Abstract
Diel cycle is of enormous biological importance as it imposes daily oscillation in environmental conditions, which temporally structures most ecosystems. Organisms developed biological time-keeping mechanisms – circadian clocks – that provide a significant fitness advantage over competitors by optimising the synchronisation of their biological activities. While circadian clocks are ubiquitous in Eukaryotes, they are so far only characterised in Cyanobacteria within Prokaryotes. However, growing evidence suggests that circadian clocks are widespread in the bacterial and archaeal domains. As Prokaryotes are at the heart of crucial environmental processes and are essential to human health, unravelling their time-keeping systems provides numerous applications in medical research, environmental sciences, and biotechnology. In this review, we elaborate on how novel circadian clocks in Prokaryotes offer research and development perspectives. We compare and contrast the different circadian systems in Cyanobacteria and discuss about their evolution and taxonomic distribution. We necessarily provide an updated phylogenetic analysis of bacterial and archaeal species that harbour homologs of the main cyanobacterial clock components. Finally, we elaborate on new potential clock-controlled microorganisms that represent opportunities of ecological and industrial relevance in prokaryotic groups such as anoxygenic photosynthetic bacteria, methanogenic archaea, methanotrophs or sulphate-reducing bacteria.
GRAPHICAL ABSTRACT
Introduction
The Earth’s rotation around its axis causes the 24-h day and night cycle known as the diel cycle. This recurrent phenomenon induces daily fluctuations of solar radiation and/or temperature, which regulate the functioning of most ecosystems. One of the most significant adaptations to the diel cycle is the emergence of circadian clocks in both Eukaryotes and Prokaryotes (Saini et al. Citation2019). Circadian clocks are endogenous molecular systems that entrain to environmental zeitgeber (i.e. external cue such as light) and allow temporal coordination of intracellular processes. These clocks follow a 24-h period oscillation that remains in a constant condition (free-running) and over a range of physiological temperatures (temperature-compensated) (Golden and Canales Citation2003). Although circadian clocks are ubiquitous in Eukaryotes, prokaryotic circadian clocks were so far only characterised in Cyanobacteria (for a review see Johnson and Rust Citation2021). The cyanobacterial clock is based on the KaiABC proteins complex that acts as an oscillator that synchronises with environmental signals and subsequently controls rhythmic gene expression (Kondo Citation2007; Markson et al. Citation2013).
Unravelling the time-keeping mechanisms of non-cyanobacterial species is crucial regarding the central role Prokaryotes play in all ecosystems. This fundamental knowledge could lead to the incorporation of their temporal structures in broad technological applications, which will undoubtedly offer great research and development opportunities. Over the past decades, a growing body of evidence suggested that time-controlled mechanisms are widespread in Prokaryotes (Schmelling et al. Citation2021). Pioneer bioinformatical analyses revealed that homologs of the Kai proteins were present across a broad range of bacterial and archaeal phyla (Dvornyk et al. Citation2003; Loza-Correa et al. Citation2010; Schmelling et al. Citation2017). Microbial community-wide diel rhythms are observed in various biomes and are attributed to non-cyanobacterial species (Ottesen et al. Citation2014; Hörnlein et al. Citation2020; Géron et al. Citation2021). Moreover, rhythmic activities are found in prokaryotic species harbouring Kai protein homologs such as purple bacteria (Van Praag et al. Citation2000; Min et al. Citation2005; Ma et al. Citation2016), non-photosynthetic bacteria (Paulose et al. Citation2019; Sartor et al. Citation2019; Eelderink-Chen et al. Citation2021) and extremophile archaea (Whitehead et al. Citation2009; Maniscalco et al. Citation2014).
In this review, we discuss the numerous applications of biological rhythms of Prokaryotes in research and development from human health to the industry. We present the known circadian systems in Prokaryotes, and we describe their properties and evolutionary paths. With the development of next-generation sequencing the number of complete, draft and metagenome-assembled genomes exponentially increased since the last phylogenetic analyses of Kai protein homologs (Dvornyk et al. Citation2003; Loza-Correa et al. Citation2010; Schmelling et al. Citation2017). Therefore, we provide an updated analysis of the presence of Kai proteins among bacterial and archaeal species, and we elaborate on new potentially clock-controlled candidates in taxonomic groups such as anoxygenic photosynthetic bacteria or methanogenic archaea.
Research focus on prokaryotic time-keeping systems: rationales and impacts
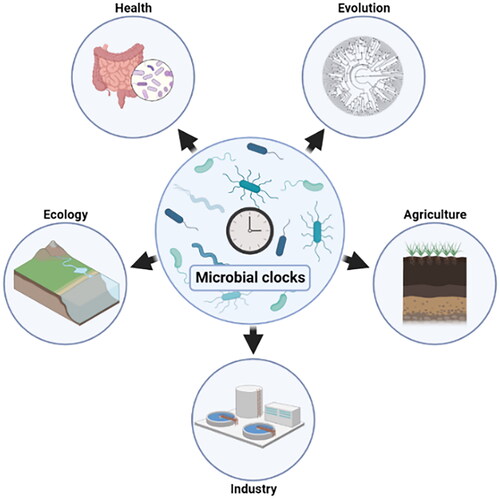
Prokaryotes are essential to humans for numerous reasons (). As a matter of fact, microorganisms outnumber human cells by tenfold and are crucial in food degradation, nutrient absorption, or defense against pathogens (Venkova et al. Citation2018). Prokaryotes also play a significant role in symbiosis with plants by providing nutrients as well as growth hormones (Backer et al. Citation2018). They also degrade organic matter and contribute to biogeochemical cycles (Madsen Citation2011). Moreover, prokaryotes are central in numerous biotechnological applications (Caplice and Fitzgerald Citation1999; Demain and Sanchez Citation2009; Akinsemolu Citation2018). In that context, research focus on the chronobiology (i.e. the study of the temporal biology of organisms) of prokaryotes in natural and artificial environments would provide important translational opportunities in the medical, ecological, and industrial fields, which could promote and maintain our health and well-being.
Figure 1. Schematic representation of chronobiology-based applications of Prokaryotes. The metabolism of humans and plants is controlled by a circadian clock that impacts their commensal microbial communities. In return, the microbiome fluctuates in terms of taxonomy, functioning and localisation, which influences their host’s physiology. A better understanding on potential time-keeping mechanisms in the microorganisms associated with plants and animals will allow more accurate models and open new perspectives on optimising these relationships. In addition, the industrial and technological fields would benefit from the integration of the natural rhythmic physiology of Prokaryotes into utilisation and manufacturing processes.

The human clock scales with environmental changes and affects the behaviour including timing and type of food intake () (Matenchuk et al. Citation2020). Consequently, the gut microbiome experiences daily oscillations leading to taxonomic, functional, and spatial variations inducing local and systemic effects on the host (Deaver et al. Citation2018; Wu et al. Citation2018; Godinho-Silva et al. Citation2019; Nobs et al. Citation2019; Saran et al. Citation2020). Although it is unknown if the gut microbiota possesses an endogenous circadian clock, there is growing evidence supporting the interconnection between the host’s and the microbiota’s circadian rhythms, their impact on host physiology, and their association with diseases such as diabetes (Thaiss et al. Citation2014; Kaczmarek et al. Citation2017; Kim et al. 2019; Choi et al. Citation2021; Heinemann et al. Citation2021). Furthermore, circadian rhythms of host immunity are interlinked to bacterial and viral infections (Curtis et al. Citation2014; Pearson et al. Citation2021). With the prevalence of antibiotic-resistance bacteria threatening global health, food security, and development, chronotherapy (i.e. time-specific therapy) capitalising on the circadian rhythms of both the host and the infectious agents could lead to a more efficient treatment strategy (Pearson et al. Citation2021). Similarly, plant’s physiology is regulated by the circadian clock, which likely generates a rhythmic environment for the root-associated microorganisms (i.e. the rhizobacteria). The rhizobacteria feed on root exudates and, in exchange, provide great benefits such as plant growth promotion, nutrient accessibility (e.g. nitrogen fixation), or protection from predators and phytopathogens () (Gould et al. Citation2018; Hubbard et al. Citation2018; Lu et al. Citation2021). In that context, the root-colonizing bacteria could use circadian programs to coordinate their metabolism with rhythms of the host to maximise the advantage of the bidirectional interaction (Asif et al. Citation2019). Plant growth-promoting rhizobacteria are of high economic importance and are commercialised to improve biomass production and quality of numerous crops (Saharan and Nehra Citation2011). Therefore, deciphering the time-controlled interaction between rhizobacteria and plants could be a promising source of valuable discoveries that can have immediate applications in the sustainable agriculture field (Maddur Puttaswamy Citation2019).
Chronobiology research focussing on anoxygenic photosynthetic organisms (purple and green (non)-sulphur bacteria), sulphate-reducing bacteria, methanogenic Archaea, or methanotrophs also represent interesting applications for both fields of ecology and microbiology-based industries (). Anoxygenic photosynthetic bacteria are often encountered in microbial mats where they interact with Cyanobacteria that possess a well-defined circadian clock (Prieto-Barajas et al. Citation2018). As they can use light as an energy source, anoxygenic photosynthetic bacteria would greatly benefit from a time-keeping mechanism that would allow them to synchronise with a both biotic and abiotic external signals oscillating daily (Hörnlein et al. Citation2020). Because of microbial mat communities displaying a highly versatile metabolism that allows them to use a broad range of substrate, there is considerable interest in industrial uses of mats, especially for water treatment and for cleaning up pollution (Abed et al. Citation2020; George et al. Citation2020). Deciphering the response to diel changes in methanogenic archaeal, methanotrophic bacterial and sulphate reducing bacterial communities is crucial as they are deeply involved in methane and sulphur cycles. Methane is the second most important greenhouse gas on Earth and methanogenic archaea produce about 70% through the methanogenesis process (Conrad Citation2009). Therefore, time-specific models of these microorganisms in their natural (e.g. microbial mats) or artificial environments (e.g. wastewater treatment plants) could provide insights to better understand their ecological functions and allow optimisation of resource management (Wang et al. Citation2017; Enzmann et al. Citation2018). Finally, Prokaryotes are widely used in biocompounds production, bioremediation, or treatment of organic and industrial wastes () (Abdel-Aziz et al. Citation2017; Alizadeh-Sani et al. Citation2018; Kallscheuer et al. Citation2019). More insights into their rhythmic physiologies would allow the integration of their temporal programs into industrial processes, which could induce metabolic adjustments and lead to improved qualitative and quantitative production (Sartor et al. Citation2019).
The circadian systems in Prokaryotes: the clock versus the hourglass
Prior to mid-1980, Prokaryotes were thought to have neither the resource nor the need to possess circadian systems. As many bacteria can divide several times over a 24h-cycle, it was believed that cellular functions would not be coupled to a biological clock. This dogma, known as the “circadian-infradian” rule, was refuted in 1986 when Huang and colleagues discovered circadian rhythmicity of nitrogen fixation and amino acid uptake in the cyanobacteria Synechococcus sp. RF-1 (Huang et al. Citation1990; Chen et al. Citation1991). Subsequently, Synechococcus elongatus PCC 7942 emerged as the model organism for studying the mechanism of the clock using luciferase reporter assay (Kondo et al. Citation1993). The cyanobacterial circadian clock uses a three-protein KaiABC system that forms a core oscillator that determines diel patterns of gene expression involved in cell division, and metabolic switches () (Mori et al. Citation1996; Dong et al. Citation2010; Pattanayek et al. 2014; Puszynska and O’Shea Citation2017). In contrast to the circadian clockwork in Eukaryotes, which is based on a series of interlocking transcriptional-translational feedback loops, the cyanobacterial circadian clock is simpler and mainly relies on the KaiC phosphorylation cycle ().
Figure 2. Circadian clocks in Cyanobacteria. (a) Schematic representation of the two circadian clock systems in the Synechococcus elongatus PCC 7942 and Prochlorococcus marinus MED4 models. The phosphorylation/dephosphorylation cycle of KaiC is represented with the different key components of the output pathway that subsequently control the gene expression. The proteins KaiA and CikA, labelled in grey, are missing from the Prochlorococcus clock model. (b) Difference between the stability of oscillation provided by a free-running clock in S. elongatus and an hourglass-like timer in P. marinus in the absence of light/dark cycle.

Throughout the course of day and night, KaiC, the central clock component, synchronises with environmental signals and undergoes a phosphorylation and dephosphorylation cycle () (Nishiwaki et al. Citation2000, Citation2004; Nishiwaki and Kondo Citation2012). At dawn, unphosphorylated KaiC has a loose and unstacked structure, exposing A-Loops where KaiA can bind as the morning progresses (Kim et al. Citation2008). Throughout the day, KaiA stimulates KaiC autokinase activity and thus, threonine and serine become phosphorylated. By dusk, both threonine and serine residues of KaiC are fully phosphorylated and KaiC becomes stiff and stacked, which hides the binding site for KaiA and exposes a B-Loop that constitutes a binding site for KaiB. KaiB undergoes a fold-switching between its free versus KaiC-bound forms (). In its free state, KaiB oligomerizes to constitute a tetramer, while in its KaiC-bound form, KaiB is in a fold-switched state which results in major structural reorganisation (Chang et al. Citation2015). As the night progresses, KaiA can no longer bind to KaiC and get sequestered by KaiB, subsequently activating the KaiC autophosphatase (Kitayama et al. Citation2003; Chang et al. Citation2012; Tseng et al. Citation2014). The dephosphorylated threonine and serine residues induce KaiC to return to its unphosphorylated state with exposed A-loops (Nishiwaki and Kondo Citation2012). The Kai oscillator interacts with circadian output components that subsequently controls physiological processes, such as photosynthesis, glycogen metabolism, and cell division. In S. elongatus, these output components include the histidine kinase SasA, its cognate response regulator RpaA, and the phosphatase CikA (Iwasaki et al. Citation2000; Takai et al. Citation2006). During the day, SasA binds to the phosphorylated KaiC protein, induces its autophosphorylation and transfers its phosphate group to RpaA, which induces phosphorylated RpaA to accumulate (Iwasaki et al. Citation2000; Takai et al. Citation2006). At dusk, KaiB competes with SasA for binding with KaiC and, as the night progresses, KaiB sequesters KaiA and attracts CikA (Tseng et al. Citation2014). CikA binds to the KaiBC, thus forming a complex that acts as a phosphatase that dephosphorylates RpaA (Chang et al. Citation2015). By the end of the night, the RpaA phosphorylation level is very low. Therefore, the interaction of SasA and CikA with the Kai oscillator creates a phosphorylation cycle of RpaA that peaks at dusk (Markson et al. Citation2013). In its phosphorylated form, RpaA acts as a transcription factor that activates the transcription of KaiBC genes and Class I genes and represses Class II genes (Markson et al. Citation2013). On the other hand, non-phosphorylated RpaA activates Class II genes and represses Class I genes. RpaB inhibits RpaA phosphorylation and represses KaiBC and Class I genes (Kappell and Waasbergen Citation2007; Espinosa et al. Citation2015). Remarkably, self-sustainable temperature-compensated circadian oscillations of KaiC phosphorylation can be reconstituted in vitro by supplying ATP to the three purified Kai proteins in a test tube (Nakajima et al. Citation2005). Although, KaiC phosphorylation cycle persists in absence of transcription and translation, the transcription and translation feedback loop increases the robustness of the cyanobacterial clock (Johnson et al. Citation2008).
Interestingly, the KaiABC clock mechanism of S. elongatus PCC 7942 is not generalised in Cyanobacteria. Indeed, the globally distributed marine Prochlorococcus harbours a simplified clock mechanism that lacks KaiA and where KaiC compensates with an enhanced autophosphorylation activity () (Axmann et al. Citation2009). In addition, Prochlorococcus marinus MED4 is missing the output component CikA. The clock depends on the KaiBC complex formation, which allows SasA to bind with KaiB and induces RpaA phosphorylation (Axmann et al. Citation2009). Phosphorylated RpaA binds to the KaiBC promoter and regulates its transcription, which controls the clock expression. Although the reduced KaiBC-system in Prochlorococcus provides genome-wide expression rhythms (Zinser et al. Citation2009), it differs from the circadian clock of S. elongatus in that it cannot maintain oscillation in the absence of external signals (Holtzendorff et al. Citation2008). Therefore, the Prochlorococcus clock rather works as an hourglass, requiring daily signals to reset the cycle (Mullineaux and Stanewsky Citation2009). Prochlorococcus could benefit from the reduced KaiBC-system in its habitat – the near-equatorial oceans – where a free-running clock may not be essential because of the high regularity of diel conditions (Axmann et al. Citation2009). In contrast, Synechococcus elongatus is found at higher longitude, where the seasonality and environment can often disturb the regularity of the signals which could make a clock system with stable rhythm indispensable. This hypothesis is reinforced by recent numerical simulations using a stochastic modelling approach that showed that an hourglass-like system outperforms a free-running clock in small organisms evolving in stable environment, which is the case of Prochlorococcus that is smaller by an order of magnitude than Synechococcus (Chew et al. Citation2018). The evolution of clock systems in Cyanobacteria could expand our perspective away from the specific free-running clock system of Synechococcus elongatus and lead us to focus more on environmentally driven time-keeping systems in Prokaryotes (Chew et al. Citation2018).
Evolution and distribution of the Kai proteins
Computational analyses helped to unravel the origins and the evolutionary paths of the Kai proteins within Prokaryotes (Dvornyk et al. Citation2003; Loza-Correa et al. Citation2010; Schmelling et al. Citation2017). Using the Kai protein sequences from Synechococcus elongatus PCC 7942 as a template for sequence alignment-based discovery, bioinformatical analyses revealed that the Kai proteins are not restricted to Cyanobacteria but instead are rather widespread in Prokaryotes. From an evolutionary perspective, KaiC is the oldest Kai protein, and its homologs are found in both archaea and bacteria (). Dvornyk and co-workers suggested that the predecessor of KaiC (pKaiC) was present in the Last Universal Common Ancestor (LUCA) about 3800 million years ago (Mya) (Dvornyk et al. Citation2003). During the next 300 Mya, pKaiC duplicated and subsequently fusion to form a double-domain structure that is essential for the functioning of the circadian oscillation. The origin of KaiB is estimated between ∼ 3500 and 2320 Mya, which coincides to the beginning of “the age of Cyanobacteria.” When Cyanobacteria started to produce oxygen and thus replaced the reducing geochemical environment on Earth, biological clocks could have stated significant adaptative advantages as they provide anticipative control on a wide variety of vital metabolic cycles (Dvornyk et al. Citation2003). KaiA, however, the youngest evolutionarily component of the free-running cyanobacterial pacemaker, occurs in Cyanobacteria about 1000 Mya.
Figure 3. Schematic view of the distribution of Kai protein homologs in Prokaryotes. (a) Kai evolutionary events were adapted from Dvornyk et al. (Citation2003). (b) Tree branches and Kai evolutionary paths are not on scale and do not reflect the evolution of kai genes in Prokaryotes but illustrate the distribution of current Kai protein homologs carriers in Archaea and Bacteria. The number of unique species carrying KaiC homologs alone or with KaiB and/or KaiA are represented for archaea, and bacteria. For clarity purpose, bacterial phyla were not display with the exception of Cyanobacteria.

In this review, we used a bioinformatic approach to search for all the prokaryotic KaiA, KaiB and KaiC protein sequences and update the list of archaeal and bacterial species harbouring Kai homologs. In combination with the increasing number of sequence data, reaching more than 200,000 bacterial and archaeal complete or draft genomes in 2020 (Zhang et al. Citation2020), we identified Kai protein homologs in 186 archaeal and 2824 bacterial species/strains among the protein sequences available on UniProt in December 2020 (). Briefly, we retrieved all known KaiA, KaiB and KaiC protein sequences from UniProt, which resulted in 114, 695 and 1531 protein sequences, respectively. Then, we performed sequence clustering with CD-HIT v. 4.8.1 (Fu et al. Citation2012) for KaiA, KaiB and KaiC sequences (cutoff = 70%) obtaining 31, 105, and 420 protein sequence clusters, respectively. We determined the most representative sequences for all clusters using Diamond v. 2.0.4 (Buchfink et al. Citation2015) (coverage > 80%, e-value < 1e-5) and subsequently queried them against the entire UniProt database. We obtained 375, 166,266 and 90,852 sequences. After the removal of duplicate sequences, an InterProScan Pfam search v. 5.47–82.0 was performed on all non-redundant KaiA, KaiB and KaiC protein homolog sequences. Only the protein sequences containing the kai functional protein family patterns (KaiA: PF07688, KaiB: PF07689, KaiC: PF06745) and registered as a circadian clock component (protein name) were kept for downstream analysis. This resulted in 374 KaiA, 1,127 KaiB and 4,282 KaiC protein homolog sequences, distributed across 374, 1,127, and 2,825 unique species, respectively.
Table 1. Taxonomic distribution of KaiA, KaiB, and KaiC protein homologs in Archaea and Bacteria at phylum level.
Although Kai protein homologs are present in a broad diversity of prokaryotic phyla, they are not distributed equally (). KaiC, the most essential and the oldest of the three Kai proteins is found in its functional double-domain version in 184 and 2621 archaea and bacteria species, respectively. KaiB is observed in 24 archaeal and 1103 bacterial species. KaiA is almost exclusively present in Cyanobacteria (373 species), and in one representative of Planctomycetes and Acidobacteria. Interestingly, KaiBC is represented in 22 and 677 species of archaea and bacteria, respectively (). In Archaea, KaiBC carriers are almost exclusively Euryarchaeota, while in bacteria, they are distributed across 21 phyla which included both photosynthetic and non-photosynthetic microorganisms. Given the fact that a functional KaiBC timer exist in Prochlorococcus, it could be speculated that similar time-keeping mechanisms may be widespread in Prokaryotes. While KaiABC was thought to be exclusively present in Cyanobacteria, the fact that KaiA homologs were detected in Planctomycetes and Actinobacteria worth further analyses. Sequence similarity analysis using Clustal 2.1 revealed that both sequences share almost 70% of similarity but only about 40% with the S. elongatus version of KaiA. These findings suggest that new KaiABC-like clock oscillator might exist beyond Cyanobacteria.
Towards the discovery of new prokaryotic clock systems
Over the past decades, studies revealed diel rhythmic activities beyond the well-studied Cyanobacteria among other prokaryotes such as extremophilic archaea, anoxygenic photosynthetic and heterotrophic bacteria. Here, we summarise the key findings and, based on our updated bioinformatical analyses of the screening of the Kai proteins, we open new perspectives on prokaryotic time-keeping mechanisms. These candidates include anoxygenic photosynthetic bacteria, methanotrophic bacteria, methanogen archaea, sulphate-reducing bacteria, and diverse plant- and animal-associated bacteria ().
Figure 4. Taxonomic distribution of KaiBC carriers at family level: (a) anoxygenic photosynthetic green and purple bacteria (S: sulphur bacteria, NS: non-sulphur bacteria), (b) methanotrophic bacteria and methanotroph archaea, (c) sulphate reducing bacteria, (d) plant-growth promoting bacteria (PGPRs). The numbers represent the unique species where KaiB and KaiC homologs were observed.

We identify a total of 69 species of anoxygenic photosynthetic bacteria that possess KaiBC homolog proteins (). Community-wide diel rhythms in anoxygenic photosynthetic bacterial communities were reported in numerous environmental studies and showed diel patterns in metabolic activity, diversity, or spatio-temporal distribution (Van Gemerden et al. Citation1985; Garcia-Pichel et al. Citation1994; Fourçans et al. Citation2006; Fecskeová et al. Citation2019; Piwosz et al. Citation2020). Though little experimental evidence at species level is available, diel rhythmic activity was observed in three anoxygenic photosynthetic purple bacteria: Rhodobacter sphaeroides (Min et al. Citation2005), Rhodospirillum rubrum (Van Praag et al. Citation2000), and Rhodopseudomonas palustris (Ma et al. Citation2016). In a 24 h-cyclic environment, Rhodopseudomonas exhibits KaiC-dependent fitness enhancement in rhythmic environments but not under constant conditions (Ma et al. Citation2016). Although the rhythms evidenced in these purple bacteria do not strictly meet all the criteria of a bona fide circadian clock, it was suggested that Rhodopseudomonas palustris could harbour an ancestral endogenous circadian program, based on a KaiBC oscillator, that would include some of the canonical properties of the circadian clocks (Ma et al. Citation2016). We suggest further investigation based on our bioinformatic analysis in the purple-sulphur bacteria Chromatiaceae, the purple non-sulphur bacteria Comamonadaceae, and the green non-sulphur phototrophic Chloroflexi families, as they include several species harbouring the KaiBC protein homologs and were previously associated with diel dynamics in their natural environment (Van Gemerden et al. Citation1985; Klatt et al. Citation2013; Shahraki et al. Citation2021).
The circadian cycle was previously investigated in extremophilic archaea and revealed rhythmic activity and a potential clock mechanism based on the Kai proteins. For instance, the halophilic genus Halobacterium exhibit light-dark entrained daily transcription but not sustained oscillations under constant conditions (Whitehead et al. Citation2009). It was hypothesised that simpler timing system solely driven by KaiC could exist in the halophilic Haloferax volcanii (Maniscalco et al. Citation2014). Moreover, phosphorylation assays with KaiC homologs of the hyperthermophilic Thermococcus litoralis and Pyrococcus horikoshii have shown a conserved kinase activity (Schmelling et al. Citation2017). Here, we significantly broadened the list of potential clock-controlled archaea as we evidenced KaiBC homolog proteins in 22 species of methanogen archaea and two capable of denitrifying anaerobic methane oxidation (DAMO) (). We also observed eighteen sulphate-reducing bacteria species/strains carrying KaiBC homolog proteins (). Methanogenic archaea and sulphate reducing bacteria are worthy of attention as they were previously associated with diel rhythmic activity in their natural environment, where they play an important role in methane and sulphur cycles (Jørgensen Citation1994; Steppe and Paerl Citation2002; Fourçans et al. Citation2006; Louyakis et al. Citation2017). An environmental metatranscriptomic study revealed that methanogenesis transcripts from methylotrophic methanogen archaea exhibited diel oscillations in thrombolite (Louyakis et al. Citation2017). In addition, diurnal oscillations of methane production were reported in diverse wetlands such as fens, mire, or peat bog where they are often associated with sulphate reducing bacteria (Mikkelä et al. Citation1995; Henneberger et al. Citation2017).
Deciphering how prokaryotes and eukaryotes are temporally embedded is crucial in the transition towards more holistic and sustainable medical and agricultural models. In this review, we evidenced 29 plant growth-promoting rhizobacteria that harbour KaiBC protein homologs (). These bacteria were mainly represented by the genera Bradyrhizobium and Rhizobium. Both are essential to leguminous plants as they form nodules on their root hair and perform nitrogen fixation, which further promote plant development (Sessitsch et al. Citation2002). The circadian clock was shown to alter up to 30% of the plant transcriptome including diel carbon fluxes (Michael et al. Citation2008). Consequently, the plant diel rhythmicity influences the rhizosphere community structure and function (Staley et al. Citation2017; Hubbard et al. Citation2018). Unravelling how rhizobacteria anticipate diel variations promoted by plant physiology would be of agroecological interest as the plant fitness and biomass strongly rely on optimal plant/rhizobacteria bidirectional interactions (Pérez-Jaramillo et al. Citation2016; Hubbard et al. Citation2018). Although there is so far no evidence of rhythmic activity of rhizobacteria, the soil bacteria Bacillus subtilis was shown to display temperature-compensated free-running ∼ 24h oscillations upon release to constant dark and temperature conditions (Eelderink-Chen et al. Citation2021). Interestingly, this bacterial species does not harbour any Kai protein homologs. Similarly, the gastrointestinal bacterium Klebsiella aerogenes displayed an endogenously generated, temperature-compensated circadian rhythm in swarming motility (Paulose et al. Citation2019). Thus, it remains essential to elucidate if the other prokaryotes associated with animal and plants displayed circadian rhythms would rely on the Kai-based system. Alternatively, it is worth noting that other molecules are strong candidates for circadian pacemaker: peroxiredoxin proteins are found in almost all organisms and have circadian rhythms of their redox state persisting even in the absence of clock rhythmicity in photosynthetic organisms such as Cyanobacteria (Edgar et al. Citation2012).
Conclusion
Life experiences diel cycles since its earliest stages and time-keeping mechanisms emerged as one of the most outstanding adaptations to these perpetual environmental fluctuations. Although only few bacterial species are known to possess an effective molecular circadian clock, evidence of widespread prokaryotic timing system accumulates. These new clocks might share the canonical properties of circadian clock or rather work differently depending on the evolutionary path, the internal properties of the microorganisms, or the specificities of their environment. Prokaryotes are often embedded in complex communities and mutually dependent on well-defined clock-controlled organisms. Therefore, future chronobiology studies should carefully design their experiments when using new clock candidates as growth conditions likely shape the time-keeping mechanisms.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abdel-Aziz SM, Elsoud MMA, Anise AA. 2017. Microbial biosynthesis: a repertory of vital natural products. In: Food biosynthesis: handbook of food bioengineering. London (UK): Academic Press. p. 25–54.
- Abed RM, Shanti M, Muthukrishnan T, Al-Riyami Z, Pracejus B, Moraetis D. 2020. The role of microbial mats in the removal of hexavalent chromium and associated shifts in their bacterial community composition. Front Microbiol. 11:12.
- Akinsemolu AA. 2018. The role of microorganisms in achieving the sustainable development goals. J Cleaner Prod. 182:139–155.
- Alizadeh-Sani M, Hamishehkar H, Khezerlou A, Azizi-Lalabadi M, Azadi Y, Nattagh-Eshtivani E, Fasihi M, Ghavami A, Aynehchi A, Ehsani A. 2018. Bioemulsifiers derived from microorganisms: applications in the drug and food industry. Adv Pharm Bull. 8(2):191–199.
- Asif M, Pervez A, Ahmad R. 2019. Role of melatonin and plant-growth-promoting rhizobacteria in the growth and development of plants. Clean – Soil Air Water. 47(6):1800459.
- Axmann IM, Dühring U, Seeliger L, Arnold A, Vanselow JT, Kramer A, Wilde A. 2009. Biochemical evidence for a timing mechanism in Prochlorococcus. J Bacteriol. 191(17):5342–5347.
- Backer R, Rokem JS, Ilangumaran G, Lamont J, Praslickova D, Ricci E, Subramanian S, Smith DL. 2018. Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front Plant Sci. 9:1473.
- Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 12(1):59–60.
- Caplice E, Fitzgerald GF. 1999. Food fermentations: role of microorganisms in food production and preservation. Int J Food Microbiol. 50(1–2):131–149.
- Chang Y-G, Cohen SE, Phong C, Myers WK, Kim Y-I, Tseng R, Lin J, Zhang L, Boyd JS, Lee Y. 2015. A protein fold switch joins the circadian oscillator to clock output in cyanobacteria. Science. 349(6245):324–328.
- Chang YG, Tseng R, Kuo NW, LiWang A. 2012. Rhythmic ring–ring stacking drives the circadian oscillator clockwise. Proc Natl Acad Sci U S A. 109(42):16847–16851.
- Chen TH, Chen TL, Hung LM, Huang TC. 1991. Circadian rhythm in amino acid uptake by Synechococcus RF-1. Plant Physiol. 97(1):55–59.
- Chew J, Leypunskiy E, Lin J, Murugan A, Rust MJ. 2018. High protein copy number is required to suppress stochasticity in the cyanobacterial circadian clock. Nat Commun. 9(1):1–10.
- Choi H, Rao MC, Chang EB. 2021. Gut microbiota as a transducer of dietary cues to regulate host circadian rhythms and metabolism. Nat Rev Gastroenterol Hepatol. 18(10):679–689.
- Conrad R. 2009. The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep. 1(5):285–292.
- Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LAJ. 2014. Circadian clock proteins and immunity. Immunity. 40(2):178–186.
- Deaver JA, Eum SY, Toborek M. 2018. Circadian disruption changes gut microbiome taxa and functional gene composition. Front Microbiol. 9:737.
- Demain AL, Sanchez S. 2009. Microbial drug discovery: 80 years of progress. J Antibiot (Tokyo)). 62(1):5–16.
- Dong G, Yang Q, Wang Q, Kim Y-I, Wood TL, Osteryoung KW, van Oudenaarden A, Golden SS. 2010. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell. 140(4):529–539.
- Dvornyk V, Vinogradova O, Nevo E. 2003. Origin and evolution of circadian clock genes in prokaryotes. Proc Natl Acad Sci U S A. 100(5):2495–2500.
- Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 485(7399):459–464.
- Eelderink-Chen Z, Bosman J, Sartor F, Dodd AN, Kovács ÁT, Merrow M. 2021. A circadian clock in a nonphotosynthetic prokaryote. Sci Adv. 7(2):eabe2086.
- Enzmann F, Mayer F, Rother M, Holtmann D. 2018. Methanogens: biochemical background and biotechnological applications. Amb Express. 8(1):1–22.
- Espinosa J, Boyd JS, Cantos R, Salinas P, Golden SS, Contreras A. 2015. Cross-talk and regulatory interactions between the essential response regulator RpaB and cyanobacterial circadian clock output. Proc Natl Acad Sci USA. 112(7):2198–2203.
- Fecskeová LK, Piwosz K, Hanusová M, Nedoma J, Znachor P, Koblížek M. 2019. Diel changes and diversity of pufM expression in freshwater communities of anoxygenic phototrophic bacteria. Sci Rep. 9(1):1–12.
- Fourçans A, Solé A, Diestra E, Ranchou-Peyruse A, Esteve I, Caumette P, Duran R. 2006. Vertical migration of phototrophic bacterial populations in a hypersaline microbial mat from Salins-de-Giraud (Camargue, France). FEMS Microbiol Ecol. 57(3):367–377.
- Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 28(23):3150–3152.
- Garcia-Pichel F, Mechling M, Castenholz RW. 1994. Diel migrations of microorganisms within a benthic, hypersaline mat community. Appl Environ Microbiol. 60(5):1500–1511.
- George DM, Vincent AS, Mackey HR. 2020. An overview of anoxygenic phototrophic bacteria and their applications in environmental biotechnology for sustainable resource recovery. Biotechnol Rep (Amst)). 28:e00563.
- Géron A, Werner J, Lebaron P, Wattiez R, Matallana-Surget S. 2021. Diel protein regulation of marine picoplanktonic communities assessed by metaproteomics. Microorganisms. 9(12):2621.
- Godinho-Silva C, Domingues RG, Rendas M, Raposo B, Ribeiro H, da Silva JA, Vieira A, Costa RM, Barbosa-Morais NL, Carvalho T. 2019. Light-entrained and brain-tuned circadian circuits regulate ILC3s and gut homeostasis. Nature. 574(7777):254–258.
- Golden SS, Canales SR. 2003. Cyanobacterial circadian clocks—timing is everything. Nat Rev Microbiol. 1(3):191–199.
- Gould PD, Domijan M, Greenwood M, Tokuda IT, Rees H, Kozma-Bognar L, Hall AJ, Locke JC. 2018. Coordination of robust single cell rhythms in the Arabidopsis circadian clock via spatial waves of gene expression. Elife. 7:e31700.
- Heinemann M, Ratiner K, Elinav E. 2021. Basic biology of rhythms and the microbiome. In: Circadian rhythms in bacteria and microbiomes. Cham: Springer. p. 317–328.
- Henneberger R, Cheema S, Folini M, Zeyer J. 2017. Diurnal patterns of greenhouse gas fluxes in a Swiss alpine fen. Wetlands. 37(2):193–204.
- Holtzendorff J, Partensky F, Mella D, Lennon JF, Hess WR, Garczarek L. 2008. Genome streamlining results in loss of robustness of the circadian clock in the marine cyanobacterium Prochlorococcus marinus PCC 9511. J Biol Rhythms. 23(3):187–199.
- Hörnlein C, Confurius-Guns V, Grego M, Stal LJ, Bolhuis H. 2020. Circadian clock-controlled gene expression in co-cultured, mat-forming cyanobacteria. Sci Rep. 10(1):1–17.
- Huang TC, Tu J, Chow TJ, Chen TH. 1990. Circadian rhythm of the prokaryote Synechococcus sp. RF-1. Plant Physiol. 92(2):531–533.
- Hubbard CJ, Brock MT, Van Diepen LT, Maignien L, Ewers BE, Weinig C. 2018. The plant circadian clock influences rhizosphere community structure and function. ISME J. 12(2):400–410.
- Iwasaki H, Williams SB, Kitayama Y, Ishiura M, Golden SS, Kondo T. 2000. A KaiC-interacting sensory histidine kinase, SasA, necessary to sustain robust circadian oscillation in cyanobacteria. Cell. 101(2):223–233.
- Johnson CH, Mori T, Xu Y. 2008. A Cyanobacterial Circadian Clockwork. Curr Biol. 18:R816–R825.
- Johnson CH, Rust MJ, editors. 2021. Circadian rhythms in bacteria and microbiomes. Cham, Switzerland: Springer.
- Jørgensen BB. 1994. Sulfate reduction and thiosulfate transformations in a cyanobacterial mat during a diel oxygen cycle. FEMS Microbiol Ecol. 13(4):303–312.
- Kaczmarek JL, Thompson SV, Holscher HD. 2017. Complex interactions of circadian rhythms, eating behaviors, and the gastrointestinal microbiota and their potential impact on health. Nutr Rev. 75(9):673–682.
- Kallscheuer N, Classen T, Drepper T, Marienhagen J. 2019. Production of plant metabolites with applications in the food industry using engineered microorganisms. Curr Opin Biotechnol. 56:7–17.
- Kappell AD, van Waasbergen LG. 2007. The response regulator RpaB binds the high light regulatory 1 sequence upstream of the high-light-inducible hliB gene from the Cyanobacterium synechocystis PCC 6803. Arch Microbiol. 187(4):337–342.
- Kim YI, Dong G, Carruthers CW, Golden SS, LiWang A. 2008. The day/night switch in KaiC, a central oscillator component of the circadian clock of cyanobacteria. Proc Natl Acad Sci U S A. 105(35):12825–12830.
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. 2003. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. Embo J. 22(9):2127–2134.
- Klatt CG, Liu Z, Ludwig M, Kühl M, Jensen SI, Bryant DA, Ward DM. 2013. Temporal metatranscriptomic patterning in phototrophic Chloroflexi inhabiting a microbial mat in a geothermal spring. Isme J. 7(9):1775–1789.
- Kondo T. 2007. A cyanobacterial circadian clock based on the Kai oscillator. In: Cold spring harbor symposia on quantitative biology. Vol. 72. Long Island, New York: Cold Spring Harbor Laboratory Press. p. 47–55.
- Kondo T, Strayer CA, Kulkarni RD, Taylor W, Ishiura M, Golden SS, Johnson CH. 1993. Circadian rhythms in prokaryotes: luciferase as a reporter of circadian gene expression in cyanobacteria. Proc Natl Acad Sci U S A. 90(12):5672–5676.
- Louyakis AS, Mobberley JM, Vitek BE, Visscher PT, Hagan PD, Reid RP, Kozdon R, Orland IJ, Valley JW, Planavsky NJ. 2017. A study of the microbial spatial heterogeneity of Bahamian thrombolites using molecular, biochemical, and stable isotope analyses. Astrobiology. 17(5):413–430.
- Loza-Correa M, Gomez-Valero L, Buchrieser C. 2010. Circadian clock proteins in prokaryotes: hidden rhythms? Front Microbiol. 1:130.
- Lu T, Zhang Z, Li Y, Zhang Q, Cui H, Sun L, Peijnenburg WJGM, Peñuelas J, Zhu L, Zhu Y-G. 2021. Does biological rhythm transmit from plants to rhizosphere microbes? Environ Microbiol. 23(11):6895–6906.
- Ma P, Mori T, Zhao C, Thiel T, Johnson CH. 2016. Evolution of KaiC-dependent timekeepers: a proto-circadian timing mechanism confers adaptive fitness in the purple bacterium Rhodopseudomonas palustris. PLoS Genet. 12(3):e1005922.
- Maddur Puttaswamy R. 2019. Circadian rhythms in plant-microbe interaction: for better performance of bioinoculants in the agricultural fields. In: Biofertilizers for sustainable agriculture and environment. Cham: Springer; p. 317–332.
- Madsen EL. 2011. Microorganisms and their roles in fundamental biogeochemical cycles. Curr Opin Biotechnol. 22(3):456–464.
- Maniscalco M, Nannen J, Sodi V, Silver G, Lowrey P, Bidle K. 2014. Light-dependent expression of four cryptic archaeal circadian gene homologs. Front Microbiol. 5:79.
- Markson JS, Piechura JR, Puszynska AM, O’Shea EK. 2013. Circadian control of global gene expression by the cyanobacterial master regulator RpaA. Cell. 155(6):1396–1408.
- Matenchuk BA, Mandhane PJ, Kozyrskyj AL. 2020. Sleep, circadian rhythm, and gut microbiota. Sleep Med Rev. 53:101340.
- Michael TP, Mockler TC, Breton G, McEntee C, Byer A, Trout JD, Hazen SP, Shen R, Priest HD, Sullivan CM. 2008. Network discovery pipeline elucidates conserved time-of-day–specific cis-regulatory modules. PLoS Genet. 4(2):e14.
- Mikkelä C, Sundh I, Svensson BH, Nilsson M. 1995. Diurnal variation in methane emission in relation to the water table, soil temperature, climate and vegetation cover in a Swedish acid mire. Biogeochemistry. 28(2):93–114.
- Min H, Guo H, Xiong J. 2005. Rhythmic gene expression in a purple photosynthetic bacterium, Rhodobacter sphaeroides. FEBS Lett. 579(3):808–812.
- Mori T, Binder B, Johnson CH. 1996. Circadian gating of cell division in cyanobacteria growing with average doubling times of less than 24 hours. Proc Natl Acad Sci U S A. 93(19):10183–10188.
- Mullineaux CW, Stanewsky R. 2009. The rolex and the hourglass: a simplified circadian clock in prochlorococcus? J Bacteriol. 191(17):5333–5335.
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. 2005. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 308(5720):414–415.
- Nishiwaki T, Kondo T. 2012. Circadian autodephosphorylation of cyanobacterial clock protein KaiC occurs via formation of ATP as intermediate. J Biol Chem. 287(22):18030–18035.
- Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. 2000. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci USA. 97(1):495–499.
- Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A. 2004. Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci Usa. 101(38):13927–13932.
- Nobs SP, Tuganbaev T, Elinav E. 2019. Microbiome diurnal rhythmicity and its impact on host physiology and disease risk. EMBO Reports. 20(4):e47129.
- Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R, Schuster SC, Scholin CA, DeLong EF. 2014. Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science. 345(6193):207–212.
- Pattanayak GK, Phong C, Rust MJ. 2014. Rhythms in energy storage control the ability of the cyanobacterial circadian clock to reset. Curr Biol. 24(16):1934–1938.
- Paulose JK, Cassone CV, Graniczkowska KB, Cassone VM. 2019. Entrainment of the circadian clock of the enteric bacterium Klebsiella aerogenes by temperature cycles. Iscience. 19:1202–1213.
- Pearson JA, Voisey AC, Boest-Bjerg K, Wong FS, Wen L. 2021. Circadian rhythm modulation of microbes during health and infection. Front Microbiol. 12:721004.
- Pérez-Jaramillo JE, Mendes R, Raaijmakers JM. 2016. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol. 90(6):635–644.
- Piwosz K, Vrdoljak A, Frenken T, González-Olalla JM, Šantić D, McKay RM, Spilling K, Guttman L, Znachor P, Mujakić I. 2020. Light and primary production shape bacterial activity and community composition of aerobic anoxygenic phototrophic bacteria in a microcosm experiment. Msphere. 5(4):e00354.
- Prieto-Barajas CM, Valencia-Cantero E, Santoyo G. 2018. Microbial mat ecosystems: structure types, functional diversity, and biotechnological application. Electron J Biotechnol. 31:48–56.
- Puszynska AM, O’Shea EK. 2017. Switching of metabolic programs in response to light availability is an essential function of the cyanobacterial circadian output pathway. eLife. 6:e23210.
- Saharan BS, Nehra V. 2011. Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res. 21(1):30.
- Saini R, Jaskolski M, Davis SJ. 2019. Circadian oscillator proteins across the kingdoms of life: structural aspects. BMC Biol. 17(1):1–39.
- Saran AR, Dave S, Zarrinpar A. 2020. Circadian rhythms in the pathogenesis and treatment of fatty liver disease. Gastroenterology. 158(7):1948–1966.e1.
- Sartor F, Eelderink-Chen Z, Aronson B, Bosman J, Hibbert LE, Dodd AN, Kovács ÁT, Merrow M. 2019. Are there circadian clocks in non-photosynthetic bacteria? Biology. 8(2):41.
- Schmelling NM, Lehmann R, Chaudhury P, Beck C, Albers SV, Axmann IM, Wiegard A. 2017. Minimal tool set for a prokaryotic circadian clock. BMC Evol Biol. 17(1):1–20.
- Schmelling NM, Scheurer N, Köbler C, Wilde A, Axmann IM. 2021. Diversity of timing systems in cyanobacteria and beyond. In: Circadian Rhythms in Bacteria and Microbiomes. Cham: Springer; p. 179–202.
- Sessitsch A, Howieson JG, Perret X, Antoun H, Martinez-Romero E. 2002. Advances in Rhizobium research. Crit Rev Plant Sci. 21(4):323–378.
- Shahraki AH, Chaganti SR, Heath DD. 2021. Diel dynamics of freshwater bacterial communities at beaches in Lake Erie and Lake St. Clair, Windsor, Ontario. Microb Ecol. 81(1):1–13.
- Staley C, Ferrieri AP, Tfaily MM, Cui Y, Chu RK, Wang P, Shaw JB, Ansong CK, Brewer H, Norbeck AD. 2017. Diurnal cycling of rhizosphere bacterial communities is associated with shifts in carbon metabolism. Microbiome. 5(1):65.
- Steppe TF, Paerl HW. 2002. Potential N2 fixation by sulfate-reducing bacteria in a marine intertidal microbial mat. Aquat Microb Ecol. 28(1):1–12.
- Takai N, Nakajima M, Oyama T, Kito R, Sugita C, Sugita M, Kondo T, Iwasaki H. 2006. A KaiC-associating SasA–RpaA two-component regulatory system as a major circadian timing mediator in cyanobacteria. Proc Natl Acad Sci USA. 103(32):12109–12114.
- Thaiss CA, Zeevi D, Levy M, Zilberman-Schapira G, Suez J, Tengeler AC, Abramson L, Katz MN, Korem T, Zmora N. 2014. Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 159(3):514–529.
- Tseng R, Chang YG, Bravo I, Latham R, Chaudhary A, Kuo NW, LiWang A. 2014. Cooperative KaiA–KaiB–KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J Mol Biol. 426(2):389–402.
- Van Gemerden H, Montesinos E, Mas J, Guerrero R. 1985. Diel cycle of metabolism of phototrophic purple sulfur bacteria in Lake Cisó (Spain) 1. Limnol Oceanogr. 30(5):932–943.
- Van Praag E, Degli Agosti R, Bachofen R. 2000. Rhythmic activity of uptake hydrogenase in the prokaryote Rhodospirillum rubrum. J Biol Rhythms. 15(3):218–224.
- Venkova T, Yeo CC, Espinosa M. 2018. The good, the bad, and the ugly: multiple roles of bacteria in human life. Front Microbiol. 9:1702.
- Wang Q, Wei W, Gong Y, Yu Q, Li Q, Sun J, Yuan Z. 2017. Technologies for reducing sludge production in wastewater treatment plants: state of the art. Sci Total Environ. 587-588:510–521.
- Whitehead K, Pan M, Masumura KI, Bonneau R, Baliga NS. 2009. Diurnally entrained anticipatory behavior in archaea. PloS One. 4(5):e5485.
- Wu G, Tang W, He Y, Hu J, Gong S, He Z, Wei G, Lv L, Jiang Y, Zhou H. 2018. Light exposure influences the diurnal oscillation of gut microbiota in mice. Biochem Biophys Res Commun. 501(1):16–23.
- Zhang Z, Wang J, Wang J, Wang J, Li Y. 2020. Estimate of the sequenced proportion of the global prokaryotic genome. Microbiome. 8(1):1–9.
- Zinser ER, Lindell D, Johnson ZI, Futschik ME, Steglich C, Coleman ML, Wright MA, Rector T, Steen R, McNulty N. 2009. Choreography of the transcriptome, photophysiology, and cell cycle of a minimal photoautotroph, Prochlorococcus. PLoS One. 4(4):e5135.
