ABSTRACT
Introduction: Coronaviruses encode a helicase that is essential for viral replication and represents an excellent antiviral target. However, only a few coronavirus helicase inhibitors have been patented. These patents include drug-like compound SSYA10-001, aryl diketo acids (ADK), and dihydroxychromones. Additionally, adamantane-derived bananins, natural flavonoids, one acrylamide derivative [(E)-3-(furan-2-yl)-N-(4-sulfamoylphenyl)acrylamide], a purine derivative (7-ethyl-8-mercapto-3-methyl-3,7-dihydro-1 H-purine-2,6-dione), and a few bismuth complexes. The IC50 of patented inhibitors ranges between 0.82 μM and 8.95 μM, depending upon the assays used. Considering the urgency of clinical interventions against Coronavirus Disease-19 (COVID-19), it is important to consider developing antiviral portfolios consisting of small molecules.
Areas covered: This review examines coronavirus helicases as antiviral targets, and the potential of previously patented and experimental compounds to inhibit the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) helicase.
Expert opinion: Small molecule coronavirus helicase inhibitors represent attractive pharmacological modalities for the treatment of coronaviruses such as SARS-CoV and SARS-CoV-2. Rightfully so, the current emphasis is focused upon the development of vaccines. However, vaccines may not work for everyone and broad-based adoption of vaccinations is an increasingly challenging societal endeavor. Therefore, it is important to develop additional pharmacological antivirals against the highly conserved coronavirus helicases to broadly protect against this and subsequent coronavirus epidemics.
KEYWORDS:
1. Introduction
Helicases are ubiquitous nucleic acid unwinding enzymes. These biological motors couple the chemical energy of nucleotide triphosphate hydrolysis (NTPase) to mechanical energy that translocates through nucleic acids, unwinding the helical structure as it progresses, thus the term ‘helicase.’ Efficient genome replication, recombination and repair require single stranded DNA (ssDNA) or single stranded RNA (ssRNA) as a template that is largely devoid of secondary structures [Citation1]. Helicases in situ generate ssDNA or ssRNA, and due to this crucial role during genome replication, repair and recombination, defects in helicase function can lead to many genetic disorders. Notable examples of helicase-associated disorders include Bloom’s syndrome, Werner’s syndrome, and X-chromosome-linked α-thalassemia [Citation2–9].
Similar to their hosts, viruses rely on nucleic acid unwinding for efficient replication of their genetic material. While many viruses encode helicases, notable exceptions such as HIV-1 do not encode helicases, relying upon host cell helicases to facilitate their replication. Some viruses encode helicase as full-length individual proteins, whereas others encode helicase domains within other functional proteins. All identified CoVs encode a full-length helicase [Citation10–13], whereas other highly pathogenic viruses such as flaviviruses (hepatitis C, dengue and Zika viruses) encode a singular multifunctional protein encoding protease and helicase activities within a single mature protein [Citation14,Citation15].
To date, seven human coronaviruses (hCoVs) have been discovered. These are hCoV-229E, NL63, OC43, HKUI1, Severe Acute Respiratory Syndrome coronavirus (SARS-CoV), Middle East Respiratory Syndrome coronavirus (MERS-CoV), and most recently SARS-CoV-2. Of these, hCoV-229E, NL63, OC43, and HKUI1 cause mild self-limiting respiratory infections, whereas SARS-CoV, MERS-CoV and SARS-CoV-2 cause highly infectious and deadly diseases.
CoVs are large (15–30 kb) positive (+) single-stranded RNA viruses. The polyadenylated and capped RNA genome has open reading frames (ORFs) that encode pp1a and pp1ab through a − 1 ribosomal frameshift during translation [Citation16,Citation17]. These large polyproteins are proteolytically cleaved to generate mature CoV non-structural proteins (nsp) as well as the structural and accessory proteins [Citation10,Citation18–24]. The polyproteins are processed by a virus-encoded papain-like proteinases (PLpro; within nsp3) [Citation25] and nsp5 (3 CLpro) [Citation10,Citation16,Citation19,Citation26–29] to yield up to 16 nsps with diverse functions and up to 10 structural proteins that facilitate viral assembly [Citation30–34]. The singular helicase protein of hCoVs is encoded by nsp13 [Citation12]. nsp13, in concert with additional nsp proteins, assembles into a replication-transcription complex that binds to the 3ʹ untranslated region of the genomic RNA and generates (-) strand RNA as well as subgenomic RNAs [Citation30–34]. A recent review has covered patented inhibitors of different CoV targets [Citation35], here we have focused on the patented inhibitors of nsp13.
2. SARS-CoV-2 life cycle and steps of nsp13 inhibition
SARS-CoV-2 infection initiates with the attachment of the virus through the cell surface receptor ACE2 interacting with the receptor binding domain (RBD) of Spike protein (S-protein). The host protease TMPRSS2 acts synergistically during the initial step of viral attachment to the cell [Citation36] as it cleaves the viral S-protein into S1 and S2, triggering endocytosis of the viral particle (). The pH of the cytoplasm induces endosomal acidification and subsequent release of the viral genome into the cytoplasm. The host ribosomal machinery translates the (+) sense viral RNA genome into polyproteins pp1ab and pp1b. The autocleavage of viral proteases, Mpro and 3Clpro, is followed by the cleavage of pp1ab and pp1b polyproteins into ~16 non-structural proteins (nsps) and ~10 structural proteins. The replicative nsps form a complex that copies the (+) sense viral RNA genome into the (-) sense RNA genome. The nascent (-) sense RNA is then used by a replicase complex comprised of nsps within the double membrane vesicles (DMVs), and generates the (+) sense genomic and subgenomic RNAs. The subgenomic RNAs are translated into the structural proteins that assemble at the Endoplasmic Reticulum Golgi Intermediate Compartment (ERGIC). The N-protein coated RNA genome and structural protein containing cell membrane form the virus like particle (VLP), which egresses from the cell as a mature and infectious virion.
Figure 1. The life-cycle of coronavirus. This figure shows the life-cycle coronaviruses. This figure also shows the steps where the nsp13 inhibitors are expected to exert the inhibitory steps. It is clear from the figure that the nsp13 inhibitors can act at the (+) sense RNA or (-) sense RNA synthesis or at both steps. Since, it is not well defined which step the replicase complex is engulfed within the double membrane vesicles (DMVs), we have shown only one step within the DMVs
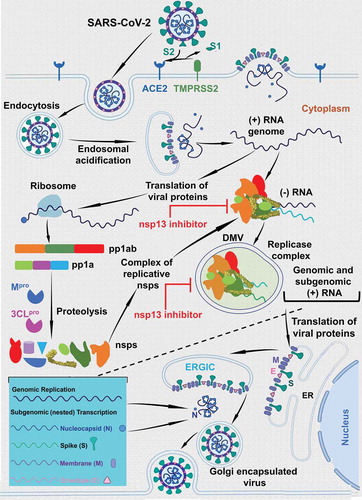
The published reports have shown that nsp13 is essential for replication of the viral genome [Citation12]. nsp13 also interacts with other nsps including nsp7, nsp8, nsp9, nsp12, and nsp14 [Citation37–42]. Recent structural studies have confirmed that nsp13 form complex with nsp7, nsp8, and nsp12 as well as with nsp7, nsp8, nsp9, and nsp12 [Citation36,Citation43]. Additionally, recent data show that SARS-CoV-2 nsp13 interacts with host factors involving in different signaling pathways [Citation44–46]. A definitive role of nsp13 during (-) sense RNA or (+) sense RNA or both steps is still not established. However, SSYA10–001, a patented nsp13 inhibitor, inhibited (+) sense, but not (-) sense, RNA synthesis in a SARS-CoV replicon assays, suggesting that nsp13 is required for the (+) strand synthesis. While it is clear that nsp13 is a pleiotropic factor, more data are needed to establish a definitive role of nsp13 in viral RNA synthesis.
3. Structure-function relationship of CoV helicases
Using conserved sequence motifs, helicases have been classified into six superfamilies (SF1–SF6) [Citation12,Citation47,Citation48]. The nsp13 of hCoV and Mouse Hepatitis Virus (MHV) (a well-studied CoV) belong to the SF1 superfamily [Citation12,Citation48,Citation49]. While, the function of nsp13 CoV during viral replication is not well understood, studies have shown that it is essential for viral replication (see below). Biochemical and molecular biological studies have shown that nsp13 interacts with nsp7, nsp8 and nsp12 [Citation37–40], and a recent cryoEM structure showed that SARS-CoV-2 nsp13 associates with nsp12 when complexed with nsp8 [Citation49]. nsp8 appears to function as an interaction hub for SARS-CoV nsps [Citation39]. Nsp13 is a multifunctional protein that has 5′-3′ directed helicase [Citation12,Citation49–51], ATPase and RNA 5′-triphosphatase activities [Citation52].
The crystal structure of MERS-CoV nsp13 was the first full-length high-resolution structure reported for any hCoV [Citation53]. Subsequent crystal structure of SARS-CoV nsp13 [Citation54] showed that the nsp13s from the two viruses share a high degree of structural similarity. These structures also revealed that nsp13 consists of multiple domains including: a zinc binding domain (ZBD); and an SF1 helicase core connected by the ‘Stalk’ and 1B domains (). The catalytic functions of nsp13 are conducted by the SF1 helicase core formed by two RecA-like domains (RacA1 and RecA2) (). The N-terminal ZBD is also essential for nsp13 function [Citation55] as several ZBD mutations inactivate ATPase and/or helicase activities of equine arteritis virus (EAV) nsp10 (the functional equivalent to SARS-CoV-2 nsp13). Inactivation of ZBD blocks sgRNA (segmented RNA) and genomic RNA synthesis as well as the production of infectious progeny [Citation55,Citation56]. These reports suggest that the ZBD is required for either the structural integrity of nsp13 or for the efficient replication of the virus or both.
Figure 2. Structure of SARS-CoV nsp13. This figure shows the overall structure and domain organization of SARS-CoV nsp13. This figure was generated by PyMol [Citation57]
![Figure 2. Structure of SARS-CoV nsp13. This figure shows the overall structure and domain organization of SARS-CoV nsp13. This figure was generated by PyMol [Citation57]](/cms/asset/0bafbd1d-5bd1-492e-906d-c44890143ed6/ietp_a_1884224_f0002_oc.jpg)
The structures of SARS-CoV, MERS-CoV and SARS-CoV-2 nsp13 not only provide valuable insights into nsp13 macromolecular organization, but also pave the way for identifying potential small molecule inhibitor pockets (). Examples of these pockets, computed by the SiteID program of SYBYL (version X, Certara, St. Louis, MO), are shown as yellow space-filled atoms (). Majority of binding pockets are in the close vicinity of the highly conserved helicase motifs (). This is not surprising since these motifs are supposed to bind ATP and/or RNA substrate. The structure was solved in the absence of substrates, it is possible that the binding sites of these substrates remained unoccupied. Additionally, potential small molecule binding pockets were detected close to K508 and Y277. Importantly, mutations K508A and Y277A conferred resistance to SSYA10–001.
Figure 3. Panel A shows potential small molecule inhibitor binding pockets (yellow space-filled atoms) in the structure of SARS-CoV as determined by SiteID program (Tripos Associates, St. Louis, MO). These binding sites were calculated from the crystal structure of SARS-CoV nsp13 (PDB file 6JYT [Citation47]) after changing selenomethionine residues to methionine. Panel B shows the close up of conserved motifs, computed small molecule binding pockets and SSYA10–001 binding site residues K508 and Y277 (shown as ball-and-sticks). The dotted line represents (42 Å) the distance between two Cα-atoms of Y227 and K508. Residues that interact with K508 from Motifs IV and V are also shown in this panel. The carbon, nitrogen and oxygen atoms are colored as white, blue and red, respectively. This figure was generated by Sybyl X (Certara, St. Louis, MO)
![Figure 3. Panel A shows potential small molecule inhibitor binding pockets (yellow space-filled atoms) in the structure of SARS-CoV as determined by SiteID program (Tripos Associates, St. Louis, MO). These binding sites were calculated from the crystal structure of SARS-CoV nsp13 (PDB file 6JYT [Citation47]) after changing selenomethionine residues to methionine. Panel B shows the close up of conserved motifs, computed small molecule binding pockets and SSYA10–001 binding site residues K508 and Y277 (shown as ball-and-sticks). The dotted line represents (42 Å) the distance between two Cα-atoms of Y227 and K508. Residues that interact with K508 from Motifs IV and V are also shown in this panel. The carbon, nitrogen and oxygen atoms are colored as white, blue and red, respectively. This figure was generated by Sybyl X (Certara, St. Louis, MO)](/cms/asset/b171cac2-97e3-4bc0-b1a6-8409061491d8/ietp_a_1884224_f0003_oc.jpg)
4. CoV helicases as antiviral targets
CoV nsp13 is a highly conserved protein as there is only a single amino acid difference between nsp13s of SARS-CoV (I570) and SRS-CoV-2 (V570). A phylogenetic tree derived from the sequence alignment of nsp13s of seven hCoVs, one bat (RaTG13) and MHV () shows that (i) SARS-CoV-2 is closely related to bat CoV RaTG13 (purple shaded node) as reported previously [Citation58–60], and (ii) all six SF1 helicase motifs are highly conserved. A structural analysis showed that all six conserved motifs are sequestered at the interface of RecA1 and RecA2 domains and form nucleotide/RNA binding sites [Citation53]. Additionally, many potential small molecule binding sites are near RecA1/RecA2 interface (), and within the vicinity of conserved motifs (), it can be easily postulated that (i) CoV nsp13 are attractive antiviral targets and (ii) the compounds binding in the pockets near conserved motifs may serve as pan-coronavirus inhibitors.
Figure 4. Phylogenetic analyses of CoVs and conservation of motifs among selected CoVs. The dendrogram shows the evolutionary relationship among hCoVs, MHV and RaTG13 using amino acid sequence of nsp13. Note that SARS-VoV-2 is closely related to RaTG13 (purple highlighted node). This figure also shows the sequence alignment of six conserved nsp13 motifs together with SSYA10–001 binding site residues marked as asterisks (*). The amino acid sequence is color coded. Dash (–) represents the gap in the sequences of nsp13, and AA represents amino acids. The figure was generated using the R package ggtree [Citation61]. The R programing language [Citation62] was used to generate this figure, . The in-house generated R codes for these figures are available upon request
![Figure 4. Phylogenetic analyses of CoVs and conservation of motifs among selected CoVs. The dendrogram shows the evolutionary relationship among hCoVs, MHV and RaTG13 using amino acid sequence of nsp13. Note that SARS-VoV-2 is closely related to RaTG13 (purple highlighted node). This figure also shows the sequence alignment of six conserved nsp13 motifs together with SSYA10–001 binding site residues marked as asterisks (*). The amino acid sequence is color coded. Dash (–) represents the gap in the sequences of nsp13, and AA represents amino acids. The figure was generated using the R package ggtree [Citation61]. The R programing language [Citation62] was used to generate this figure, Figures 6 and 7. The in-house generated R codes for these figures are available upon request](/cms/asset/cf8f57f8-aca4-4939-8647-3ecbf2d96ee6/ietp_a_1884224_f0004_oc.jpg)
5. Patented nsp13 inhibitors
5.1. SSYA10–001
One of the most characterized CoV nsp13 patented inhibitor is SSYA10–001 (US20140005241A1; 2016). SSYA10–001 is a 1,2,4-triazole compound () that was identified through a screening of a library of drug-like compounds using fluorescence resonance energy transfer (FRET) based helicase assay [Citation63]. SSYA10–001 functionally inhibited helicase and ATPase activities of SARS-CoV nsp13 in a noncompetitive manner with respect to ATP and nucleic acid substrate [Citation63]. Subsequent cell-culture assays showed that SSYA10–001 inhibits SARS-CoV, MERS-CoV and MHV with comparable IC50 [Citation64]. Therefore, it was termed as a pan-CoV inhibitor [Citation64]. To identify compounds that share structural and physico-chemical properties of SSYA10–001, we conducted a PubChem search using SSTA10–001 as template. This search resulted in the identification of seven compounds (). Below we discuss structural and physico-chemical properties of these compounds with respect to SSYA10–001, however, they are yet to be tested for anti-CoV activity.
Figure 5. Structure of SSYA10–001 and related compounds. The compounds were searched from PubChem database using SSYA10–001 as template. The compounds were scored using Tanimoto fingerprints [Citation66]
![Figure 5. Structure of SSYA10–001 and related compounds. The compounds were searched from PubChem database using SSYA10–001 as template. The compounds were scored using Tanimoto fingerprints [Citation66]](/cms/asset/17f8fdc6-048f-499d-af91-a60e6e6b9ee4/ietp_a_1884224_f0005_b.gif)
Table 1. List of compounds in PubChem sharing structural similarity with SSYA10–001
Clustering of compounds by structural or property similarity is a powerful approach for correlating compound features with biological activities. This technique also provides a glimpse of structure activity relationship when combined with binding/inhibition of the target protein. A heatmap derived from the structure-based hierarchical clustering of compounds in generated by ChemMine Tools [Citation65] is shown in . To generate structure-based clustering of these compounds, the atom pair descriptors (features) for each compound are generated and used to calculate a similarity matrix based on the common and unique features observed among all compound pairs using the Tanimoto coefficient [Citation66]. The Tanimoto coefficient ranges between 0 and 1 with a higher value representing greater similarity. In subsequent clustering steps, the similarity matrix is converted into a distance matrix (Z-Score) by subtracting the Tanimoto similarity values from 1. The Z-scores were finally used to clustering the compounds (). From this heatmap, it is clear that compound CPD-850 () is most dissimilar to the other compounds, while CPD-815 and CPD-062 are very similar with respect to structural properties, and a high degree of similarity exists between SSYA10–001, CPD-815 and CPD-062 ().
Figure 6. A structural property comparison of SSYA10-100 and related compounds. This heatmap and dendrogram show the relationship among SSYA10–001 and seven compounds identified from PubChem. ChemMine Tool [Citation65] and in-house R code (available upon request) was used to generate structural relationships among the compounds. Similarity scale (range of Z-scores) shown at the top of figure refers to as the most similar structure being blue or zero Z-score, and most dissimilar structure as red or Z-score equal to 1
![Figure 6. A structural property comparison of SSYA10-100 and related compounds. This heatmap and dendrogram show the relationship among SSYA10–001 and seven compounds identified from PubChem. ChemMine Tool [Citation65] and in-house R code (available upon request) was used to generate structural relationships among the compounds. Similarity scale (range of Z-scores) shown at the top of figure refers to as the most similar structure being blue or zero Z-score, and most dissimilar structure as red or Z-score equal to 1](/cms/asset/73e96456-0cfb-437c-a772-64b5b1c31b16/ietp_a_1884224_f0006_oc.jpg)
A heatmap generated using physicochemical property-based clustering of compounds in is shown in . Selected Open Babel descriptors [Citation67] (HBD, number of H-bond donors; HBA, number of H-bond acceptors; ArBD, aromatic bonds; and MW, molecular weight) were used to generate the heatmap and dendrogram using the ChemMineR package (Cheminformatics Toolkit for R) [Citation68] (). This heatmap highlights the correlations between the compounds in . For example, ‘ArBD’ (aromatic bond) feature reveals that 6 out of 8 compounds have comparable aromatic bonds, a measure of hydrophobicity. Additionally, the computed MW and logP of compounds is closely related. The dendrogram at the top of the figure shows that the hydrophobicity of the compounds is closely related to the molecular weight of the compounds. Additionally, horizontal dendrograms show that the identified compounds can be grouped, and prioritized for experimental evaluation. For example, CPD-241 is closely related to SSYA10–001, although CPD-241 is significantly hydrophobic (as seen from logP) compared to SSYA10–001. To get better insight about the solubility of all compounds compared to SSYA10–001, the plotted logP values are shown in . It is clear that except CPD-241, each compound has a better or comparable solubility profile compared to SSYA10–001. Additionally, the docking of these compounds at the two possible binding sites in the structure of nsp13 coupled with the experimental data can provide SAR strategies to develop more potent inhibitors.
Figure 7. A comparison of physico-chemical properties of SSYA10–100 and related compounds. Panel A shows the comparison of physico-chemical properties of SSYA10–001 with the compounds in . The physico-chemical properties were determined using Open Babel chemical toolbox [Citation67]. These properties include H-bond donor (HBD), aromatic bonds (ArBD), H-bond acceptors (HBA), and molecular weight (MW). For simplicity, Open Babel descriptors HBA1 and HBA2 were added and represented as HBA. Additionally, Open Babel descriptor ‘abonds’ was changed to ‘ArBD’ for clarity. Panel B shows the bar plot of computed octanol/water partition coefficient (logP) of the compounds. The scale, height and the color of the bar represent the range of logP. This figure was generated using an in-house R code (available upon request)
![Figure 7. A comparison of physico-chemical properties of SSYA10–100 and related compounds. Panel A shows the comparison of physico-chemical properties of SSYA10–001 with the compounds in Table 1. The physico-chemical properties were determined using Open Babel chemical toolbox [Citation67]. These properties include H-bond donor (HBD), aromatic bonds (ArBD), H-bond acceptors (HBA), and molecular weight (MW). For simplicity, Open Babel descriptors HBA1 and HBA2 were added and represented as HBA. Additionally, Open Babel descriptor ‘abonds’ was changed to ‘ArBD’ for clarity. Panel B shows the bar plot of computed octanol/water partition coefficient (logP) of the compounds. The scale, height and the color of the bar represent the range of logP. This figure was generated using an in-house R code (available upon request)](/cms/asset/02554bd3-416c-4b57-9f75-5cfe26256ce9/ietp_a_1884224_f0007_oc.jpg)
5.2. Dihydroxychromone and aryl diketoacids
Two Korean patents include compounds based on 3, 5-dihydroxychromone and aryl diketoacids () (KR20100029528A; 2010 and KR20110006083A; 2011). These patents arose from the identification of natural flavonoids myricetin, scutellarein and quercetin as potent inhibitors of SARS-CoV nsp13 [Citation69,Citation70] (). The compounds described in these patents contain common scaffold 3, 5-dihydroxychromone. Using this scaffold, modified quercetin derivatives were developed that showed anti-nsp13 activity. Modifications R1 = R2 = H and R3 = 3-CN-Benzyl, and R3 = 4-Cl-Benzyl were the most potent nsp13 inhibitors with IC50 ~ 3.0 µM.
Figure 8. Structures of naturally occurring flavonoids and patented 3,5-dihydroxychromone compounds. Green shaded oval represents dihydroxychromone chemical group, which was used in the patents (KR20110006083A; 2011)
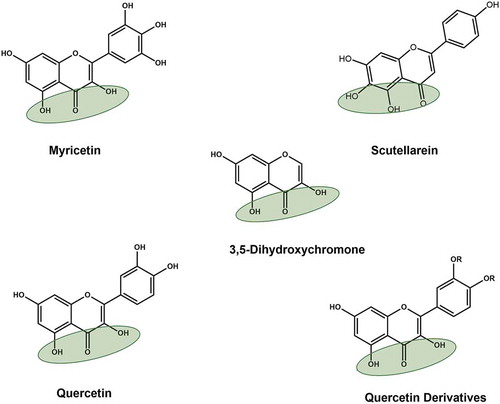
Figure 9. Structural comparison of diketoacid group containing compounds. A well-known HIV-1 integrase inhibitors dolutegravir, which has a diketoacid group is included as a reference compound. The other two structures represent patented inhibitors of SARS-CoV nsp13 inhibitors. 1-ADK refers to a patented compound 1 containing ADK group (KR20110006083A; 2011)
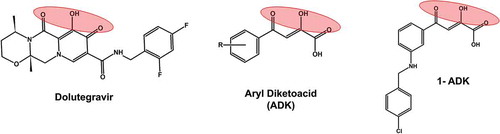
The diketoacids are known to chelate two divalent cations, and thereby inhibit enzymes that use metal ions (such as Mg2+ and Mn2+) as cofactors. For example, all approved HIV-1 integrase inhibitors are diketoacid compounds (e.g., Dolutegravir, ) [Citation71]. The inventors of Korean patent (KR20100029528A; 2010) exploited the divalent cation chelating property of diketoacids and developed a series of compounds that inhibited nsp13 in in vitro assays. The most potent inhibitor was a compound with R1 = H, R2 = NHCH2(4-Cl-phenol), and R3 = H (1-ADK, ). This compound had an IC50 = 0.96 μM, and EC50 = 0.82 μM against SARS-CoV.
5.3. Mechanism of SSYA10–001, dihydroxychromone and aryl diketoacids inhibition of nsp13
The SSYA10–001 mechanism of nsp13 inhibition, the functional impact of mutations within the predicted SSYA10–001 binding site was examined. Two nsp13 mutations, Y277A and K508A, conferred resistance to SSYA10–001, as their estimated IC50s for helicase function were 12 and 50 μM, respectively, compared to 5.9 μM for WT SARS-CoV nsp13 [Citation64]. Based on these results and the computational modeling, it was concluded that Y277 and K508 were part of the binding pocket for SSYA10–001 within SARS-CoV nsp13 [Citation64]. However, the crystal structure of SARS nsp13 showed that the two residues (Y277 and K508) are nearly 42 Å apart (), suggesting that the two residues are not part of the same binding pocket. Hence, the precise SSYA10–001 binding pocket remains unknown. It is possible that SYA10–001 (i) binds at one site and exerts a long-range effect on the other site or (ii) there exist two binding sites for SSYA10–001.
In the crystal structure of SARS-CoV nsp13 (PDB file 6JYT [Citation54]), K508 is exposed to the solvent and does not appear to contribute to the nearest small-molecule binding pocket. However, K508 forms a salt-bridge with D542 (motif IV), which in turn forms a salt-bridge with K569 (motif V) (). Mutation K508A may disrupt the positions of D542 and K569. Importantly, D542 is next to Y543, whereas K569 is next to I570. Both residues (Y543 and I570) hydrophobically interact with W506, which is part of the proposed SSYA10–001 binding pocket [Citation64]. Hence, it appears that the resistance to SSYA10–001 due to K508A mutation is an indirect effect structural alteration in its vicinity. Mutation Y277A is resistant to SSYA10–001 although ~twofold less compared to K508A. The SiteID program of Sybyl X (Certara, St. Louis, MO) predicted a small pocket near Y277. However, the size of this pocket appeared too small to bind SSYA10–001. At this point, the structure nsp13 does not provide a plausible mechanism for SSYA10–001 resistance of Y277A. A crystal structure of nsp13/SSYA10–001 is needed to resolve this issue. The molecular mechanism of inhibition of other patented nsp13 inhibitors (aryl diketoacid and 3,5-dihydroxychromone) has not been investigated and the binding sites for these inhibitors are currently unknown. However, based on the metal binding properties of these compounds, it is quite possible that these compounds bind at or near motifs I and II.
6. Miscellaneous nsp13 inhibitors
In addition to above mentioned patented nsp13 inhibitors, several compounds that inhibited CoV nsp13 activities have been reported [Citation69,Citation70,Citation72–80]. These include adamantane-derived bananins, bismuth complexes, thioxopyrimidine derivatives, an acrylamide derivative [(E)-3-(furan-2-yl)-N-(4-sulfamoylphenyl)acrylamide], a purine derivative (7-ethyl-8-mercapto-3-methyl-3,7-dihydro-1 H-purine-2,6-dione), and RNA aptamers [Citation81]. Of all tested adamantane-derived bananins, bananin was the most potent with an IC50 of 2.3 µM and 3.0 µM for ATPase and helicase activities of SARS-CoV nsp13, respectively [Citation66]. An acrylamide derivative [(E)-3-(furan-2-yl)-N-(4-sulfamoylphenyl)acrylamide] showed an IC50 for ATPase activity comparable to bananin, but the IC50 for helicase activity was 4.4-fold greater (13.0 µM) [Citation72]. The inhibitory profile of a purine derivative (7-ethyl-8-mercapto-3-methyl-3,7- dihydro-1 H-purine-2,6-dione) was poorer than bananin (ATPase IC50 = 8.66 ± 0.26 μM and helicase IC50 = 41.6 ± 2.3 μM) [Citation73]. Some of the bismuth complexes had IC50 values for ATPase activity of SARS-CoV nsp13 less than 1 µM and IC50 values for helicase activity less than 3 µM [Citation75]. However, these compounds have not been pursued since their discovery in 2007 until very recently it was shown that they inhibit SARS-CoC-2 nsp13 [Citation82]. Very recently, another compound N‐(4‐((4,6‐dioxo‐2‐thioxotetrahydropyrimidin‐5(2 H)‐ylidene)methyl)phenyl)acetamide showed good potency for the inhibition of ATPase activity of SARS-CoV nsp13 (IC50 = 1.19 ± 0.16 μM) [Citation80]. However, the IC50 (32.9 ± 1.0 μM) for helicase function was similar to that of 7-ethyl-8-mercapto-3-methyl-3,7- dihydro-1 H-purine-2,6-dione [Citation80].
7. Expert opinion
Here, we present a description and analysis of the currently patented inhibitors of nsp13 as potential small molecule inhibitors of SARS-CoV or MERS-CoV. Considering the high sequence homology between nsp13 of SARS-CoV-2 and SARS-CoV (98.9%), we suggest that it is extremely likely that the compounds discussed in the review will inhibit SARS-CoV-2 nsp13. Mechanistically, among the patented molecules, SSYA10–001 is the best characterized nsp13 inhibitor. The existing analogs of SSYA10–001 have not been tested for efficacy against SARS-CoV or SARS-CoV-2 nsp13 inhibition, however, since they are structurally related and may bind the same nsp13 region, it is possible that one or more analogs may show better efficacy than SSYA10–001.
Despite the critical role of helicases, drug-discovery against this class of enzymes has lagged behind when compared to other viral targets, including nucleic acid polymerases. However, the clinical success of herpes simplex virus (HSV) helicase/primase inhibitors Amenamevir (ASP2151) and Pritelivir (AIC316) demonstrates the potential of helicases as small molecule antiviral targets [Citation83–85]. Following the encouraging results of clinical trials of Pritelivir [Citation86], the Food and Drug Administration (FDA) approved this compound for the treatment of acyclovir-resistant mucocutaneous herpes simplex virus (HSV) infections in immunocompromised adults [Citation87]. Efforts have been made to identify the inhibitors of helicase from other viruses as reported in prior reviews [Citation88,Citation89]. For example, the hepatitis C virus NS3 helicase is an attractive target for small molecule antiviral compounds [Citation90] as several lead compounds have been identified. One class of compounds, triazoles, inhibit HCV helicase activity and impair cellular HCV RNA replication and appear to hold immense potential. Unfortunately, these compounds failed to enter the clinic.
Due to their essential function(s) during the viral life cycle, helicases are clearly attractive antiviral targets regardless of the pathogen. While some helicase-targeting small molecules inhibitors have entered clinical trials, drug development against helicases remains challenging. One of these challenges is to design specific nsp13 inhibitors that compete with the natural substrate (ATP) and bind at the ATP binding site. As with many viral functional proteins and enzymes, the ATPase function of nsp13 is common to a broad array of cellular enzymes. Therefore, not only is binding essential, but substrate specificity amongst a sea of similar substrates adds an additional layer of complexity. For example, protein phosphatase 2A (PP2A), which is involved in many cellular functions, also conducts ATP hydrolysis by binding ATP through two metal ions [Citation91]. Hence, any inhibitor that chelates metal ions can bind PP2A-like enzymes resulting into adverse toxicity profiles. Additionally, a two metal ions mechanism originally proposed for the 3ʹ – 5ʹ exonuclease function of E. coli DNA polymerase I [Citation92] is used by many cellular polymerases and primases. Therefore, the compounds targeting ATPase function of nsp13 through metal chelating property can potentially bind to these enzymes and interfere normal cellular function. This type of challenge is not unique to drugs targeting viral proteins/enzymes, as similar issues emerged during the discovery of anti-HIV compounds. Several nucleoside reverse transcriptase inhibitors (NRTIs) inhibit polymerase γ, a mitochondrial DNA polymerase and exert mitochondrial toxicity [Citation93–96].
The structures of viral components such as nucleic acid polymerases, proteases and integrases have helped overcome the specificity challenges. For example, prior to the structure of HIV-1 integrase being solved, the structures of Prototype foamy virus integrase complexed with DNA was used as a molecular scaffold to identify potent integrase inhibitors [Citation96–98].
Currently patented nsp13 inhibitors have limitations. First, the potency of these compounds is in the micromolar range. Second, the pharmacokinetics data for all patented compounds are not known. And most importantly, ADK (aryl diketo acid) and dihydroxychromones contain chemical moieties that can inhibit the function of many cellular enzymes as they can potentially scavenge metal ions required for many cellular enzymes. These limitations can be overcome by the combination of structure-based drug design techniques, medicinal chemistry, and virological assays. Thus, the reported structures of SARS-CoV [Citation54] and MERS-CoV [Citation53] and recently reported structure of SARS-CoV-2 in complex with nsp12, nsp7 and nsp8 [Citation49] as well as the structures of helicases from other CoVs [Citation99,Citation100] provide important opportunities to design specific CoV inhibitors. The resistance profile, most likely obtained through the virus passage assays can aid in the identification of inhibitor binding site residues that can be used to exploit the structures of nsp13. Such strategies have been previously used to design non-nucleoside reverse transcriptase inhibitors (NNRTIs) and the mechanism of their resistance [Citation101–105]. Additional challenges in designing nsp13 inhibitors is the constant evolution of SARS-CoV-2 virus. The diversity in viral genome is known to compromise the efficacy of antiviral drugs [Citation106,Citation107]. Continuously evolving SARS-CoV-2 including two mutations (P504L and Y541C) [Citation108] can also limit the efficacy patented and investigational nsp13 inhibitors.
Acknowledgment
KS, FG and TPQ acknowledge the computational facilities of the Molecular Interactions Core, University of Missouri, Columbia, MO 65211.
Article highlights
Overview of currently patented coronavirus helicase inhibitors.
Overview of other compounds that inhibit nsp13.
Inclusion of compounds that reflect structure activity relationship (SAR) of patented compound SSYA10–001.
Overview of a strategy to identify new compounds related to the patented inhibitors.
Challenges in designing nsp13 specific antiviral compounds.
Expert opinion on future directions of the field. We suggested that SAR can be used to identify the compounds that may be better inhibitors of nsp13 than the patented compounds.
This box summarizes key points contained in the article.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Declaration of interest
K Singh serves as a consultant for Sanctum Therapeutics Corporation, Sunnyvale, CA, USA and has served as a consultant for Shift Pharmaceuticals, Overland Park, KS, USA. CL Lorson is the co-founder and CSO of Shift Pharmaceuticals, Overland Park, KS, USA that has licensed SSYA10-001. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Additional information
Funding
References
- Patel SS, Picha KM Structure and function of hexameric helicases. Annu Rev Biochem. 2000, 69, 651–697.
- Ellis NA, Groden J, Ye TZ, et al. The bloom’s syndrome gene product is homologous to RecQ helicases. Cell 1995, 83, 655–666.
- Karow JK, Chakraverty RK, Hickson ID The Bloom’s syndrome gene product is a 3ʹ-5ʹ DNA helicase. J Bio Chem. 1997, 272, 30611–30614.
- Yu CE, Oshima J, Fu YH, et al. Positional cloning of the Werner’s syndrome gene. Science 1996, 272, 258–262.
- Shen JC, Gray MD, Oshima J, et al. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998, 26, 2879–2885.
- Gray MD, Shen JC, Kamath-Loeb AS, et al. The Werner syndrome protein is a DNA helicase. Nat Genet. 1997, 17, 100–103.
- Nakura J, Ye L, Morishima A, et al. Helicases and aging. Cell Mol Life Sci. 2000, 57, 716–730.
- Cunniff C, Bassetti JA, Ellis NA Bloom’s syndrome: clinical spectrum, molecular pathogenesis, and cancer predisposition. Mol Syndromol. 2017, 8, 4–23.
- Luo J Wrn protein and Werner syndrome. N Am J Med Sci (Boston) 2010, 3, 205–207.
- Snijder EJ, Bredenbeek PJ, Dobbe JC, et al. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J Mol Biol. 2003, 331, 991–1004.
- Gorbalenya AE, Enjuanes L, Ziebuhr J, et al. Nidovirales: evolving the largest RNA virus genome. Virus Res. 2006, 117, 17–37.
- Ivanov KA, Thiel V, Dobbe JC, et al. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J Virol. 2004, 78, 5619–5632.
- Gorbalenya AE, Snijder EJ, Spaan WJ severe acute respiratory syndrome coronavirus phylogeny: toward consensus. J Virol. 2004, 78, 7863–7866.
- Frick DN Step-by-step progress toward understanding the hepatitis C virus RNA helicase. Hepatology 2006, 43, 1392–1395.
- Frick DN Helicases as antiviral drug targets. Drug News Perspect 2003, 16, 355–362.
- Rota PA, Oberste MS, Monroe SS, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399.
- Marra MA, Jones SJ, Astell CR, et al. The genome sequence of the SARS-associated coronavirus. Science 2003, 300, 1399–1404.
- Herold J, Siddell S, Ziebuhr J Characterization of coronavirus RNA polymerase gene products. Methods Enzymol. 1996, 275, 68–89.
- Thiel V, Ivanov KA, Putics A, et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003, 84, 2305–2315.
- Ziebuhr J The coronavirus replicase. Curr Top Microbiol Immunol. 2005, 287, 57–94.
- Ziebuhr J Molecular biology of severe scute respiratory syndrome coronavirus. Curr Opin Microbiol 2004, 7, 412–419.
- Putics A, Filipowicz W, Hall J, et al. ADP-ribose-1”-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J Virol. 2005, 79, 12721–12731.
- Perlman S, Netland J Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 2009, 7, 439–450.
- Navas-Martin SR, Weiss S Coronavirus replication and pathogenesis: implications for the recent outbreak of Severe Acute respiratory syndrome (SARS), and the challenge for vaccine development. J Neurovirol. 2004, 10, 75–85.
- Harcourt BH, Jukneliene D, Kanjanahaluethai A, et al. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J Virol. 2004, 78, 13600–13612.
- Ziebuhr J, Snijder EJ, Gorbalenya AE Virus-encoded proteinases and proteolytic processing in the nidovirales. J Gen Virol. 2000, 81, 853–879.
- Fan K, Wei P, Feng Q, et al. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J Biol Chem. 2004, 279, 1637–1642.
- Anand K, Ziebuhr J, Wadhwani P, et al. Coronavirus main proteinase (3Clpro) structure: basis for design of anti-SARS drugs. Science 2003, 300, 1763–1767.
- Yang H, Yang M, Ding Y, et al. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc Natl Acad Sci U S A 2003, 100, 13190–13195.
- Bernini A, Spiga O, Venditti V, et al. Tertiary structure prediction of SARS coronavirus helicase. Biochem Biophys Res Commun. 2006, 343, 1101–1104.
- Minskaia E, Hertzig T, Gorbalenya AE, et al. Discovery of an RNA virus 3ʹ->5ʹ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc Natl Acad Sci U S A 2006, 103, 5108–5113.
- Ricagno S, Egloff MP, Ulferts R, et al. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci U S A 2006, 103, 11892–11897.
- Bhardwaj K, Sun J, Holzenburg A, et al. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. J Mol Biol. 2006, 361, 243–256.
- Sawicki SG, Sawicki DL, Siddell SG A contemporary view of coronavirus transcription. J Virol. 2007, 81, 20–29.
- Kumar V, Jung YS, Liang PH Anti-SARS coronavirus agents: a patent review (2008 – present). Expert Opin Therap Patents 2013, 23, 1337–1348.
- Yan L, Ge J, Zheng L, et al. Cryo-Em structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell 2021, 184, 184–193 e110.
- von Brunn A, Teepe C, Simpson JC, et al. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One 2007, 2, e459.
- Pan J, Peng X, Gao Y, et al. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One 2008, 3, e3299.
- Prentice E, McAuliffe J, Lu X, et al. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J Virol. 2004, 78, 9977–9986.
- Imbert I, Snijder EJ, Dimitrova M, et al. The SARS-coronavirus plnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 2008, 133, 136–148.
- Adedeji AO, Marchand B, Te Velthuis AJ, et al. Mechanism of nucleic acid unwinding by SARS-CoV helicase. PLoS One 2012, 7, e36521.
- Romano M, Ruggiero A, Squeglia F, et al. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 2020, 9. 9
- Yan L, Zhang Y, Ge J, et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat Commun. 2020, 11, 5874.
- Xia H, Cao Z, Xie X, et al. Evasion of type i interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234.
- Appelberg S, Gupta S, Svensson Akusjarvi S, et al. Dysregulation in akt/mtor/hif-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg Microbes Infect. 2020, 9, 1748–1760.
- Yuen CK, Lam JY, Wong WM, et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020, 9, 1418–1428.
- Singleton MR, Wigley DB Modularity and specialization in superfamily 1 and 2 helicases. J Bacteriol. 2002, 184, 1819–1826.
- Gorbalenya AE, Koonin EV Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 1989, 17, 8413–8440.
- Chen J, Malone B, Llewellyn E, et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 2020, 182, 1560–1573 e1513.
- Lee NR, Kwon HM, Park K, et al. Cooperative translocation enhances the unwinding of duplex DNA by SARS coronavirus helicase nsp13. Nucleic Acids Res. 2010, 38, 7626–7636.
- Tanner JA, Watt RM, Chai YB, et al. The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5ʹ to 3ʹ viral helicases. J Biol Chem 2003, 278, 39578–39582.
- Flores MJ, Sanchez N, Michel B A fork-clearing role for uvrd. Mol Microbiol. 2005, 57, 1664–1675.
- Hao W, Wojdyla JA, Zhao R, et al. Crystal structure of Middle East respiratory syndrome coronavirus helicase. PLoS Pathog 2017, 13, e1006474.
- Jia Z, Yan L, Ren Z, et al. Delicate structural coordination of the severe acute respiratory syndrome coronavirus nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019, 47, 6538–6550.
- Seybert A, Posthuma CC, van Dinten LC, et al. A complex zinc finger controls the enzymatic activities of nidovirus helicases. J Virol. 2005, 79, 696–704.
- van Dinten LC, van Tol H, Gorbalenya AE, et al. The predicted metal-binding region of the arterivirus helicase protein is involved in subgenomic mRNA synthesis, genome replication, and virion biogenesis. J Virol. 2000, 74, 5213–5223.
- DeLano WL An open-source molecular graphics tool. Ccp4 Newsl Protein Crystallogr 2002, 40, 82–92.
- Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273.
- Boni MF, Lemey P, Jiang X, et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nature Microbiol. 2020, 5, 1408–1417.
- Neogi U, Hill KJ, Ambikan AT, et al. Feasibility of known RNA polymerase inhibitors as anti-SARS-CoV-2 drugs. Pathogens 2020, 9. 9
- Yu G, Lam TT, Zhu H, et al. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol. 2018, 35, 3041–3043.
- Team RC R: a language and environment for statistical computing. R foundation for statistical computing, vienna, austria. Isbn 3–900051–07–0. http://www.R-project.org 2013.
- Adedeji AO, Singh K, Calcaterra NE, et al. Severe Acute Respiratory Syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase. Antimicrob Agents Chemother. 2012, 56, 4718–4728.
- Adedeji AO, Singh K, Kassim A, et al. Evaluation of SSYA10–001 as a replication inhibitor of severe acute respiratory syndrome, mouse hepatitis, and Middle East respiratory syndrome coronaviruses. Antimicrob Agents Chemother. 2014, 58, 4894–4898.
- Backman TW, Cao Y, Girke T Chemmine tools: an online service for analyzing and clustering small molecules. Nucleic Acids Res 2011, 39, W486–491.
- Rogers DJ, Tanimoto TT A computer program for classifying plants. Science 1960, 132, 1115–1118.
- O’Boyle NM, Banck M, James CA, et al. Open BABEL: an open chemical toolbox. J Chem inform. 2011, 3, 33.
- Cao Y, Charisi A, Cheng LC, et al. Chemminer: a compound mining framework for R. Bioinformatics 2008, 24, 1733–1734.
- Yu MS, Lee J, Lee JM, et al. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsp13. Bioorg Med Chem Lett. 2012, 22, 4049–4054.
- Keum YS, Jeong YJ Development of chemical inhibitors of the SARS coronavirus: viral helicase as a potential target. Biochem Pharmacol. 2012, 84, 1351–1358.
- Di Santo R, Fermeglia M, Ferrone M, et al. Simple but highly effective three-dimensional chemical-feature-based pharmacophore model for diketo acid derivatives as hepatitis C virus RNA-dependent RNA polymerase inhibitors. J Med Chem. 2005, 48, 6304–6314.
- Lee JM, Cho JB, Ahn HC, et al. A novel chemical compound for inhibition of SARS coronavirus helicase. J Microbiol Biotechnol. 2017, 27, 2070–2073.
- Cho JB, Lee JM, Ahn HC, et al. Identification of a novel small molecule inhibitor against SARS coronavirus helicase. J Microbiol Biotechnol. 2015, 25, 2007–2010.
- Tanner JA, Zheng BJ, Zhou J, et al. The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chem Biol. 2005, 12, 303–311.
- Yang N, Tanner JA, Wang Z, et al. Inhibition of SARS coronavirus helicase by bismuth complexes. Chem Commun (Camb) 2007, 4413–4415.
- Lee C, Lee JM, Lee NR, et al. Investigation of the pharmacophore space of severe acute respiratory syndrome coronavirus (SARS-CoV) ntpase/helicase by dihydroxychromone derivatives. Bioorg Med Chem Lett. 2009, 19, 4538–4541.
- Shum KT, Tanner JA Differential inhibitory activities and stabilisation of DNA aptamers against the SARS coronavirus helicase. Chembiochem. 2008, 9, 3037–3045.
- Yang N, Tanner JA, Zheng BJ, et al. Bismuth complexes inhibit the SARS coronavirus. Angew Chem Int Ed Eng. 2007, 46, 6464–6468.
- Lee C, Lee JM, Lee NR, et al. Aryl diketoacids (adk) selectively inhibit duplex DNA-unwinding activity of SARS coronavirus NTPase/helicase. Bioorg Med Chem Lett. 2009, 19, 1636–1638.
- Lee JM, Cho JB, Ahn HC, et al. Selective inhibition of enzymatic activities of severe acute respiratory syndrome coronavirus helicase with a thioxopyrimidine derivative. Bull Korean Chem Soc. 2016, 37, 2066–2068.
- Jang KJ, Lee NR, Yeo WS, et al. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase. Biochem Biophys Res Commun. 2008, 366, 738–744.
- Shu T, Huang M, Wu D, et al. SARS-coronavirus-2 nsp13 possesses ntpase and RNA helicase activities that can be inhibited by bismuth salts. Virol Sin. 2020, 35, 321–329.
- Chono K, Katsumata K, Kontani T, et al. Asp2151, a novel helicase-primase inhibitor, possesses antiviral activity against varicella-zoster virus and herpes simplex virus types 1 and 2. J Antimicrob Chemother. 2010, 65, 1733–1741.
- Katsumata K, Weinberg A, Chono K, et al. Susceptibility of herpes simplex virus isolated from genital herpes lesions to Asp2151, a novel helicase-primase inhibitor. Antimicrob Agents Chemother. 2012, 56, 3587–3591.
- Tyring S, Wald A, Zadeikis N, et al. Asp2151 for the treatment of genital herpes: a randomized, double-blind, placebo- and valacyclovir-controlled, dose-finding study. J Infect Dis. 2012, 205, 1100–1110.
- Wald A, Corey L, Timmler B, et al. Helicase-primase inhibitor pritelivir for HSV-2 infection. N Eng J Med. 2014, 370, 201–210.
- Aicuris. A double-blind randomized placebo controlled dose-finding trial to investigate different doses of a new antiviral drug in subjects with genital HSV type 2 infection. Clin trial. 2011; NCT01047540. available from: http://clinicaltrials.Gov/ct2/results?Term=nct01047540&
- Weller SK, Kuchta RD The DNA helicase-primase complex as a target for herpes viral infection. Expert Opin Ther Targets 2013, 17, 1119–1132.
- Datta A, Brosh RM Jr. New insights into DNA helicases as druggable targets for cancer therapy. Front Mol Biosci. 2018, 5, 59.
- Belon CA, Frick DN Helicase inhibitors as specifically targeted antiviral therapy for hepatitis C. Future Virol. 2009, 4, 277–293.
- Guo F, Stanevich V, Wlodarchak N, et al. Structural basis of PP2A activation by PTPA, an ATP-dependent activation chaperone. Cell Res. 2014, 24, 190–203.
- Beese LS, Steitz TA Structural basis for the 3ʹ-5ʹ exonuclease activity of Escherichia coli DNA polymerase i: a two metal ion mechanism. Embo J. 1991, 10, 25–33.
- Copeland WC The mitochondrial DNA polymerase in health and disease. Subcell Biochem. 2010, 50, 211–222.
- Lewis W, Simpson JF, Meyer RR Cardiac mitochondrial DNA polymerase-gamma is inhibited competitively and noncompetitively by phosphorylated zidovudine. Circ Res. 1994, 74, 344–348.
- Lewis W, Day BJ, Copeland WC Mitochondrial toxicity of NRTI antiviral drugs: an integrated cellular perspective. Nat Rev Drug Discov. 2003, 2, 812–822.
- Engelman A, Cherepanov P Retroviral integrase structure and DNA recombination mechanism. Microbiol Spectr. 2014, 2, 1–22.
- Maskell DP, Renault L, Serrao E, et al. Structural basis for retroviral integration into nucleosomes. Nature 2015, 523, 366–369.
- Engelman A, Cherepanov P The structural biology of HIV-1: mechanistic and therapeutic insights. Nat Rev Microbiol. 2012, 10, 279–290.
- Deng Z, Lehmann KC, Li X, et al. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res. 2014, 42, 3464–3477.
- Tang C, Deng Z, Li X, et al. Helicase of type 2 porcine reproductive and respiratory syndrome virus strain HV reveals a unique structure. Viruses 2020, 12. 12
- Ruiz FX, Hoang A, Das K, et al. Structural basis of HIV-1 inhibition by nucleotide-competing reverse transcriptase inhibitor indopy-1. J Med Chem. 2019, 62, 9996–10002.
- Singh K, Flores JA, Kirby KA, et al. Drug resistance in non-B subtype HIV-1: impact of HIV-1 reverse transcriptase inhibitors. Viruses 2014, 6, 3535–3562.
- Das K, Bauman JD, Clark AD Jr., et al. High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A 2008, 105, 1466–1471.
- Janssen PA, Lewi PJ, Arnold E, et al. In search of a novel anti-HIV drug: multidisciplinary coordination in the discovery of 4-[[4-[[4-[(1e)-2-cyanoethenyl]-2,6-dimethylphenyl]amino]-2- pyrimidinyl]amino]benzonitrile (R278474, rilpivirine). J Med Chem. 2005, 48, 1901–1909.
- Singh K, Marchand B, Rai DK, et al. Biochemical mechanism of HIV-1 resistance to rilpivirine. J Biol Chem. 2012, 287, 38110–38123.
- Haggblom A, Svedhem V, Singh K, et al. Virological failure in patients with HIV-1 subtype c receiving antiretroviral therapy: an analysis of a prospective national cohort in Sweden. Lancet HIV 2016, 3, e166–174.
- van Domselaar R, Njenda DT, Rao R, et al. HIV-1 subtype C with PYxE insertion has enhanced binding of Gag-p6 to host cell protein ALIX and increased replication fitness. J Virol. 2019, 93. 93
- Kannan SR, Spratt AN, Quinn TP, et al. Infectivity of SARS-CoV-2: there is something more than D614G? J Neuroimmune Pharmacol. 2020. 15 574–577
