8p11(eight-p-eleven) myeloproliferative syndrome (EMS), which is an uncommon neoplasm characterized by translocations involving the fibroblast growth factor receptor-1 (FGFR1). This entity is included in myeloid/lymphoid neoplasms with eosinophilia and tyrosine kinase gene fusions (MLN-TK) in the World Health Organization (WHO) classification [Citation1]. Seventeen FGFR1 translocation partners are reported. The three most prevalent are zinc finger MYM-type containing 2 (ZMYM2), centriolin (CNTRL) and FGFR1 oncogene partner (FGFR1OP). Their N-terminal self-association motifs are fused to the C-terminal tyrosine kinase domain of FGFR1 resulting in constitutive activation of the FGFR1 kinase and downstream signaling pathways [Citation2].
The clinical course is aggressive with rapid progression to acute leukemia. Most people are resistant to tyrosine kinase-inhibitors (TKIs). In persons with CNTRL::FGFR1 the complete remission rate is only 20% with chemotherapy [Citation3] and 1-year survival is only 40% [Citation4].
Olverembatinib is a novel oral ATP-binding site inhibitor targeting BCR::ABL1 [Citation5]. People with BCR::ABL1T315I respond better to olverembatinib compared with BCR::ABL1 [Citation6]. Olverembatinib inhibits fms-like tyrosine kinase 3 (FLT3), FGFR1 and platelet-derived growth factor receptor α (PDGFRα) kinase activity in vitro [Citation7]. However, olverembatinib has not been used to treat people with FGFR1 fusions. We present a person with MLN and a CNTRL::FGFR1 fusion who achieved transient hematological and cytogenetic responses after receiving olverembatinib.
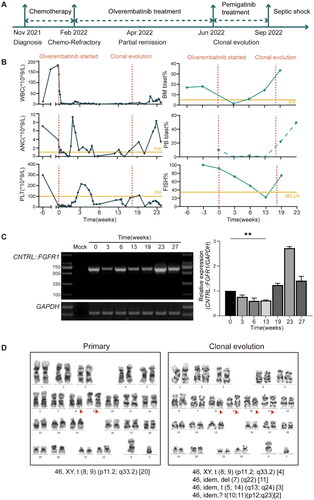
The clinical course is shown in . This patient was a 36-year-old man presented with lymphadenopathy and splenomegaly. Complete blood count demonstrated elevated white blood cell (WBC) at 47.85 × 109/L with normal hemoglobin (12.5 g/dL) and platelet count (103 × 109/L). Bone marrow aspirate revealed 9.1% B-cell lymphoblasts with immature features (CD34+, HLA-DR+, CD19+, CD10+, cytoplasmic CD79a+, CD38 dim, CD81 dim, CD20-), 0.6% T-cell lymphoblasts with cortical-T phenotype (cytoplasmic CD3+, CD7+, CD38+, CD4+, CD2+, CD99+, TDT+, CD5 dim), and 5.24% aberrant myeloid blasts (CD34+, CD117+, HLA-DR+, CD33+, CD13+, CD15-). Conventional cytogenetics showed an abnormal karyotype, 46, XY, t (8;9) (p11.2; q33.2) [20] (). FGFR1/D8Z2 probe confirmed an FGFR1 rearrangement with positive FGFR1 signals of 100% (Supplementary Figure S1 (A)). We further validated the CNTRL::FGFR1 fusion transcript with breakpoints located at 40th exon of CNTRL (Refseq NM_007018.6) and 10th exon of FGFR1(Refseq NM_023110.3) (Supplementary Figure S1 (C)).
Figure 1. Clinical presentation of the patient with the CNTRL::FGFR1 fusion. (A) Schematic representation of the treatment course of the patient with the CNTRL::FGFR1 fusion. (B) Peripheral WBC counts, ANC, PLT, peripheral blood and bone marrow blast cell percentage, FISH positive signal before and during treatment with olverembatinib. (C) QRT-PCR analysis from patient bone marrow samples before and during therapy with olverembatinib. A fragment of a 635 bp unique CNTRL::FGFR1 fusion sequence was amplified. A mock RT-PCR reaction with water was included as controls. (D) Cytogenetic analyses showed the reciprocal translocation t(8;9) (p11.2;q33.2) and complex abnormalities. **p < 0.01. PR: partial remission; McyR: major cytogenetic response; WBC: white blood cell; ANC: absolute neutrophil counts; PLT: platelet counts; FISH: fluorescence in situ hybridization; QRT-PCR: Quantitative real-time polymerase chain reaction.
Then he was treated with a 4-drug (vindesine, idarubicin, cyclophosphamide, prednisone) induction. After two cycles, bone marrow demonstrated 11.6% B-cell lymphoblasts, 2.7% myeloid blasts and 0.3% T-cell lymphoblasts by multiparameter flow cytometric measurable residual disease (MRD). Thereafter hyper CVAD-A and B regimens were given as reinduction. Patient in the recovery period showed rapidly increased WBC (181.7 × 109/L) with 10% myeloid blasts. Bone marrow revealed 0.33% B-cell lymphoblasts, 1.6% myeloid blasts and 4.2% T-cell lymphoblasts. The percentage of positive FGFR1 signals was 92%.
Subsequent he was started on induction therapy with combination of olverembatinib(40 mg orally every other day) plus 5-drugs (etoposide, vindesine, cytarabine, cyclophosphamide, dexamethasone) regimen. Upon taking olverembatinib, hepatosplenomegaly and lymphadenopathy rapidly resolved. A rapid decrease in WBC was achieved. Bone marrow aspirate demonstrated 0.79% B-cell lymphoblasts and 0.34% T-cell lymphoblasts. The percentage of positive FGFR1 signals reduced to 50%. He achieved complete hematologic remission (CHR) with 4 weeks (). Thereafter he continued the same regimen one more cycle. End-of-consolidation bone marrow revealed 9.14% T-cell lymphoblasts and 0.05% B-cell lymphoblasts. There was a major cytogenetic response (MCyR) (the percentage of positive FGFR1 probe signals reduced from 92% to 22%) (). QRT-PCR revealed a reduction of CNTRL::FGFR1 transcript in bone marrow (). There was no adverse event during treatment.
However, two months later, bone marrow aspirate revealed hypercellularity and an increased proportion of blasts (Supplementary Figure S1(B)). Cytogenetic analysis suggested clonal evolution with additional chromosomal aberrations (ACA), 46, XY, t (8;9) (p11.2; q33.2) [3]/46, idem, del (7) (q22) [1]/46, idem,? t(5; 14) (q13; q24) [2]/46, XY [23]. Next generation sequencing (NGS) revealed several pathogenic mutations including RUNX1-c.C958T, PHF6-c.615dupT, and HLA-A-c.627_628insCC with variant allelic fraction (VAF) of 43.80%, 77.90% and 10.00%, respectively. To find out if there are mutations within kinase domain, we amplified CNTRL::FGFR1 transcript with entire kinase domain. Sequence analysis showed the fusion gene had acquired K656E and V561M mutations (Supplementary Figure S1(D)). Subsequent he received salvage therapy with pemigatinib(9 mg orally once a day) plus a 5-drug induction containing high dose cytarabine (mitoxantrone, vindesine, cytarabine, etoposide, dexamethasone). However, the patient rapidly progressed to blast phase with 24.56% T-cell lymphoblasts, and 0.25% B-cell lymphoblasts. ACA clone was expanded (). He died from septic shock two months later.
Although MLN-TK with alternations involving PDGFRα and platelet-derived growth factor receptor β (PDGFRβ) usually respond exquisitely to imatinib, some cases may still develop drug resistance due to acquired mutations in the kinase domain. Our previous report described a 28-year-old man with FIP1-like-1(FIP1L1)::PDGFRα fusion who acquired a PDGFRα T674I mutation one year after receiving imatinib [Citation8]. In addition, we also report for the first time a 46-year-old male patient with tuberous sclerosis complex subunit 1(TSC1)::PDGFRβ fusion who achieved molecular remission for up to 15 years after imatinib treatment. In vitro experiments revealed that Ba/F3 cells expressing TSC1::PDGFRβT681I were refractory to imatinib and nilotinib [Citation9]. PDGFRα T674I and PDGFRβ T681I are analogous to T315I in BCR::ABL1, which directly affect the binding of TKI [Citation10]. Therefore, we explored whether olverembatinib and FGFRs inhibitors (pemigatinib) could target these gatekeeper mutations.
We constructed stably expressing CNTRL::FGFR1, CNTRL::FGFR1K656E, CNTRL::FGFR1V561M, FGFR1OP::FGFR1, TSC1::PDGFRβ, TSC1::PDGFRBT681I, FIP1L1::PDGFRα, FIP1L1::PDGFRαT674I in Ba/F3 cells. These cells were all able to proliferate without IL-3. Ba/F3-CNTRL::FGFR1, CNTRL::FGFR1K656E and FGFR1OP::FGFR1 were efficiently inhibited by much lower concentrations of olverembatinib and pemigatinib with IC50 around 1 nM (, ), which is lower than olverembatinib in Ba/F3-BCR::ABLT315I, with IC50 of 7.1 nM [Citation11]. When compared with CNTRL::FGFR1, CNTRL::FGFR1V561M were 1000 times more resistant to pemigatinib and 10 times more resistant to olverembatinib and ponatinib, but olverembatinib was still the most sensitive one with IC50 of 11.86 nM. CNTRL::FGFR1K656E had no significant change in IC50 compared with CNTRL::FGFR1. These indicate olverembatinib profoundly inhibited CNTRL::FGFR1, FGFR1OP::FGFR1, CNTRL::FGFR1K656E and CNTRL::FGFR1V561M in Ba/F3 models (Supplementary Table S1). TSC1::PDGFRβT681I and FIP1L1::PDGFRαT674I was highly resistant to imatinib and dasatinib. The IC50 for TSC1::PDGFRβT681I was 10.68 nM and 23.79 nM for olverembatinib and ponatinib, respectively, and the IC50 for FIP1L1::PDGFRαT674I was 1.57 nM and 1.45 nM. These imply that olverembatinib could overcome FIP1L1::PDGFRα and TSC1::PDGFRβ gatekeeper mutations.
Figure 2. Olverembatinib inhibits cell function in Ba/F3 cells expressing CNTRL::FGFR1. (A) BaF3-CNTRL::FGFR1, CNTRL::FGFR1K656E, CNTRL::FGFR1V561M exhibited differential sensitivity to TKIs. (B) Olverembatinib induced significant apoptosis in CNTRL::FGFR1 expressing cells after 48 h. BaF3-CNTRL::FGFR1, CNTRL::FGFR1K656E were treated with 10 nM of olverembatinib, ponatinib and pemigatinib, while BaF3-CNTRL::FGFR1V561M was treated with 100 nM of the three drugs. (C) Effect of olverembatinib, ponatinib and pemigatinib on cell cycle distribution in Ba/F3 cells after 24 h. The concentration of the drugs is the same as apoptotic assays. (D) Olverembatinib dose-dependently inhibits phosphorylation of FGFR1, STAT5 and STAT3. Cells were treated with the olverembatinib at the different concentrations for 4 h. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
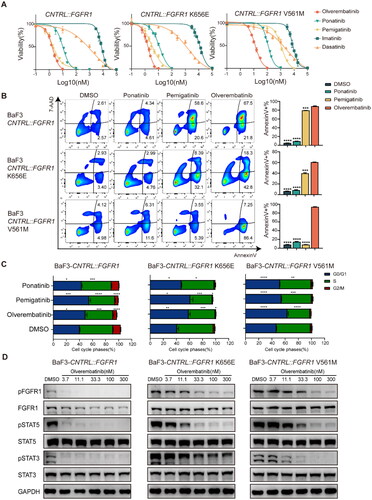
To further ascertain the efficacy of olverembatinib, cell cycle and apoptosis assay were performed in Ba/F3 models. At the same concentration as pemigatinib and ponatinib, olverembatinib caused a significant arrest in cell cycle progression in G0/G1 phase and an increased proportion in apoptosis. Although pemigatinib increased apoptosis rate and cell cycle arrest in Ba/F3-CNTRL::FGFR1, it was still less effective than olverembatinib in the presence of the other two mutations (). Consistent with these findings, olverembatinib dose-dependently inhibited tyrosine phosphorylation of CNTRL::FGFR1 and its downstream targets STAT5, STAT3 with a concentration of approximately 3 nM, whereas inhibition of K656E and V561M required higher concentrations of olverembatinib (30 and 100 nM, respectively) (). Olverembatinib also decreased the total level of FGFR1 protein in Ba/F3-CNTRL::FGFR1. Under the same conditions, imatinib and dasatinib could not inhibit the phosphorylation of signal pathway while ponatinib and pemigatinib could also efficiently inhibit the signal pathway only in CNTRL::FGFR1 and CNTRL::FGFR1K656E (Supplementary Figure S3(A-C)). As for CNTRL::FGFR1V561M, only olverembatinib can strongly inhibit its signal pathway at lower nanomolar concentration.
Our EMS patient achieved CHR and MCyR within 8 weeks with olverembatinib. FGFR1 K656E and V561M mutations were detected at relapse, along with the acquisition of new ACA. Our results showed that compared with CNTRL::FGFR1, the K656E mutation had no significant effect on drug sensitivity to TKIs and pemigatinib, but enhanced phosphorylation levels in signaling pathways. This is not the same as the research that BaF3-FGFR1 K656E were insensitive to pemigatinib with 9.3 fold increase in IC50 [Citation12]. Accordingly, same mutations of fusion genes and single genes may possess different mechanisms that ultimately affect drug sensitivity. V561M mutation conferred resistance to pemigatinib while still be sensitive to olverembatinib. This is consistent with previous study that pemigatinib had less potency against FGFR1 V561M [Citation13] and olverembatinib can overcome V561M mutation of fusion gene and single gene of FGFR1 [Citation12]. Thus, clonal evolution to acquire new ACA and other somatic mutations may be the main cause of relapse. Additional chromosomal translocations like t(10;11)(p12;q23) which leads to the formation of mixed lineage leukemia(MLL)::AF10 fusion. Acute myeloid leukemia(AML) patients harboring MLL::AF10 have a particularly poor prognosis compared to those not carrying it [Citation14]. Although clones harboring CNTRL::FGFR1 fusion preserved sensitivity, clones with ACA were resistant to olverembatinib. Previous study found that multiple oncogenes and several signaling pathways contribute cooperatively to the pathogenesis of CNTRL::FGFR1 in their mouse model [Citation15]. These indicate CNTRL::FGFR1 may induce the genomic instability to alter multiple oncogenes, disrupting signaling pathways to drive the disease progression. So, earlier treatment with olverembatinib may have the potential to achieve complete remission for EMS patients.
In conclusion, olverembatinib is a potent inhibitor against CNTRL::FGFR1, CNTRL::FGFR1K656E, and CNTRL::FGFR1V561M. That may be efficacious in treatment for patients with FGFR1 rearrangements. Given the first approval of olverembatinib in China for treatment of chronic phase CML (CML-CP) and accelerated phase CML (CML-AP) with mutation T315I, our findings warrant that olverembatinib can serve as a bridge to allogeneic hematopoietic stem cell transplantation which is the only curative option for EMS patients to prolong survival [Citation3].
Author contributions
Zhijian Xiao designed the study. Yiru Yan performed the experiments. Yiru Yan, Shiqiang Qu, Jinqin Liu, Chengwen Li, and Yujiao Jia, acquired the data, analyzed, and interpreted the data, performed statistical analysis; Xiao Yan, Zefeng Xu, Tiejun Qin, Lijuan Pan, Qingyan Gao, Bing Li and Meng Jiao participated in clinical information collection. Yiru Yan, Shiqiang Qu, Robert Peter Gale and Zhijian Xiao drafted this article. All authors read and approved the final manuscript.
Patient consent
The patient gave written informed consent compliant with the Declaration of Helsinki.
Supplemental Material
Download MS Word (1.5 MB)Acknowledgments
The authors acknowledge Ascentage Pharma (Jiangsu, China) for providing olverembatinib and all physicians, and laboratory researchers for their assistance.
Disclosure statement
RPG is a consultant to NexImmune Inc, Nanexa Pharma, Ascentage Pharm Group and Antengene Biotech LLC. He is Medical Director of FFF Enterprises Inc., Partner in AZAC Inc., is on the Board of Directors of the Russian Foundation for Cancer Research Support and is a member of the Scientific Advisory Board of StemRad Ltd. All other authors have no conflicts of interest to declare.
Data availability statement
The data that support the findings of this studc from the corresponding author upon reasonable request.
Additional information
Funding
References
- Khoury JD, Solary E, Abla O, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022; 36(7):1703–1719. doi: 10.1038/s41375-022-01613-1.
- Reiter A, Gotlib J. Myeloid neoplasms with eosinophilia. Blood. 2017;129(6):704–714. doi: 10.1182/blood-2016-10-695973.
- Chen M, Wang K, Cai X, et al. Myeloid/lymphoid neoplasm with CEP110-FGFR1 fusion: an analysis of 16 cases show common features and poor prognosis. Hematology. 2021; 26(1):153–159. doi: 10.1080/16078454.2020.1854493.
- Umino K, Fujiwara SI, Ikeda T, et al. Clinical outcomes of myeloid/lymphoid neoplasms with fibroblast growth factor receptor-1 (FGFR1) rearrangement. Hematology. 2018; 23(8):470–477. doi: 10.1080/10245332.2018.1446279.
- Dhillon S. Olverembatinib: first approval. Drugs. 2022; 82(4):469–475. doi: 10.1007/s40265-022-01680-9.
- Jiang Q, Li Z, Qin Y, et al. Olverembatinib (HQP1351), a well-tolerated and effective tyrosine kinase inhibitor for patients with T315I-mutated chronic myeloid leukemia: results of an open-label, multicenter phase 1/2 trial. J Hematol Oncol. 2022; 15(1):113. doi: 10.1186/s13045-022-01334-z.
- Wang Y, Zhang L, Tang X, et al. GZD824 as a FLT3, FGFR1 and PDGFRα inhibitor against leukemia in vitro and in vivo. Transl Oncol. 2020; 13(4):100766. doi: 10.1016/j.tranon.2020.100766.
- Qu SQ, Wang Y, Sun XJ. [FIP1L1-PDGFRA positive chronic eosinophilic leukemia with imatinib-resistant T674I mutant of PDGFRA gene: a case report and literature review]. Zhonghua Xue Ye Xue Za Zhi. 2013; 34(2):159–161.
- Zhang Y, Qu S, Wang Q, et al. A novel fusion of PDGFRB to TSC1, an intrinsic suppressor of mTOR-signaling pathway, in a chronic eosinophilic leukemia patient with t(5;9)(q32;q34). Leuk Lymphoma. 2018; 59(10):2506–2508. doi: 10.1080/10428194.2018.1427855.
- Alves R, Gonçalves AC, Rutella S, et al. Resistance to tyrosine kinase inhibitors in chronic myeloid leukemia-from molecular mechanisms to clinical relevance. Cancers. 2021; 13(19):4820. doi: 10.3390/cancers13194820.
- Ren X, Pan X, Zhang Z, et al. Identification of GZD824 as an orally bioavailable inhibitor that targets phosphorylated and nonphosphorylated breakpoint cluster region-Abelson (Bcr-Abl) kinase and overcomes clinically acquired mutation-induced resistance against imatinib. J Med Chem. 2013; 56(3):879–894. doi: 10.1021/jm301581y.
- Jiang K, Tang X, Guo J, et al. GZD824 overcomes FGFR1-V561F/M mutant resistance in vitro and in vivo. Cancer Med. 2021; 10(14):4874–4884. doi: 10.1002/cam4.4041.
- Lin Q, Chen X, Qu L, et al. Characterization of the cholangiocarcinoma drug pemigatinib against FGFR gatekeeper mutants. Commun Chem. 2022; 5(1):100. doi: 10.1038/s42004-022-00718-z.
- Dreyling MH, Schrader K, Fonatsch C, et al. MLL and CALM are fused to AF10 in morphologically distinct subsets of acute leukemia with translocation t(10;11): both rearrangements are associated with a poor prognosis. Blood. 1998; 91(12):4662–4667.
- Ren M, Qin H, Kitamura E, et al. Dysregulated signaling pathways in the development of CNTRL-FGFR1-induced myeloid and lymphoid malignancies associated with FGFR1 in human and mouse models. Blood. 2013; 122(6):1007–1016. doi: 10.1182/blood-2013-03-489823.
