhard A, Zinecker H, Dorador C, Imhoff JF. 2011. Abenquines A–D: aminoquinone derivatives produced by Streptomyces sp. strain DB634. J Antibiot. 64(12):763–768.
- Sengupta S, Pramanik A, Ghosh A, Bhattacharyya M. 2015. Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiol. 15:170. .
- Sepcic K, Zalar P, Gunde-Cimerman N. 2010. Low water activity induces the production of bioactive metabolites in halophilic and halotolerant fungi. Mar Drugs. 9(1):43–58.
- Shand R, Price O’Connor O. 1999. Halocins: protein antibiotics from hypersaline environments. In: Oren A, editor. Microbiology and biogeochemistry of hypersaline environments. Boca Raton (FL): CRC Press. [accessed 2017 Mar 20]. https://books.google.co.uk/books?hl=en&lr=&id=rBk1hHU1zx8C&oi=fnd&pg=PA295&dq=halocins:+protein+antibiotics+from+hypersaline+environments.+in+Microbiology+and+biogeochemistry+of+hypersaline+environments.&ots=SKvUZkjrCk&sig=fpTwNykfaSP343Ks3eZm-JWkQPM#v=on.
- Shand RF. 2006. 29 detection, quantification and purification of halocins: peptide antibiotics from haloarchaeal extremophiles. Methods Microbiol. 35:703–718.
- Shand RF, Leyva KJ. 2007. Peptide and protein antibiotics from the domain Archaea: halocins and sulfolobicins. In: Riley MA, Chavan MA, editors. Bacteriocins. Berlin/Heidelberg, Germany: Springer; p. 93–109.
- Shand RF, Leyva KJ. 2008. Archaeal antimicrobials: an undiscovered country. In: Blum P, editor. Archaea: new models for prokaryotic biology. Norfolk: Caister Academic Press.
- Shand R, Price L, O’Connor E. 1998. Halocins: protein antibiotics from hypersaline environments. In: Oren A, editor. Microbiology and biogeochemistry of hypersaline environments. Boca Raton: CRC Press; p. 295–306.
- Sharma AK, Gohel S, Singh SP. 2012. Actinobase: database on molecular diversity, phylogeny and biocatalytic potential of salt tolerant alkaliphilic actinomycetes. Bioinformation. 8(11):535–538.
- Shiomi K, Iinuma H, Naganawa H, Hamada M, Hattori S, Nakamura H, Takeuchi T, Iitaka Y. 1990. New antibiotic produced by micromonospora globosa. J Antibiot. 43(8):1000–1005.
- Simmons TL, Andrianasolo E, McPhail K, Flatt P, Gerwick WH. 2005. Marine natural products as anticancer drugs. Mol Cancer Ther. 4(2):333–342.
- Simpkin VL, Renwick MJ, Kelly R, Mossialos E. 2017. Incentivising innovation in antibiotic drug discovery and development: progress, challenges and next steps. J Antibiot. 70(12):1087–1096. .
- Singh LS, Baruah I, Bora TC. 2006. Actinomycetes of Loktak habitat: isolation and screening for antimicrobial activities. Biotechnology. 5(2):217–221.
- Soppa J, Oesterhelt D. 1989. Halobacterium sp. GRB: a species to work with!? Can J Microbiol. 35(1):205–209.
- Sorokin DY, Tourova TP, Lysenko AM, Muyzer G. 2006. Diversity of culturable halophilic sulfur-oxidizing bacteria in hypersaline habitats. Microbiology. 152(Pt 10):3013–3023.
- Stewart EJ. 2012. Growing unculturable bacteria. J Bacteriol. 194(16):4151–4160.
- Sun C, Li Y, Mei S, Lu Q, Zhou L, Xiang H. 2005. A single gene directs both production and immunity of halocin C8 in a haloarchaeal strain AS7092. Mol Microbiol. 57(2):537–549.
- Sun Y, Tomura T, Sato J, Iizuka T, Fudou R, Ojika M. 2016. Isolation and biosynthetic analysis of haliamide, a new PKS-NRPS hybrid metabolite from the marine myxobacterium Haliangium ochraceum. Molecules. 21(1):59.
- Suthindhiran K, Kannabiran K. 2009. Cytotoxic and antimicrobial potential of actinomycete species Saccharopolyspora salina VITSDK4 isolated from the Bay of Bengal Coast of India. American J of Infectious Diseases. 5(2):90–98.
- Swan BK, Ehrhardt CJ, Reifel KM, Moreno LI, Valentine DL. 2010. Archaeal and bacterial communities respond differently to environmental gradients in anoxic sediments of a California hypersaline lake, the Salton Sea. Appl Environ Microbiol. 76(3):757–768.
- Tanaka Y, Hanada S, Manome A, Tsuchida T, Kurane R, Nakamura K, Kamagata Y. 2004. Catellibacterium nectariphilum gen. nov., sp. nov., which requires a diffusible compound from a strain related to the genus Sphingomonas for vigorous growth. Int J Syst Evol Microbiol. 54(Pt 3):955–959.
- Tang K, Zhang X-H. 2014. Quorum quenching agents: resources for antivirulence therapy. Mar Drugs. 12(6):3245–3282.
- Teasdale ME, Donovan KA, Forschner-Dancause SR, Rowley DC. 2011. Gram-positive marine bacteria as a potential resource for the discovery of quorum sensing inhibitors. Mar Biotechnol. 13(4):722–732.
- Teasdale ME, Liu J, Wallace J, Akhlaghi F, Rowley DC. 2009. Secondary metabolites produced by the marine bacterium Halobacillus salinus that inhibit quorum sensing-controlled phenotypes in gram-negative bacteria. Appl Environ Microbiol. 75(3):567–572.
- Thaker MN, Wang W, Spanogiannopoulos P, Waglechner N, King AM, Medina R, Wright GD. 2013. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol. 31(10):922–927.
- Thumar JT, Dhulia K, Singh SP. 2010. Isolation and partial purification of an antimicrobial agent from halotolerant alkaliphilic Streptomyces aburaviensis strain Kut-8. World J Microbiol Biotechnol. 26(11):2081–2087.
- Tian S, Yang Y, Liu K, Xiong Z, Xu L, Zhao L. 2014. Antimicrobial metabolites from a novel halophilic actinomycete Nocardiopsis terrae YIM 90022. Nat Prod Res. 28(5):344–346.
- Tian S-Z, Pu X, Luo G, Zhao L-X, Xu L-H, Li W-J, Luo Y. 2013. Isolation and characterization of new p-Terphenyls with antifungal, antibacterial, and antioxidant activities from halophilic actinomycete Nocardiopsis gilva YIM 90087. J Agric Food Chem. 61(12):3006–3012.
- Tommonaro G, Abbamondi GR, Iodice C, Tait K, De Rosa S. 2012. Diketopiperazines produced by the halophilic archaeon, Haloterrigena hispanica, activate AHL bioreporters. Microb Ecol. 63(3):490–495.
- Torreblanca M, Meseguer I, Rodriguez-Valera F. 1989. Halocin H6, a bacteriocin from Haloferax gibbonsii. J Gen Microbiol. 135(10):2655–2661.
- Torreblanca M, Meseguer I, Ventosa A. 1994. Production of halocin is a practically universal feature of archaeal halophilic rods. Lett Appl Microbiol. 19(4):201–205.
- Tresner HD, Hayes JA, Backus EJ. 1968. Differential tolerance of streptomycetes to sodium chloride as a taxonomic aid. Appl Microbiol. 16(8):1134–1136.
- Tulp M, Bohlin L. 2005. Rediscovery of known natural compounds: nuisance or goldmine? Bioorg Med Chem. 13(17):5274–5282.
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W, Jensen PR, Moore BS. 2007. Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci U S A. 104(25):10376–10381.
- Vartoukian SR, Palmer RM, Wade WG. 2010. Cultivation of a synergistetes strain representing a previously uncultivated lineage. Environ Microbiol. 12(4):916–928.
- Vasavada SH, Thumar JT, Singh SP. 2006. Secretion of a potent antibiotic by salt-tolerant and alkaliphilic actinomycete Streptomyces sannanensis strain RJT-1. Curr Sci. 91:1393–1397.
- Velho-Pereira S, Furtado I. 2012. Antibacterial activity of halophilic bacterial bionts from marine invertebrates of Mandapam-India. Indian J Pharm Sci. 74(4):331–338.
- Velmurugan S, Raman K, Thanga Viji V, Donio MBS, Adlin Jenifer J, Babu MM, Citarasu T. 2013. Screening and characterization of antimicrobial secondary metabolites from Halomonas salifodinae MPM-TC and its in vivo antiviral influence on Indian white shrimp Fenneropenaeus indicus against WSSV challenge. J King Saud Univ Sci. 25(3):181–190.
- Von Nussbaum F, Brands M, Hinzen B, Weigand S, Häbich D. 2006. Antibacterial natural products in medicinal chemistry – Exodus or revival? Angew Chem Int Ed Engl. 45(31):5072–5129.
- Wang G, Dai S, Chen M, Wu H, Xie L, Luo X, Li X. 2010. Two diketopiperazine cyclo(PRO-PHE) isomers from marine bacteria Bacillus subtilis sp. 13-2. Chem Nat Compd. 46(4):583–585.
- Wang S, Zheng Z, Zou H, Li N, Wu M. 2019. Characterization of the secondary metabolite biosynthetic gene clusters in archaea. Comput Biol Chem. 78:165–169.
- Wang W, Zhu T, Tao H, Lu Z, Fang Y, Gu Q, Zhu W. 2007. Two new cytotoxic quinone type compounds from the halotolerant fungus Aspergillus variecolor. J Antibiot. 60(10):603–607.
- Watve MG, Tickoo R, Jog MM, Bhole BD. 2001. How many antibiotics are produced by the genus Streptomyces? Arch Microbiol. 176(5):386–390.
- Weber T. 2014. In silico tools for the analysis of antibiotic biosynthetic pathways. Int J Med Microbiol. 304(3–4):230–235.
- Wegmann K, Ben-Amotz A, Avron M. 1980. Effect of temperature on glycerol retention in the halotolerant algae dunaliella and asteromonas. Plant Physiol. 66(6):1196–1197.
- Weiss C, Figueras E, Borbely AN, Sewald N. 2017. Cryptophycins: cytotoxic cyclodepsipeptides with potential for tumor targeting. J Pept Sci. 23(7–8):514–531.
- Wilson ZE, Brimble MA. 2009. Molecules derived from the extremes of life. Nat Prod Rep. 26(1):44–71.
- Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 74(11):5088–5090.
- Xiang W, Guo J, Feng W, Huang M, Chen H, Zhao J, Zhang J, Yang Z, Sun Q. 2008. Community of extremely halophilic bacteria in historic dagong brine well in southwestern China. World J Microbiol Biotechnol. 24(10):2297–2305.
- Yoshida M, Matsubara K, Kudo T, Horikoshi K. 1991. Actinopolyspora mortivallis sp. nov., a moderately halophilic actinomycete. Int J Syst Bacteriol. 41(1):15–20.
- Zalar P, De Hoog GS, Schroers H-J, Frank JM, Gunde-Cimerman N. 2005. Taxonomy and phylogeny of the xerophilic genus Wallemia (Wallemiomycetes and Wallemiales, cl. et ord. nov.). Antonie Van Leeuwenhoek. 87(4):311–328.
- Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, et al. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 541(7637):353–358.
- Zhao L-X, Huang S-X, Tang S-K, Jiang C-L, Duan Y, Beutler JA, Henrich CJ, McMahon JB, Schmid T, Blees JS, et al. 2011. Actinopolysporins A–C and tubercidin as a Pdcd4 stabilizer from the halophilic actinomycete Actinopolyspora erythraea YIM 90600. J Nat Prod. 74(9):1990–1995. [doi].
- Zheng Y, Saitou A, Wang C-M, Toyoda A, Minakuchi Y, Sekiguchi Y, Ueda K, Takano H, Sakai Y, Abe K, et al. 2019. Genome features and secondary metabolites biosynthetic potential of the class Ktedonobacteria. Front Microbiol. 10:893.

