Abstract
Amyloidosis is a disease group caused by pathological aggregation and deposition of peptides in diverse tissue sites. Apart from the fibril protein, amyloid deposits frequently enclose non-fibrillar constituents. In routine diagnostics, we noticed the presence of complement 9 (C9) in amyloid. Based on this observation, we systematically explored the occurrence of C9 in amyloid. Apolipoprotein E (apoE), caspase 3 and complement 3 (C3) served as controls. From the Amyloid Registry Kiel, we retrieved 118 formalin-fixed and paraffin-embedded tissue samples, including eight different amyloid- and 18 different tissue types. The expression patterns were assessed immunohistochemically in relation to amyloid deposits. A literature search on proteomic data was performed. Amyloid deposits stained for C9 and apoE in 117 (99.2%) and 112 of 118 (94.9%) cases, respectively. A homogeneous immunostaining of the entire amyloid deposits was found in 75.4% (C9) and 61.9% (apoE) of the cases. Caspase 3 and C3 were present only in 22 (19.3%) of 114 and 20 (36%) of 55 assessable cases, respectively. Caspase 3 and C3 immunostaining rarely covered substantial areas of the amyloid deposits. The literature search on proteomic data confirmed the frequent detection of apoE and the occurrence of C9 and C3 in amyloid deposits. No data were found regarding caspase 3. Our findings demonstrate the ubiquitous, spatial and specific enrichment of C9 in amyloid deposits irrespective of amyloid-, organ- or tissue type. Our findings lend support to the hypothesis that amyloidosis might activate the complement cascade, which could lead to the formation of the membrane attack complex and cell death.
Introduction
Amyloidosis constitutes a large group of diseases caused by misfolding of peptides and proteins, which generate insoluble and toxic protein aggregates. These aggregates are oriented in a β-pleated sheet structure assembling into non-branching fibrils of up to 10–12 nm in diameter [Citation1,Citation2]. To date, 36 different autologous, physiological peptides and proteins have been identified to cause local and/or systemic amyloid deposits in different tissues and organs, both extracellularly and intracellularly [Citation2,Citation3]. The diagnosis is made histologically by Congo red staining in combination with polarization microscopy when a characteristic green–yellow–orange birefringence is found [Citation4,Citation5]. Immunohistochemistry (IHC) or proteomics are mandatory in order to identify the amyloid protein and the underlying disease [Citation6]. In the last 10 years, the proteomic approach for amyloid typing was further established and developed. With a sample preparation method like laser microdissection (LMD) followed by liquid chromatography (LC) and tandem mass spectrometry (LC–MS/MS), the amyloidogenic as well as other marker proteins can be detected in subcutaneous fat aspirate (SFA) [Citation7] or formalin-fixed, paraffin-embedded (FFPE) tissue samples [Citation8].
Apart from the amyloid protein, amyloid deposits often enclose various other non-amyloidogenic but seemingly disease-specific constituents, such as serum amyloid P-component (SAP), apolipoprotein E (apoE), highly sulfated glycosaminoglycans, laminin, vitronectin, clusterin, among other [Citation1,Citation2,Citation7–9].
Apolipoprotein E is synthesized primarily by the liver and also by the peripheral and central nervous system [Citation10]. As the main component of lipoproteins, it mediates the binding of the lipoproteins in plasma to their specific receptors on the surface of the target cells [Citation11]. Apolipoprotein E immunoreactivity in amyloid deposits with Aβ-protein in senile plaques has already been shown [Citation10,Citation12]. Due to its polymorphism, there are three isoforms (apoE1, apoE2, apoE4), having isoform-specific effects on Aβ metabolism [Citation11]. ApoE4 binds to Aβ more rapidly and effectively when certain conditions prevail [Citation13].
Proteomic analyses of amyloid occasionally report the presence of members of the complement system. The complement system, located in blood serum of humans, is one of the principal, immunologically relevant effector systems that comprise over 14 soluble proteins, a number of receptors and several soluble membrane proteins performing regulatory functions. The major target of the complement components is the cell membrane. A direct attack that requires participation of all nine complement components (C1–C9) leads to an irreversible damage and subsequently ultrastructural lesion [Citation14,Citation15]. The terminal pathway is the assembly of C5b, C6, C7, C8 and multiple copies of complement 9 (C9) into the membrane attack complex (MAC). This MAC functions as a transmembrane channel [Citation16,Citation17], causing loss of membrane integrity and finally leads to cell death by necrosis [Citation18].
In surgical pathology of cardiac biopsies, C9 is used as a surrogate marker for single cell myocyte necrosis [Citation19,Citation20]. During the assessment of cardiac biopsies in routine diagnostic pathology, we noticed that C9 not only stained single cell necrosis in the heart but also interstitial amyloid deposits. Intrigued by this observation and findings made by proteomic analyses, we aimed to systematically explore the presence of C9 in diverse types of amyloid. As C9 could also highlight single cell necrosis induced by the presence of amyloid, we also explored the occurrence of apoptosis by caspase 3 immunostaining and used apoE and complement 3 (C3) as a control. We aimed to test the following hypotheses: (1) C9 is specifically and spatially enriched in amyloid deposits regardless of amyloid type; (2) the presence of amyloid may lead to either cell necrosis and/or apoptosis of cells adjacent to and enclosed by the amyloid deposits. The study is supported by a literature search relating proteomic analyses of the presence of specific proteins such as apoE, caspase 3, C3 and C9.
Materials and methods
Ethics statement
This project was approved by the local ethics committee of the University Hospital in Kiel conforming to the Declaration of Helsinki (D581-585/15; D 469/18) and of the Friedrich-Alexander University (FAU) Erlangen-Nürnberg (reference number 4415).
Patients
From the Amyloid Registry of the Department of Pathology, Christian-Albrechts-University Kiel, we retrieved a series of 118 patients (77 men (65.3%) and 41 women (34.7%); median age 68.5 years) (study cohort). The samples were obtained from 2016 through 2018 and included eight different types of amyloid, i.e. AA (n = 10), AIns (4), ALκ (21), ALλ (42), ATTR (35), Aβ (3), mixed (1) and unclassified (2) amyloidosis. The specimens were obtained from the heart (n = 28), kidney (21), synovialis and retinaculum flexorum of the wrist (17), skin (14), adipose and connective tissue (7), gastrointestinal tract (7), liver (6), lung (5) and other (13) (Supplementary Table S1). The specimens were randomly selected and the only inclusion criterion was the histologically confirmed presence of amyloid.
Control tissue (first set of controls) was obtained from five patients with focal segmental glomerulosclerosis (FSGS) and nine with hemorrhagic infarcts/infarctions from various tissues and across different age groups (<1 year up to 91 years). Amyloidosis was excluded in all control samples by Congo red staining and polarization microscopy.
Another set of 22 controls (second set of controls) was obtained from the archive of the Department of Nephropathology, Institute of Pathology, FAU Erlangen-Nürnberg with renal AA- (six men, two women; mean age ± SD: 62.2 ± 9.5 years), AL- (eight men, three women; mean age ± SD: 73.6 ± 9.3 years) and AFib amyloidosis (three men; mean age ± SD: 66.7 ± 18.2 years). The latter had been confirmed by genetic testing (data not shown).
Histology
All samples had been FFPE. Serial sections were cut from each paraffin block and stained with hematoxylin and eosin and Congo red. The presence of amyloid was confirmed when a typical green–yellow–orange birefringence was found in Congo red stained tissue sections.
Immunohistochemistry
Immunohistochemistry was carried out with commercially available monoclonal antibodies directed against AA amyloid (dilution 1:2000), β amyloid (1:50; both DAKO, Hamburg, Germany), insulin (1:500; BioGenex, San Ramon, CA), apoE (1:10,000), C3 (1:400; both Abcam, Berlin, Germany), C1q (1:75,000), C3c (1:75,000; both DAKO, Hamburg, Germany), C3d (1:1,000; Abcam, Cambridge, UK), C5b-9 (1:100; DAKO, Hamburg, Germany), MASP-2 (1:200; Sigma-Aldrich, Taufkirchen, Germany), C9 (1:400; Biozol, Eching, Germany) and monoclonal rabbit antibodies directed against caspase 3 (1:100; Cell Signaling, Danvers, MA) and polyclonal rabbit antibodies directed against amyloid P-component (1:2000), kappa-light chain (1:100,000), lambda-light chain (1:14,000; all DAKO, Hamburg, Germany) and non-commercially available polyclonal rabbit antibodies directed against apolipoprotein AI (1:1000), transthyretin (TTR3, 1:2000), lambda-light chain-derived amyloid proteins (AL1 antibody, 1:250), anti-lambda-light chain peptides (AL3, 1:250; AL7, 1:200) and kappa-light chain peptides (AK3, 1:1000; all Pineda, Berlin, Germany) [Citation6]. Immunostaining was done on FFPE sections with the Bond Max Leica immunostainer using the Bond Polymer Refine Detection Kit (Leica Biosystems, Wetzlar, Germany) (study cohort) or with the Ventana Benchmark immunostainer and ‘ultraView Universal DAB detection Kit’ (Roche Diagnostics Deutschland GmbH, Mannheim, Germany) (second set of controls) as described previously [Citation21].
Antigen retrieval was carried out with Leica ER1-Bond Epitope Retrieval Solution 1 (C9, C3), Leica ER2-Bond Epitope Retrieval Solution 2 (amyloid P-component, kappa-light chain, lambda-light chain, TTR3, apoE, caspase 3), Enzyme 1 (AL7; all Leica Biosystems, Wetzlar, Germany), pronase E digestion and backing (C1q, C3c, C5b-9) or cooking in target retrieval solution pH 6 (DAKO, Hamburg, Germany) for 2.5 min using a pressure cooker (C3d, MASP-2) according to the manufacturer’s instructions. Immunohistochemical classification of amyloid was carried out and had been validated as described in detail elsewhere [Citation6,Citation22].
In brief, identification of the amyloid was considered to be positive when there was a strong and homogenous immunostaining of the entire amyloid deposits. Uneven and weak staining of some deposits was not assumed to be proof of the amyloid protein. If the staining was clearly positive with more than one antibody against different amyloid precursor proteins, the case was categorized as mixed amyloidosis. AL amyloid not otherwise specified (n.o.s.) was characterized by positive staining with antibodies directed against λ- and κ-light chain and negative immunostaining for the other amyloid proteins tested.
On slide positive and negative controls using a tissue microarray with AA-, ALλ- and ATTR amyloid as well as non-neoplastic liver tissue were used on each staining round.
Evaluation of immunostaining
Serial sections were used throughout this study. Initially, we confirmed the presence of amyloid in the first serial section. Next, the distribution of amyloid deposits was compared with the immunostaining of the putative amyloid proteins. Then, the spatial distribution of the classified amyloid proteins was compared to the spatial distribution of the apoE, caspase 3, C3 and C9 immunostaining. The intensity of immunostaining was graded as absent (0), weak (1+), moderate (2+) or strong (3+). A staining intensity of 1+ to 3+ was considered positive.
The colocalization of the target proteins with the amyloid was documented in steps of 5%. In addition, cellular immunostaining of both caspase 3 and C9 was documented in relation to the amyloid deposits.
Literature search
Proteomic data on apoE, caspase 3, C3 and C9 published between 1 January 2008 and 22 July 2020 were retrieved from PubMed. Keywords were proteomics, amyloidosis, amyloid, mass spectrometry and LMD. Since only a few workgroups published comparable data generated by optional LMD prior to LC and tandem mass spectrometry (MS/MS) (e.g. Vrana et al. [Citation8] or Lavatelli and Vrana [Citation23]), we screened the literature again for the affiliated author’s names as well as the search terms ‘amyloid’, or ‘amyloidosis’. Records were limited to those in English language. Data were further analyzed if FFPE tissue or SFA sample material was analyzed by LMD LC–MS/MS and the amyloid was histologically confirmed. Protein data must include the (1) protein name and/or comprehensible identifier, (2) the number of total peptide spectra as a semiquantitative measurement for protein abundance and (3) a protein probability score over 95%. Duplicate protein data were excluded.
Bioinformatics and statistics
The protein names of interest and/or identifiers were compared to the UniProt database [Citation24] and their accession number was retrieved. The extracted protein data were loaded into R version 4.0.0 [Citation25] and merged into one dataset for further processing, particularly using the packages tidyverse version 1.3.0 [Citation26], ggplot 2 version 3.3.2 [Citation27] and ggpubr version 0.3.0 [Citation28]. If more than one sample was reported per case, the spectra number’s average was calculated and classified into the categories 0, 1–3, 4–10, 11–20 and >21 number of spectra according to D'Souza et al. [Citation29]. Then, the median of the spectral class was determined after grouping cases by protein, organ and amyloid type and plotted as balloonplot using color scale from viridis version 0.5.1 package [Citation30].
Data collection and creation of tables were performed using IBM SPSS statistic version 25 (International Business Machines, Armonk, NY).
Results
One hundred and eighteen resection and biopsy samples with amyloid deposits were included in the study cohort. Eight different amyloid types and 18 different histoanatomical locations were examined. Supplementary Table S1 summarizes the amyloid- and tissue types of the case series. AL- and ATTR amyloid were the most common types of amyloid studied, while cardiac and kidney biopsies were the most frequent organ sites.
As shown in , amyloid deposits homogeneously stained for C9 and apoE (). Furthermore, C9 was present within the amyloid deposits in 117 of 118 (99.2%) cases. Only a single case with ATTR amyloid in a cardiac biopsy showed no colocalized C9 immunoreactivity. Apolipoprotein E was present in 112 of 118 (94.9%) specimens. It was not detected in two cases of each ALκ, ALλ and ATTR, respectively. Caspase 3 was assessable in 114 cases and present in 22 (19.3%). Complement 3 was studied in 55 cases and found in 20 (36%) ( and ).
Figure 1. Immunostaining of apolipoprotein E, caspase 3, complement 3 and 9 in relation to amyloid (study cohort). (A–H) Comparison between immunohistochemistry and the enrichment of investigated proteins using the example of renal ALκ amyloidosis. Serial sections: hematoxylin eosin staining (A), Congo red staining in bright light (B), immunostaining with antibodies directed against κ-light chain (C), Congo red staining in fluorescence light (D; same section as in B), immunostaining with antibodies directed against apolipoprotein E (E), complement 9 (F), caspase 3 (G) and complement 3 (H); (I) caspase 3 immunoreactive cardiomyocytes (arrows) within amyloid deposits (asterisk). (J) Complement 9 immunoreactive cardiomyocytes (arrow head) surrounded by complement 9 immunoreactive amyloid deposits. Hematoxylin counterstain.
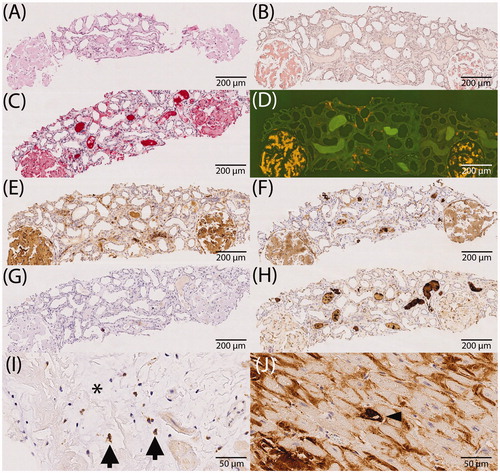
Table 1. Presence of positivity of the respective protein within amyloid deposits differentiated by amyloid type.
Table 2. Presence of positivity of the respective protein within amyloid deposits differentiated by organ-/tissue type.
In order to provide an impression of the extent of immunolabeling, we evaluated the percentage area of amyloid deposits stained with antibodies directed against apoE, caspase 3, C3 and C9. and illustrate the percentage area covered by C9 immunolabeling. Interestingly, immunolabeling of the entire amyloid deposits (i.e. 100%) was noted in 89 (75.4%) cases. In 109 (92.4%) cases, C9 immunoreactivity covered at least 80% of the Congo red positive amyloid area. Focused on amyloid type, all cases with AA-, AIns-, Aβ- and mixed amyloid showed labeling in at least 80% of the amyloid area. This degree of coverage was observed in 19 out of 21 (90%) ALκ cases, 38 out of 42 (90%) ALλ, 33 out of 35 (94%) ATTR and one out of two unclassifiable amyloidosis cases (). Based on histoanatomical localization, the following numbers demonstrate an agreement of at least 80% in spatial enrichment of C9 within amyloid deposits: all specimens obtained from the kidney, skin, synovialis and ligament of carpal tunnel, gastrointestinal tract, liver and lung (each 100%); heart, 21 out of 28 (75%); adipose and connective tissue, six out of seven (86%); and other; 12 out of 13 (92%) cases (). Interstitial immunostaining of C9 was not present outside the amyloid deposits in any case. All in all, the average accordance for all samples was more than 90%.
Figure 2. Accordance by type of amyloidosis. The proportion of immunostained area of complement 9, apolipoprotein E, caspase 3 and complement 3 in relation to the entire amyloid deposits (i.e. 100%). Presentation according to the amyloid type. Each bar corresponds to one case.
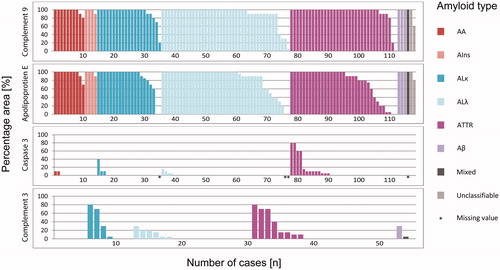
Figure 3. Accordance by histoanatomical localization. The proportion of immunostained area of complement 9, apolipoprotein E, caspase 3 and complement 3 in relation to the entire amyloid deposits (i.e. 100%). Presentation according to the organ-/tissue-type. Each bar corresponds to one case.
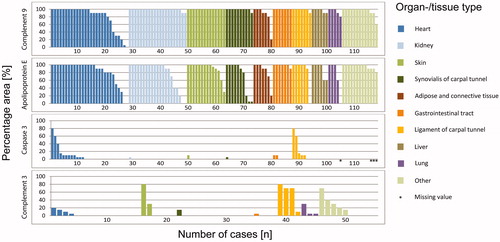
We then assessed the area percentage of amyloid labeled for apoE. In 73 (61.9%) cases, apoE immunostaining of the entire amyloid area was shown. In 94 (79.7%) specimens at least 80% of the Congo red stained area was apoE positive ( and ). The average amyloid deposition area covered by apoE immunostaining was over 80%.
Caspase 3 immunostaining is depicted in and . Immunostaining of the amyloid deposits was only noted in 22 (19.3%) out of 114 cases. None of the cases showed a complete coverage with the amyloid deposition area. The maximum accordance of 80% was detected in only two (1.8%) cases with ATTR amyloidosis. Thus, 107 (93.9%) samples, including those without staining, showed labeling of a maximum of 10% of the amyloid area. The average accordance of caspase 3 immunolabeling with amyloid deposits was less than 5%.
Finally, and illustrate the results of C3 immunolabeling. There was no case with staining of the entire amyloid area. For the majority of samples (n = 35 (64%) cases), no C3 immunostaining was observed. The maximum accordance of 80% was only noted in two (4%) cases, one biopsy of the ligament of carpal tunnel with ATTR and one biopsy of skin with ALκ. Regarding all specimens analyzed for C3, the average accordance was about 10%.
Collectively, these data demonstrate that C9 and apoE were ubiquitously, spatially and specifically enriched in the amyloid deposits of diverse origin irrespective of organ- or tissue-site. In contrary, caspase 3 and C3 were rarely found within amyloid deposits and usually only covered a fraction of the area.
Subsequently, we examined the cellular detection of C9 and caspase 3 within or adjacent to the amyloid deposits (). and demonstrate the distribution of C9 and caspase 3 single cells according to the type of amyloidosis and the histoanatomical localization. In total, 23 out of 118 (19.5%) cases were found to enclose any C9 positive single cells. Out of these, 16 (13.6%) cases also exhibited amyloid-associated single positive cells, i.e. the cells were located directly in interstitial amyloid deposits or nearby (). Any caspase 3 single positive cells were found in 25 out of 114 (21.9%) cases. Only 18 (15.8%) out of these cases contained amyloid-associated single positive cells. Regarding cardiac biopsies, C9 as well as caspase 3 were positively tested in amyloid-associated single cells in 17.9% of cases. In kidney biopsies, C9 was enriched in single amyloid-associated cells in 9.5% of cases and no positively stained cells for caspase 3 were found in any case (). With regard to the amyloid type, we investigated ALκ, ALλ and ATTR for amyloid-associated single positive cells. C9 positive single cells were found in 19.0% of ALκ amyloidosis cases, in 12% of ALλ and in 14% of ATTR cases, whereas caspase 3 was present in single cells in 15% of ALκ, in 10% of ALλ and in 6% of ATTR cases ().
Table 3. Prevalence of caspase 3-/complement 9 single positive cells differentiated by organ-/tissue type.
Table 4. Prevalence of caspase 3-/complement 9 single positive cells differentiated by amyloid type.
Since amyloid may be contaminated by serum proteins, we next assessed C9 immunostaining in five kidney biopsies with FSGS (positive control) and nine tissue specimens from hemorrhagic infarcts (negative control; first set of controls). Expectedly, a mesangial C9 staining in the majority of glomeruli affected by glomerulosclerosis was observed in FSGS and has been described previously (Supplementary Figure 1), while hemorrhagic infarcts only showed a faint and diffuse immunostaining (Supplementary Figure 1) without a spatial enrichment in any histoanatomical structure.
Since testing for complement components in nephropathology is common, we then explored the detection of commonly used complement markers in 22 kidney biopsies with AA- (n = 8), AFib- (n = 3) and AL amyloidosis (n = 11), respectively (second set of controls; Supplementary Figure 2). All renal biopsies showed Congo red positive amyloid deposits (Supplementary Figure 2A–C) but these were in most cases negative for C1q, the initiator of the classic complement activation pathway (Supplementary Figure 2D–F). MASP-2, an activator of the lectin-mediated complement cascade, was barely detectable in AA- and AL amyloidosis, but was detectable in AFib amyloidosis (Supplementary Figure 2G–I). Interestingly, the stable C3 cleavage product C3c is primarily seen in AA amyloidosis (Supplementary Figure 2J–L) while C3d and C9 were clearly detectable in all forms of renal amyloidosis of the second set of controls (Supplementary Figure 2M–R).
The literature search, finally, revealed 48 articles containing proteomic data on apoE, C3 or C9 usable for data processing (Supplementary Table S2). We found peptide spectra for apoE in all 214 amyloidosis cases tested, C9 spectra were reported in 15 out of 20 (75%) cases and C3 spectra in 36 out of 44 cases (82%) (, Supplementary Table S2). We did not retrieve any comparable proteomic data referring to caspase 3. Most cases (26.6%, n = 57) that were apoE positive were diagnosed with AL amyloidosis. Another 24 cases (11.2%) presented with a mixed AL amyloidosis (AL/AH, n = 22; AL/ATTR, n = 2), followed by ATTR (11.7%, n = 25) and ALECT2 amyloidosis (10.3%, n = 22). Apolipoprotein E was positive in 86 (40.2%) cases investigating kidney samples, which was by far the most frequently reported tissue. In this regard, kidney was the tissue reported most often with 86.4% of cases positive for C3, and 95.0% of cases positive for C9. The amyloid type reported the most often in C9 and C3 positive cases was AFib (C9, n = 5; C3, n = 8). Our reviewed data presented a varying number of peptide spectra in apoE, C3 and C9 positive amyloid deposits. Most apoE positive cases were identified with more than 21 spectra (). This indicated a relatively high abundance, as the most abundant proteins in a sample were usually reported with 21 up to more than 100 spectral counts (Supplementary Table S2). Single samples were detected with a median spectral number of 11–20, for example in renal ALECT2 amyloidosis. The minority of all cases resulted in an apoE spectral number below 10 (single cases of AL/ATTR, lymph node; ATTR/AGel, colon/rectum; ATTR, synovium) or even below three peptide spectra in single AGel and ATTR/AGel deposits in SFA. A variable spectral count was measured in C3 positive amyloid deposits. The spectral number measured for kidney samples was >21 in AApoCII, AFib and AGel cases or was mainly 4–10 for other amyloid types. C9 spectral counts had a lower relative abundance in all cases analyzed, varying between 4 and 20.
Figure 4. Proteomics data obtained by literature search. Occurrences of apolipoprotein E (A), complement 9 (B) and complement 3 (C) in amyloid tissue sections are depicted for amyloid and organ type. Proteomic data were reviewed and extracted from amyloidosis cases analyzed by LMD LC–MS/MS (method described in [Citation9–11]). The total number of patients tested is indicated by data point area. Positive cases are colored/grey scaled depending on the number of spectras’ median measured, whereas negative cases are depicted by black areas. The number of spectra refers to the total number of peptide spectra identified for a protein and is considered a semiquantitative measurement of protein abundance. Proteins were identified with a >95% probability. For three cases, samples were derived from more than one organ type: *heart and gastrointestinal tract (GI-tract; not otherwise specified); **lung and skin; ***heart, subcutaneous fat aspirate (SFA) and large intestine.
![Figure 4. Proteomics data obtained by literature search. Occurrences of apolipoprotein E (A), complement 9 (B) and complement 3 (C) in amyloid tissue sections are depicted for amyloid and organ type. Proteomic data were reviewed and extracted from amyloidosis cases analyzed by LMD LC–MS/MS (method described in [Citation9–11]). The total number of patients tested is indicated by data point area. Positive cases are colored/grey scaled depending on the number of spectras’ median measured, whereas negative cases are depicted by black areas. The number of spectra refers to the total number of peptide spectra identified for a protein and is considered a semiquantitative measurement of protein abundance. Proteins were identified with a >95% probability. For three cases, samples were derived from more than one organ type: *heart and gastrointestinal tract (GI-tract; not otherwise specified); **lung and skin; ***heart, subcutaneous fat aspirate (SFA) and large intestine.](/cms/asset/3d7eee5e-3f53-4018-9d58-de857ab70d84/iamy_a_1932799_f0004_c.jpg)
Discussion
The pathogenesis of amyloid is a multifactorial process. While a disease-specific misfolding of a particular amyloid precursor protein forms the amyloid fibril, non-fibrillar constituents including SAP, apoE, matrix components like collagen and highly sulfated glycosaminoglycans are regularly found across all amyloid types [Citation1,Citation2,Citation8].
Our immunohistochemical results clearly demonstrated the ubiquitous, spatial and specific occurrence of C9 and apoE in the amyloid deposits irrespective of amyloid type or tissue site, and the irregular presence of caspase 3 and C3. With this study, we could even show that the enrichment of C9 in 99.2% of all cases analyzed is more consistent in comparison to the one of apoE in 94.9%. The analysis of proteomic data partially confirms these observations by IHC. Every single case studied by proteomics enclosed apoE, wherefore it has been previously declared an amyloid signature protein, e.g. for SFA samples [Citation7] or FFPE tissue [Citation8]. Its occurrence is independent of the amyloid- and tissue type. The limited proteomic data on C9 and C3 at least show that so far both components could be identified in amyloid deposits. For instance, they confirm the occurrence of C3 and C9 in different types of renal amyloidosis, as illustrated by . In IHC, we found C9 in all cases of renal AA (n = 4) and AL amyloidosis (n = 13) in the study cohort and in all 22 cases of renal AA-, AFib- and AL amyloidosis of the second set of controls. Likewise, the proteomic data from Sethi et al. showed two cases with AA amyloidosis, both positive for C9 with small spectral numbers, whereas the analysis of AL amyloidosis showed C9 positive amyloid deposits only in one of three cases [Citation31]. Other amyloid types mentioned in the C9 plot were not studied by IHC in our series. Hence, we observed an enrichment of C9 by IHC in amyloid deposits, whereas proteomic analysis of amyloid specimens for diagnostic purposes rarely reported complement pathway components. A reason for that could be that the diagnostic approach of amyloid analysis by LMD LC–MS/MS mainly focuses on the fibril forming protein and the amyloid signature proteins. Amyloid components like complement factors might have not been detected due to their absence or low abundance, or they were simply overlooked in the past and therefore, not published as the top 10–20 most abundant or relevant amyloid constituents. Most recent findings by Kourelis et al. support our IHC findings, as they identified C9 and C3 besides complement factor H and complement factor H related peptide 5 as differentially expressed proteins in ATTR amyloidosis of the heart by LMD LC–MS/MS. Complement factor H related peptide 1 was differentially expressed in heart samples of both AL and ATTR amyloidosis [Citation32]. Additionally, Brambilla et al. have shown that C3 was up-regulated in ATTR patients’ fat aspirate samples, compared to non-amyloid affected fat samples, and identified C9 in several amyloidosis cases [Citation33]. Supplementary, it has been reviewed by Krance et al. that C3 and C9 tend to be more abundant in patients with Alzheimer’s disease compared to case controls without neurodegenerative diseases [Citation34].
The literature search revealed the occurrence of C3 within amyloid deposits in 36 out of 44 reported cases. In relation to the amyloid types mentioned in the C3 plot (), we examined only cases with AL- and ATTR deposits for C3 occurrence by IHC in the study cohort. With regards to AL amyloidosis, three cases of renal and five cases of cardiac biopsies were analyzed immunohistochemically. None of the kidney samples showed any C3 staining, though in two cardiac samples a lower C3 occurrence of 5% and 20% was found. Our literature search on proteomic data revealed peptide spectra for C3 in two out of five kidney samples and no C3 was found in the only investigated cardiac case [Citation23,Citation31,Citation35]. With regard to ATTR amyloidosis, we examined seven cardiac biopsies by IHC, two of them with coverage of 10% and 15%, and five without C3 enrichment. In this respect, the proteomic analysis described one cardiac case, which enclosed C3 [Citation36]. However, it is interesting to note that cleavage products of C3, i.e. C3c and C3d, were found in the second set of controls. These findings support our hypothesis that C9 and other complement pathway components occur independently of amyloidosis- and tissue type. They might play an important role in amyloid pathogenesis.
It is known that C9 plays a crucial role in cell lysis, as it is an essential component of the MAC. An activation of a sequence of proteins, i.e. the complement system, leads to the formation of a multiprotein pore that inserts into the cell membrane and causes cell death. MAC is the common end of three biochemical pathways, i.e. the classical, lectin and alternative complement pathway. All three pathways have in common the cleavage and activation of C3. Thus, C3 represents the cascade’s center and following its activation the executive functions of the complement system are initiated. The importance of the complement cascade is not only evident in terms of amyloidosis, but also in renal diseases, such as the atypical hemolytic uremic syndrome (aHUS) and the C3 glomerulopathies (C3G) [Citation37]. These diseases are caused by a dysregulated and uncontrolled activation of the complement system, i.e. aHUS by pathogenic mutations in different complement genes among others [Citation38], and C3G mainly by the presence of C3 nephritic factor [Citation39], an autoantibody directed against the C3 convertase of the alternative pathway, the C3bBb. In addition, there are various studies suggesting an involvement of an active complement pathway in FSGS. For instance, they demonstrate higher levels of complement activation fragments in the plasma and urine, both in animal [Citation40] and human models [Citation41,Citation42]. Strassheim et al. were able to demonstrate a colocalization of glomerular IgM and C3 by immunofluorescence microscopy [Citation40]. A recent study even showed a correlation between plasma and urine C5b-9 levels with proteinuria [Citation41]. Our IHC observations on FSGS confirm these results, as we also detected a specific and regular glomerular C9 enrichment. Hence, all these findings indicate the participation of complement activation in renal disease, now including also amyloidosis, and their progressing damage mediated by MAC. Since C9 is present also in other, complement-mediated diseases, e.g. FSGS, the utility of C9-immuostaining as a general diagnostic marker for renal amyloidosis is challenging.
Both necrosis and apoptosis are forms of cell death. Caspase 3 is an effector caspase in the execution part of cell apoptosis [Citation43]. Its cleaved and activated form is used as a surrogate marker for apoptosis [Citation44]. Therefore, the occurrence of necrosis and apoptosis nearby amyloid deposits was under investigation. Due to the limited number of examined cases per sampling site, statements can be made at most about heart and kidney. In 17.9% of cases, amyloid-associated C9 and caspase 3 single positive cells were found in cardiac biopsies, a slightly higher frequency compared to the average across all tissue sites of 13.6% (C9) and 15.8% (caspase 3). In kidney biopsies, the percentage of amyloid-associated cells was even lower. Only 9% of cases hold C9 positive cells and none of them contained any caspase 3 positive cells. Regarding to ALκ, ALλ and ATTR, the percentage also remains at a lower level. Thus, single cell necrosis and apoptosis were detected but it seems to be a rare event. However, this may have been caused by a sampling error: a tissue biopsy is only a spatially restricted ‘snap shot’ in an ongoing and lasting disease. In time scales of months and years, cell loss due to single cell necrosis and apoptosis can be substantial, and is a frequent finding in advanced disease stages.
Limitations of the study are the shortage of proteomic data for C9 and C3. Proteomic data on C9 implicate a medium to low measured abundance compared to apoE in amyloid deposits (). The number of spectra for a protein is a label-free semiquantitative method to determine the abundance of a protein, which has been reviewed elsewhere [Citation45]. Our reviewed literature often included only the top 10 or 20 proteins per analysis with regard to their probability score or number of spectra. Therefore, it is unclear whether C9 spectra numbers’ abundance and/or identity scores were too low for many reported amyloidosis cases or if data on complement components were simply not shown.
In conclusion, this study provides evidence for the specific and spatial enrichment of C9 within diverse types of amyloid. Whether amyloid itself or the disease mechanisms leading to amyloid formation activate the complement cascade, is currently unknown. We occasionally found amyloid-associated, C9 positive cells supporting the contention that amyloid can lead to single cell necrosis. In the past it was shown that the classical pathway of the complement cascade can be activated by SAP via C1q [Citation46] and also by Aβ fibrils under physiological conditions [Citation47]. Furthermore, previous studies proved a hypothesis that stands for soluble oligomers, which are intermediates in amyloid formation and cause metabolic changes, toxicity and cell death [Citation48,Citation49]. Especially annular pore-like oligomers, made by amyloid forming proteins like Aβ or serum amyloid A, demonstrate this channel-like/pore-like activity [Citation50]. The MAC, including its numerous copies of C9, also acts as a transmembrane channel. To find out more about amyloid pathogenesis, a possible association between C9 and amyloid oligomers should be pursued. Therefore, it must be examined if there is an increased incidence of C9 positive cells showing cell death during the different amyloid load and formation stages, e.g. in cell culture experiments or animal models. Additionally, in future studies, it also needs to be investigated whether there is proof of binding mechanisms between C9 and mature amyloid fibrils.
Based on our findings, we propose to introduce C9 as a possible new biomarker for clinical practice and diagnostics of amyloidosis. Thus, this study suggests the subsequent proteomics analyses should also focus on C9.
| Abbreviations | ||
| ApoE | = | apolipoprotein E |
| C3 | = | complement 3 |
| C9 | = | complement 9 |
| Cas3 | = | caspase 3 |
| FFPE | = | formalin-fixed paraffin-embedded |
| LC | = | liquid chromatography |
| LMD | = | laser microdissection |
| MAC | = | membrane attack complex |
| MS/MS | = | tandem mass spectrometry |
| SFA | = | subcutaneous fat aspirate |
Supplemental Material
Download Zip (22.4 MB)Acknowledgements
The authors thank Mrs. Sandra Krüger and Miriam Reutelshöfer for their technical assistance.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349(6):583–596.
- Benson MD, Buxbaum JN, Eisenberg DS, et al. Amyloid nomenclature 2020: update and recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid. 2020;27(4):217–222.
- Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68.
- Howie AJ, Brewer DB. Optical properties of amyloid stained by Congo red: history and mechanisms. Micron. 2009;40(3):285–301.
- Puchtler H, Sweat F, Levine M. On the binding of Congo red by amyloid. J Histochem Cytochem. 1962;10(3):355–364.
- Schönland SO, Hegenbart U, Bochtler T, et al. Immunohistochemistry in the classification of systemic forms of amyloidosis: a systematic investigation of 117 patients. Blood. 2012;119(2):488–493.
- Brambilla F, Lavatelli F, Di Silvestre D, et al. Reliable typing of systemic amyloidoses through proteomic analysis of subcutaneous adipose tissue. Blood. 2012;119(8):1844–1847.
- Vrana JA, Gamez JD, Madden BJ, et al. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood. 2009;114(24):4957–4959.
- Mollee P, Boros S, Loo D, et al. Implementation and evaluation of amyloidosis subtyping by laser-capture microdissection and tandem mass spectrometry. Clin Proteomics. 2016;13:30.
- Namba Y, Tomonaga M, Kawasaki H, et al. Apolipoprotein E immunoreactivity in cerebral amyloid deposits and neurofibrillary tangles in Alzheimer's disease and kuru plaque amyloid in Creutzfeldt-Jakob disease. Brain Res. 1991;541(1):163–166.
- Huang Y, Mahley RW. Apolipoprotein E: structure and function in lipid metabolism, neurobiology, and Alzheimer's diseases. Neurobiol Dis. 2014;72:3–12.
- Wisniewski T, Frangione B. Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid. Neurosci Lett. 1992;135(2):235–238.
- Strittmatter WJ, Weisgraber KH, Huang DY, et al. Binding of human apoE to synthetic amyloid β peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(17):8098–8102.
- Müller-Eberhard HJ. Complement. Annu Rev Biochem. 1969;38:389–414.
- Tegla CA, Cudrici C, Patel S, et al. Membrane attack by complement: the assembly and biology of terminal complement complexes. Immunol Res. 2011;51(1):45–60.
- Podack ER, Müller-Eberhard HJ, Horst H, et al. Membrane attack complex of complement (MAC): three-dimensional analysis of MAC-phospholipid vesicle recombinants. J Immunol. 1982;128(5):2353–2357.
- Esser AF, Kolb WP, Podack ER, et al. Molecular reorganization of lipid bilayers by complement: a possible mechanism for membranolysis. Proc Natl Acad Sci U S A. 1979;76(3):1410–1414.
- Papadimitriou JC, Drachenberg CB, Shin ML, et al. Ultrastructural studies of complement mediated cell death: a biological reaction model to plasma membrane injury. Virchows Arch. 1994;424(6):677–685.
- Jenkins CP, Cardona DM, Bowers JN, et al. The utility of C4d, C9, and troponin T immunohistochemistry in acute myocardial infarction. Arch Pathol Lab Med. 2010;134(2):256–263.
- Jasra SK, Badian C, Macri I, et al. Recognition of early myocardial infarction by immunohistochemical staining with cardiac troponin-I and complement C9. J Forensic Sci. 2012;57(6):1595–1600.
- Pfister F, Vonbrunn E, Ries T, et al. Complement activation in kidneys of patients with COVID-19. Front Immunol. 2020;11:594849.
- Röcken C, Schwotzer EB, Linke RP, et al. The classification of amyloid deposits in clinicopathological practice. Histopathology. 1996;29(4):325–335.
- Lavatelli F, Vrana JA. Proteomic typing of amyloid deposits in systemic amyloidoses. Amyloid. 2011;18(4):177–182.
- UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515.
- Team RC. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2020.
- Wickham H, Averick M, Bryan J, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4(43):1686.
- Wickham H. ggplot2: elegant graphics for data analysis (use R!). Cham: Springer; 2016.
- Kassambra A. 'ggplot2' based publication ready plots. 0.3.0; 2020.]
- D'Souza A, Theis JD, Vrana JA, et al. Pharmaceutical amyloidosis associated with subcutaneous insulin and enfuvirtide administration. Amyloid. 2014;21(2):71–75.
- Garnier S, Ross N, Rudis B, et al. Default color maps from 'matplotlib'. 0.5.1; 2018.
- Sethi S, Theis JD, Vrana JA, et al. Laser microdissection and proteomic analysis of amyloidosis, cryoglobulinemic GN, fibrillary GN, and immunotactoid glomerulopathy. Clin J Am Soc Nephrol. 2013;8(6):915–921.
- Kourelis TV, Dasari SS, Dispenzieri A, et al. A proteomic atlas of cardiac amyloid plaques. JACC Cardiooncol. 2020;2(4):632–643.
- Brambilla F, Lavatelli F, Di Silvestre D, et al. Shotgun protein profile of human adipose tissue and its changes in relation to systemic amyloidoses. J Proteome Res. 2013;12(12):5642–5655.
- Krance SH, Wu C-Y, Zou Y, et al. The complement cascade in Alzheimer's disease: a systematic review and meta-analysis. Mol Psychiatry. 2019.
- Sethi S, Theis JD. Pathology and diagnosis of renal non-AL amyloidosis. J Nephrol. 2018;31(3):343–350.
- Maleszewski JJ, Murray DL, Dispenzieri A, et al. Relationship between monoclonal gammopathy and cardiac amyloid type. Cardiovasc Pathol. 2013;22(3):189–194.
- Michels MAHM, van de Kar NCAJ, Okrój M, et al. Overactivity of alternative pathway convertases in patients with complement-mediated renal diseases. Front Immunol. 2018;9:612.
- Maga TK, Nishimura CJ, Weaver AE, et al. Mutations in alternative pathway complement proteins in American patients with atypical hemolytic uremic syndrome. Hum Mutat. 2010;31(6):1445–1460.
- Sethi S, Fervenza FC, Zhang Y, et al. C3 glomerulonephritis: clinicopathological findings, complement abnormalities, glomerular proteomic profile, treatment, and follow-up. Kidney Int. 2012;82(4):465–473.
- Strassheim D, Renner B, Panzer S, et al. IgM contributes to glomerular injury in FSGS. J Am Soc Nephrol. 2013;24(3):393–406.
- Huang J, Cui Z, Gu Q-H, et al. Complement activation profile of patients with primary focal segmental glomerulosclerosis. PLoS One. 2020;15(6):e0234934.
- Thurman JM, Wong M, Renner B, et al. Complement activation in patients with focal segmental glomerulosclerosis. PLoS One. 2015;10(9):e0136558.
- Boatright KM, Salvesen GS. Mechanisms of caspase activation. Curr Opin Cell Biol. 2003;15(6):725–731.
- Gown AM, Willingham MC. Improved detection of apoptotic cells in archival paraffin sections: immunohistochemistry using antibodies to cleaved caspase 3. J Histochem Cytochem. 2002;50(4):449–454.
- Lundgren DH, Hwang S-I, Wu L, et al. Role of spectral counting in quantitative proteomics. Expert Rev Proteom. 2010;7(1):39–53.
- Bottazzi B, Doni A, Garlanda C, et al. An integrated view of humoral innate immunity: pentraxins as a paradigm. Annu Rev Immunol. 2010;28:157–183.
- Tacnet-Delorme P, Chevallier S, Arlaud GJ. Beta-amyloid fibrils activate the C1 complex of complement under physiological conditions: evidence for a binding site for A beta on the C1q globular regions. J Immunol. 2001;167(11):6374–6381.
- Quist A, Doudevski I, Lin H, et al. Amyloid ion channels: a common structural link for protein-misfolding disease. Proc Natl Acad Sci U S A. 2005;102(30):10427–10432.
- Harper JD, Wong SS, Lieber CM, et al. Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem Biol. 1997;4(2):119–125.
- Hirakura Y, Carreras I, Sipe JD, et al. Channel formation by serum amyloid A: a potential mechanism for amyloid pathogenesis and host defense. Amyloid. 2002;9(1):13–23.

