Abstract
Introduction
Methylated circulating tumour DNA (ctDNA) blood tests for BCAT1/IKZF1 (COLVERA) and SEPT9 (Epi proColon) are used to detect colorectal cancer (CRC). However, there are no ctDNA assays approved for other gastrointestinal adenocarcinomas. We aimed to characterize BCAT1, IKZF1 and SEPT9 methylation in different gastrointestinal adenocarcinoma and non-gastrointestinal tumours to determine if these validated CRC biomarkers might be useful for pan-gastrointestinal adenocarcinoma detection.
Methods
Tissue DNA methylation data from colorectal (COAD, READ), gastroesophageal (ESCA, STAD), pancreatic (PAAD) and cholangiocarcinoma (CHOL) adenocarcinoma cohorts within The Cancer Genome Atlas were used for differential methylation analyses. Clinicodemographic predictors of BCAT1, IKZF1 and SEPT9 methylation, and the selectivity of hypermethylated BCAT1, IKZF1 and SEPT9 for colorectal adenocarcinomas in comparison to other cancers were each explored with beta regression.
Results
Hypermethylated BCAT1, IKZF1 and SEPT9 were each differentially methylated in colorectal and gastroesophageal adenocarcinomas. IKZF1 was differentially methylated in pancreatic adenocarcinoma. Hypermethylated DNA biomarkers BCAT1, IKZF1 and SEPT9 were largely stable across different stages of disease and were highly selective for gastrointestinal adenocarcinomas relative to other cancer types.
Discussion
Existing CRC methylated ctDNA blood tests for BCAT1/IKZF1 and SEPT9 might be usefully repurposed for use in other gastrointestinal adenocarcinomas and warrant further prospective ctDNA studies.
Introduction
Five million new gastrointestinal cancers were diagnosed in 2020, accounting for a quarter of all newly diagnosed cancers worldwide (World Cancer Research Fund International Citation2020). Together, colorectal (9.4%), stomach (7.7%), oesophagus (5.5%) and pancreatic (4.7%) cancers contributed to over one-quarter of global cancer-related deaths (Sung et al. Citation2021). Adenocarcinomas are the most common type of gastrointestinal cancers, found as the majority of colorectal, stomach and pancreatic cancers, as well as intrahepatic cholangiocarcinoma and a rising number of oesophageal cancers (between 5 and 66%, varies by geographic location) (Massarweh and El-Serag Citation2017; Arnold et al. Citation2020). Early detection is vital in improving treatment options and outcomes as most cancers are diagnosed at a late stage. While CRC screening programs have been established across Europe, the Americas, Asia and Oceania, uptake of faecal-based screening tests remains suboptimal, and there are no other simple and non-invasive tests in use for screening of other gastrointestinal cancers (Navarro et al. Citation2017; Australian Institute of Health and Welfare Citation2023). Further, existing blood biomarkers commonly used to monitor cancer recurrence; protein biomarkers carbohydrate antigen 19-9 (CA 19-9) and carcinoembryonic antigen (CEA), are not reliable for detection of gastrointestinal adenocarcinoma (Maestranzi et al. Citation1998). There is an urgent need to improve non-invasive detection of gastrointestinal cancers to reduce disease burden and mortality.
Development of gastrointestinal adenocarcinomas involves genetic mutations and extensive epigenetic alterations which silence the expression of tumour suppressor genes and/or activate oncogenes, contributing to changes in tumour growth, initiation and metastatic potential (Laird Citation2003). DNA hypermethylation is a common epigenetic aberration which can be used as a biomarker for cancer diagnosis, treatment monitoring and prediction of response (Levenson Citation2010). DNA hypermethylation can be detected in tiny fragments of circulating tumour DNA (ctDNA) released by the tumour into the plasma, and are detected more frequently than cancer-associated mutations (Locke et al. Citation2019). DNA methylation-based ctDNA assays offer a cost-effective and minimally invasive alternative to detect cancers compared to current technologies, highlighting their value as a diagnostic tool. Quantitative methylated ctDNA assays can also reflect disease burden; reducing in abundance after tumour resection or efficacious cancer treatment and highlighting their utility in treatment monitoring, recurrence surveillance and detection of minimal residual disease (Sanz-Garcia et al. Citation2022).
While many candidate hypermethylated biomarkers have been investigated for detection of gastrointestinal adenocarcinoma, few have been translated into clinical use. Two real-time PCR-based methylated ctDNA biomarker assays in clinical use for detection of colorectal adenocarcinoma include the United States Food and Drug Administration (FDA)-approved Epi proColon (Epigenomics AG), a CRC screening test for methylated SEPT9 in patients with a history of non-adherence to other screening methods, and COLVERA (Clinical Genomics); a laboratory-developed test measuring the combination of methylated BCAT1 and/or IKZF1 with Medicare coverage for minimal residual disease and recurrence monitoring in CRC (Mitchell et al. Citation2014; Potter et al. Citation2014; Mitchell et al. Citation2016; Johnston et al. Citation2023).
Methylated SEPT9 was first identified alongside 55 other candidate CRC biomarkers using methylation-sensitive restriction enzyme discovery in colorectal tissue and blood (Lofton-Day et al. Citation2008). Methylated BCAT1 and IKZF1 were identified as candidate CRC biomarkers alongside 30 other biomarkers (including SEPT9) using data from the Illumina HumanMethylation27 (27K) BeadChip array in tandem with gene expression and genome-wide DNA methylation analyses (Mitchell et al. Citation2014). All three biomarkers underwent further clinical development based on the high frequency of DNA hypermethylation in primary colorectal cancer (CRC) tissue and low frequency of methylation in the circulating cell free-DNA in plasma from healthy controls. There are currently no FDA-approved ctDNA assays for other gastrointestinal adenocarcinomas, all of which have poorer survival outcomes than CRC (Sung et al. Citation2021). Since gastrointestinal adenocarcinomas arise from tissues within the same embryonic layer (Bhatia et al. Citation2023), methylated biomarkers may therefore be shared between different gastrointestinal adenocarcinomas. Thus, repurposing existing methylated ctDNA assays used for detection of CRC, may also detect other gastrointestinal adenocarcinomas, offering potential for rapid clinical implementation as a pan-gastrointestinal blood test.
The aim of this study was to characterize the methylation of BCAT, IKZF1 and SEPT9 in different gastrointestinal adenocarcinoma tissues and further compare this to the methylation of BCAT1, IKZF1 and SEPT9 in non-gastrointestinal tumour tissues to determine if these validated CRC biomarkers may also be useful for pan-gastrointestinal adenocarcinoma detection.
Materials and methods
Overview
The results published here are based upon data generated by the TCGA Research Network (Weinstein et al. Citation2013). Given that the methylated ctDNA blood tests for BCAT1, IKZF1 and SEPT9 were designed for colorectal adenocarcinoma, differential methylation analyses were performed on different gastrointestinal adenocarcinomas for an unbiased overview of the performance of BCAT1, IKZF1 and SEPT9 with respect to other significantly methylated probes. The association between the clinicodemographic variables and BCAT1, IKZF1 and SEPT9 hypermethylation within gastrointestinal adenocarcinomas was analyzed, and the β-values were assessed across all TCGA cancer datasets to characterize the discriminatory ability of hypermethylated BCAT1, IKZF1 and SEPT9 for gastrointestinal adenocarcinoma.
Ethics statement
All deidentified data used in this study obtained from a publicly accessible database. Ethical approval to conduct the observations within this study is not required.
DNA methylation datasets
Publicly available tissue DNA methylation data and accompanying clinical information collected from patients diagnosed with cancer were obtained from the Genomic Data Commons using the TCGAbiolinks package (Supplementary Table S1; Colaprico et al. Citation2016). DNA methylation data were harmonized prior to downloading, using pipelines described in the GDC Data User’s Guide (NCI Genomic Data Commons Citation2022). GrCh38-annotated β-values from the Illumina Infinium HumanMethylation450 (450K) BeadChip were obtained for each TCGA DNA methylation dataset. Masked probes, probes annotated to non-CpG or common SNPs, cross-reactive probes, or probes lacking DNA methylation measurements were removed prior to downstream analysis. Where applicable, measurements from replicate observations obtained from the same tissue sample were averaged.
Differential methylation analysis
Gastrointestinal adenocarcinoma datasets were subject to differential methylation analysis for both probes and regions separately. β values were converted to M-values, and differentially methylated probes were identified in normal vs. tumour separately for colorectal adenocarcinoma (TCGA-COAD and TCGA-READ), gastroesophageal adenocarcinoma (TCGA-ESCA and TCGA-STAD), pancreatic adenocarcinoma (TCGA-PAAD) and cholangiocarcinoma (TCGA-CHOL) using the empirical bayes method within limma (Ritchie et al. Citation2015), followed by differentially methylated region analysis using DMRcate (Peters et al. Citation2015) with default settings. TCGA-ESCA and TCGA-STAD datasets were combined in adenocarcinoma analyses due to a lack of normal tissue (n = 2) within the TCGA-STAD dataset. Tumours within the TCGA-ESCA dataset were divided into adenocarcinomas and squamous cell carcinomas, and squamous cell carcinomas were not classified as gastrointestinal adenocarcinomas. A probe or region was considered differentially methylated if it had a false discovery rate (FDR)-adjusted p-value < 0.05. Volcano plots were created using and ggplot2 (Wickham et al. Citation2019). Genomic regions were visualized using IGV-Web (Robinson et al. Citation2023).
Probe annotations
All probes were annotated to GrCh38 genes using TCGA metadata, and further annotated to CpG island annotations that incorporated: 1) shores: regions of up to 2 kb from a CpG island; 2) shelves: regions from 2 to 4 kb from a CpG island; and 3) open sea: the rest of the genome, from the UCSC Table Browser (Karolchik et al. Citation2004). GENCODE v36 was used for gene annotation, with promoters defined as +2 kb to −0.2 kb from the gene of interest transcription start site, as per the GenomicRanges package (Lawrence et al. Citation2013). Probes annotated to BCAT1, IKZF1 and SEPT9 and used for downstream β-value analyses are listed in Supplementary Table S2. BCAT1- and IKZF1-associated probes were identified based on overlapping amplicons produced in the COLVERAd methylated BCAT1 and IKZF1 ctDNA real-time PCR assays, using primers obtained from publicly available data (Pedersen et al. Citation2015). As no probes covered within the TCGA Illumina Infinium 450K BeadChip array overlapped amplicons produced in the FDA-approved methylated SEPT9 Epi proColon real-time PCR assay (Church et al. Citation2014), the two closest probes flanking either side of the amplicon produced in the Epi proColon test were selected as representative for SEPT9 (Supplementary Table S2). One of these probes (cg12783819) was filtered out from many comparisons due to insufficient measurements in samples subject to methylation analysis. Observations on methylation β-values were averaged within each biomarker, as up to two probes were annotated for each amplicon.
Statistical analysis
Data analyses were performed in RStudio using the tidyverse and gtsummary packages (Sjoberg et al. Citation2021; Wickham et al. Citation2019). Methylation β-values for BCAT1, IKZF1 and SEPT9 were used for all clinicodemographic and inter-cancer analyses because they are more biologically interpretable when compared to M-values and more closely correspond to the percentage methylation of a given site (Du et al. Citation2010). Where available, age data was dichotomized into under 50 years versus 50 years and older, consistent with the definition of early-onset cancer (Harrold et al. Citation2023). For observations on CRC, tumours were grouped into lesions affecting the left colon (rectum, sigmoid colon, descending colon, and splenic flexure) or right colon (transverse colon, hepatic flexure, ascending colon, and caecum). Heatmaps of methylation at BCAT1, IKZF1 and SEPT9 were visualized for all gastrointestinal adenocarcinomas within the TCGA dataset using tidyheatmaps, where data were clustered based on Euclidean distance using complete linkage (Engler Citation2022). All other visualizations were performed using ggplot2 (Wickham et al. Citation2019). Normal vs. tumour β-values were compared using a Wilcoxon rank sum test. The effect of tumour staging on methylation was compared first using a Kruskal–Wallis rank sum test, with post-hoc comparisons analyzed using Wilcoxon rank-sum test. Pearson’s Chi-squared (for categorical variables) and Wilcoxon or Kruskal–Wallis tests (for continuous variables) were used as indicated in table legends. Methylated biomarkers BCAT1, IKZF1, and/or SEPT9 were considered positive where β ≥ 0.25, interpreted as partial methylation or hypermethylation.
Beta regression was used to compare predictors of BCAT1, IKZF1 and SEPT9 methylation in colorectal adenocarcinomas by multivariable analysis. The selectivity of methylated BCAT1, IKZF1 and SEPT9 was compared between colorectal adenocarcinomas and all other tumours (inclusive of both gastrointestinal and non-gastrointestinal tumours) using univariate beta regression. Yardstick was used to create receiver-operating characteristic curves (ROC) and calculate the area under curve (AUC) for BCAT1, IKZF1 and SEPT9 methylation in (a) specified gastrointestinal adenocarcinoma tissues over all other tumour tissues available from TCGA (Supplementary Table S1), and (b) specified gastrointestinal adenocarcinoma tissue over their corresponding normal controls (Kuhn et al. Citation2023).
Results
Methylation of BCAT1, IKZF1 and SEPT9 in different gastrointestinal cancers
CpG sites within or immediately flanking those detected by BCAT1, IKZF1 and SEPT9 ctDNA blood tests were frequently hypermethylated in gastrointestinal adenocarcinomas (, Supplementary Figure S1A-D). The demographic and clinical characteristics of gastrointestinal adenocarcinomas are summarized in . BCAT1, IKZF1 and SEPT9 methylation was significantly higher in both colorectal and gastroesophageal adenocarcinoma when compared to corresponding normal tissue (; mean delta β-values (tumour – normal) ranged 0.30 − 0.42 and 0.23 − 0.33 for colorectal and gastroesophageal adenocarcinoma, respectively), even when divided into different stages (Supplementary Figure S2A-B). Pancreatic adenocarcinoma contained significantly higher methylation of BCAT1, IKZF1 and SEPT9 when compared to normal tissue (, mean β-values BCAT1; 0.09 vs. 0.25, IKZF1; 0.03 vs. 0.30, and SEPT9; 0.07 vs. 0.24 for normal vs. tumour, respectively), but only BCAT1 and IKZF1 were increased at every stage of pancreatic cancer when compared to normal tissue (Supplementary Figure S2C). Cholangiocarcinomas were infrequently methylated at BCAT1, IKZF1 and SEPT9 (Supplementary Figure S1D), and neither BCAT1, IKZF1, nor SEPT9 methylation was able to differentiate normal bile duct tissue from cholangiocarcinoma (, Supplementary Figure S2D). When each type of gastrointestinal adenocarcinoma was compared to their corresponding normal tissues, the highest AUC for BCAT1 and IKZF1 were seen for colorectal adenocarcinomas (Supplementary Figure S3, 0.94 and 0.90 for BCAT1 and IKZF1, respectively), followed by pancreatic adenocarcinoma (Supplementary Figure S3, 0.81 and 0.99 for BCAT1 and IKZF1, respectively). For methylated SEPT9, the highest area under the ROC curve when compared to corresponding normal tissue was for colorectal adenocarcinoma followed by gastroesophageal adenocarcinoma (Supplementary Figure S3, AUC 0.97 and 0.84, respectively).
Figure 1. BCAT1, IKZF1 And SEPT9 are hypermethylated in gastrointestinal adenocarcinomas. (A) Heatmap showing BCAT1, IKZF1 and SEPT9 methylation (β-value) in gastrointestinal adenocarcinomas (n = 1,093 tumours). (B) Methylation of BCAT1, IKZF1 and SEPT9 in normal tissue versus tumour for (in order from left to right) colorectal adenocarcinoma (n = 45 normal and 396 tumour), gastroesophageal adenocarcinoma (n = 18 normal and 484 tumour), pancreatic adenocarcinoma (n = 10 normal and 177 tumour) and cholangiocarcinoma (n = 9 normal and 36 tumour). ns, no significant difference; *, p < 0.05; ***, p < 0.001 by wilcoxon rank sum test. (C) Gene browser tracks showing the sites detected by the BCAT1, IKZF1 and SEPT9 test (‘amplicon’), corresponding CpG annotation and overlap of differentially methylated regions (coloured bars) called in gastrointestinal adenocarcinoma.
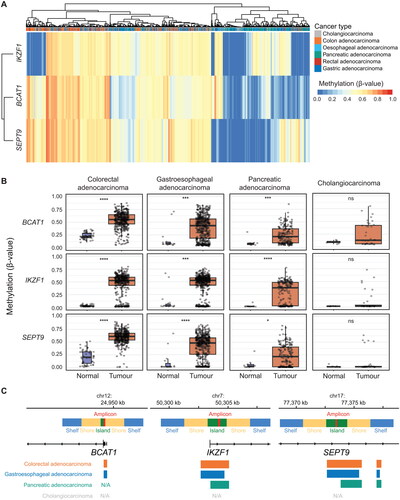
Figure 2. BCAT1, IKZF1 And SEPT9 methylation is more prevalent in gastrointestinal adenocarcinoma when compared to other cancer types. (A) Forest plots comparing methylation of BCAT1, IKZF1 and SEPT9 in different cancers relative to colorectal adenocarcinomas (n = 396 colorectal adenocarcinomas vs. 8601 other tumours). Data analyzed using beta regression. (B) Receiver operating characteristic curves summarizing the sensitivity and discriminatory ability (herein termed specificity) for BCAT1, IKZF1 and SEPT9 methylation in gastrointestinal adenocarcinomas over all other cancers described in (a).
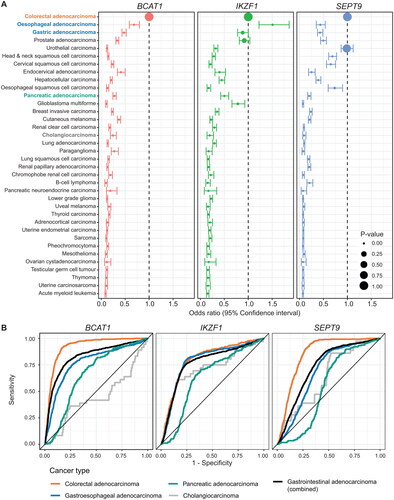
Table 1. Clinicodemographic characteristics and corresponding BCAT1, IKZF1 and SEPT9 methylation in gastrointestinal adenocarcinomas (n = 1093).
Differential methylation of BCAT1, IKZF1 and SEPT9 in gastrointestinal adenocarcinoma
Differential methylation analyses were performed on colorectal adenocarcinoma, gastroesophageal adenocarcinoma, pancreatic adenocarcinoma and cholangiocarcinoma to determine whether ctDNA-associated CRC biomarkers were independently differentially methylated in gastrointestinal adenocarcinoma across the ∼450,000 CpG sites detected in the Infinium HumanMethylation450 BeadChip array (, Supplementary Figure S4A-D). The total number of differentially methylated probes and regions within each dataset are summarized in Supplementary Table S3. BCAT1, IKZF1 and SEPT9 were differentially hypermethylated in colorectal adenocarcinoma and gastroesophageal adenocarcinoma (, Supplementary Figure S4A-B). While probes annotated to BCAT1, IKZF1 and SEPT9 were differentially hypermethylated in pancreatic adenocarcinoma, only IKZF1 was annotated to a differentially hypermethylated region which overlapped the sites detected using the ctDNA blood test (, Supplementary Figure S4C). BCAT1, IKZF1 and/or SEPT9 were neither differentially methylated probes, nor regions in cholangiocarcinoma (, Supplementary Figure S4D).
Predictors of BCAT1, IKZF1 and SEPT9 methylation in gastrointestinal adenocarcinoma
A multivariable assessment of demographic and clinical predictors for all combined gastrointestinal adenocarcinoma tissue hypermethylation in BCAT1, IKZF1 and SEPT9 are listed in , and analyses limited to CRC are available in Supplementary Table S4. Overall, individuals aged under 50 years had significantly lower levels of BCAT1 methylation compared to those aged over 50 years (, odds ratio [OR] 0.77), females had significantly lower levels of IKZF1 methylation compared to males (, OR 0.83), and SEPT9 tumour methylation was raised in individuals of Asian descent (, OR 1.46). When limited to CRC, left-sided lesions were more likely to have reduced IKZF1 methylation and increased SEPT9 methylation when compared to right-sided lesions (Supplementary Table S4, OR 0.66 and 1.15 for IKZF1 and SEPT9, respectively). BCAT1 and SEPT9 methylation was significantly lower in all other gastrointestinal adenocarcinomas when compared to colorectal adenocarcinomas. While there was no significant difference in IKZF1 methylation between colorectal and gastroesophageal adenocarcinomas (), we found that oesophageal adenocarcinomas contained higher BCAT1 and IKZF1 methylation when compared to gastric adenocarcinomas (Supplementary Table S5, OR 0.75 and 0.58 for BCAT1 and IKZF1, respectively). While IKZF1 and SEPT9 methylation remained consistent independent of cancer stage, reduced BCAT1 methylation was observed with gastrointestinal adenocarcinomas staged at II and IV (). All (100%, n = 396/396) colorectal adenocarcinomas, 92% (n = 446/484) of gastroesophageal adenocarcinomas, and 79% (n = 140/177) pancreatic adenocarcinomas were positive for at least one of either methylated BCAT1, IKZF1 or SEPT9 (, Supplementary Figure S5A). Permutations of methylated BCAT1, IKZF1, and SEPT9 positivity offered limited ability to discriminate between different gastrointestinal adenocarcinomas (Supplementary Figure S5A).
Table 2. Predictors of BCAT1, IKZF1 and SEPT9 methylation in gastrointestinal adenocarcinomas.
Selectivity of methylated BCAT1, IKZF1 and SEPT9 for gastrointestinal cancers
Given that BCAT1, IKZF1 and SEPT9 are validated ctDNA biomarkers for CRC, and that DNA shed from any tumour can contribute to the cell-free DNA within an individual, we compared methylation of BCAT1, IKZF1 and SEPT9 in colorectal adenocarcinomas (i.e. TCGA-COAD/TCGA-READ) to that of all tumour types represented within TCGA to characterize the ability of methylated BCAT1, IKZF1 and SEPT9 to differentiate between different cancers. BCAT1 methylation was most prevalent in colorectal adenocarcinoma and reduced in all other cancer types (), including commonly diagnosed cancers such as breast, prostate, and lung cancer. IKZF1 methylation was significantly more prevalent in oesophageal adenocarcinomas when compared to colorectal adenocarcinoma (OR 1.51 [95% confidence interval 1.22 − 1.85]) and did not significantly differ between colorectal, gastric adenocarcinoma, nor prostate adenocarcinoma (). SEPT9 was most frequently methylated in colorectal adenocarcinoma and bladder urothelial carcinomas () and significantly reduced in all other cancer types (). SEPT9 methylation was observed in oesophageal, head and neck, and cervical squamous cell carcinomas, albeit at a significantly reduced methylation level when compared to colorectal adenocarcinomas (). BCAT1 had the greatest performance in differentiating colorectal adenocarcinoma from all other cancer types, followed by SEPT9 (AUC 0.94 and 0.85, respectively, , Supplementary Table S6). When tumours were assessed for methylated biomarker positivity, the combination of BCAT1 + SEPT9 (with any IKZF1 result) was most selective for colorectal adenocarcinoma in all cancer types above all other permutations (Supplementary Figure S5B, AUC 0.89). Out of the three assessed biomarkers, IKZF1 had the highest AUC for gastroesophageal adenocarcinoma (AUC 0.79, , Supplementary Table S6). Similarly, IKZF1-positive tumours were most selective for gastroesophageal adenocarcinomas (AUC 0.77, Supplementary Figure 5B). The highest combined AUC for detection of gastrointestinal adenocarcinoma over all other cancer types was observed with methylated BCAT1 (AUC 0.83, , Supplementary Table S6), or when at least two methylated biomarkers were positive out of BCAT1, IKZF1 or SEPT9 using a cut-off of β ≥ 0.25 (AUC 0.78, Supplementary Figure 5B).
Discussion
The high incidence and mortality from gastrointestinal cancers across the world shows a clear need for test that is sensitive and specific. Many discovery studies report long lists of candidate biomarkers, but few of these biomarkers reach FDA approval (Paulovich et al. Citation2008). Further, commonly used blood-based tumour biomarkers for gastrointestinal adenocarcinoma are limited to protein biomarkers CA 19-9 and CEA, which are limited in use for treatment monitoring or detection or residual or recurrent disease, and are also raised in benign gastrointestinal diseases such as gastritis, peptic ulcers, duodenitis, esophagitis, diverticulitis, jaundice, pancreatitis and cirrhosis (Maestranzi et al. Citation1998). Repurposing existing CRC biomarkers already in clinical use, including the BCAT1/IKZF1 (COLVERA) or SEPT9 (Epi proColon) methylated ctDNA assays for the detection of gastrointestinal adenocarcinoma may enable rapid translation into clinical use. Herein we observed frequent and selective hypermethylation of BCAT1, IKZF1 and/or SEPT9 in colorectal adenocarcinoma, oesophageal adenocarcinoma, gastric adenocarcinoma and partially in pancreatic adenocarcinoma, highlighting the value of these existing ctDNA assays to be adapted for use as a non-invasive assay for detection of pan-gastrointestinal adenocarcinoma.
The present study assessed the methylation of BCAT1, IKZF1 and SEPT9 in a variety of normal and tumour tissues. However, these biomarkers must be detectable in plasma from people with cancer at a significantly higher rate than that of healthy individuals to warrant further development as a ctDNA assay for pan-gastrointestinal adenocarcinoma. Previous work has shown that methylated ctDNA biomarkers for BCAT1, IKZFI and SEPT9 were able to detect colorectal adenocarcinoma with a sensitivity of 65%, 68% and 70%, respectively (Church et al. Citation2014; Pedersen et al. Citation2015), and have undergone further development as tests for both screening and post-operative follow-up of colorectal adenocarcinoma (Symonds et al. Citation2016; Lamb and Dhillon Citation2017; Pedersen et al. Citation2023). Preliminary studies adapting the methylated BCAT1/IKZF1 ctDNA assay have also shown promise in oesophageal, gastric and pancreatic adenocarcinoma (Symonds et al. Citation2018A; Chan et al. Citation2022; Winter et al. Citation2022A). Similarly, we found that the combination of methylated BCAT1, IKZF1 and SEPT9 were excellent at detecting gastrointestinal adenocarcinomas, but were not effective at discriminating between different types of gastrointestinal adenocarcinoma. As such, it might be recommended that individuals who test positive to existing COLVERA (BCAT1/IKZF1) or Epi proColon (SEPT9) assays undergo bi-directional endoscopic and colonoscopic follow-up, or in cases of potential metastatic recurrence, radiological scans for investigation of gastrointestinal adenocarcinoma.
Importantly, methylated plasma ctDNA assays require low rates of methylation in the plasma circulating cell-free DNA from healthy individuals; of which ∼84% comes from cells of haematopoietic lineage, ∼0.9% from colorectal epithelial cells, ∼0.8% from pancreatic epithelial cells, and ∼0% from gastroesophageal epithelial cells (Moss et al. Citation2018). It has already been established that BCAT1, IKZF1 and SEPT9 methylation is low in white blood cells obtained from healthy individuals (Mitchell et al. Citation2016), making them suitable candidates to differentiate ctDNA from normal circulating cell-free DNA. Further studies have shown that IKZF1 and SEPT9 possess hyper-stable methylation (or lack thereof) in plasma samples from healthy individuals (Shvetsov et al. Citation2015; Zaimi et al. Citation2018). While BCAT1 and SEPT9 are partially methylated in normal colorectal cells, established ctDNA assays for BCAT1/IKZF1 and separately for SEPT9 highlight the suitability of these biomarkers to differentiate CRC from contaminating circulating cell-free DNA originating from normal gastrointestinal epithelial cells (Mitchell et al. Citation2014; Potter et al. Citation2014; Mitchell et al. Citation2016). These data also highlight the need for caution in adapting BCAT1 and SEPT9 for other non-invasive biomarker tests, such as stool-based biomarkers, due to the abundance of low-level methylation of these biomarkers present in normal gastrointestinal epithelial cells, which may be shed through normal cell turnover into faeces.
While BCAT1, IKZF1 and SEPT9 tissue methylation were discriminatory for gastrointestinal adenocarcinoma over a variety of other tumour types when assessed both in isolation or when combined, we also observed methylated IKZF1 and SEPT9 in tissue from prostate adenocarcinoma and bladder urothelial carcinoma, respectively. Previous studies have shown that ctDNA is more readily detectable in patients with gastrointestinal cancer when compared to prostate cancer (Bettegowda et al. Citation2014). Accordingly, the methylated BCAT1/IKZF1 ctDNA assay has previously shown poor sensitivity for prostate adenocarcinoma, suggesting that is unlikely to be a common contributor to an unexplained positive blood test in the context of a negative endoscopy/colonoscopy (Symonds et al. Citation2018A; Winter et al. Citation2022B). ; As methylated SEPT9 was prevalent in bladder urothelial carcinoma, it is possible that methylated SEPT9 may benefit from further development as a urine biomarker, and urine cytology may be recommended as a rule-out test for individuals who test positive for Epi proColon but negative at endoscopy/colonoscopy.
The present study identified independent predictors of BCAT1, IKZF1 and SEPT9 methylation, including age, sex and race. The relationship between age and increased methylation has been well described (Hannum et al. Citation2013). Accordingly, BCAT1 and SEPT9 methylation was reduced in gastrointestinal adenocarcinomas obtained from individuals aged under 50 years. Female sex was associated with reduced IKZF1 methylation, consistent with observations on global methylation from other studies (Zhang et al. Citation2011). Finally, people of Asian descent had higher rates of SEPT9 tumour methylation. This observation is consistent with previous findings, where SEPT9 had a greater diagnostic sensitivity for CRC in people of Asian descent (Hu et al. Citation2019), and highlights the value of using a panel of ctDNA biomarkers to compensate for potential diagnostic disparities by racial and ethnic status that may be encountered when a sole biomarker is assessed.
Panel-based methylated multi-cancer blood tests have been previously investigated for lung, gynaecological, upper and lower gastrointestinal cancers (Liu et al. Citation2020). The next-generation sequencing-based Galleri (GRAIL Inc) blood test is currently undergoing trial for asymptomatic individuals in the United Kingdom (Swanton et al. Citation2022). Galleri’s panel-based approach allows prediction of the primary cancer type for clinical follow-up, but can result in unnecessary, invasive and prolonged follow-up tests (162 days), especially in individuals with a false-positive blood test (Wise Citation2022). In contrast, methylated BCAT1, IKZF1 and SEPT9 appear largely gastrointestinal adenocarcinoma-specific, where a positive blood test can be confirmed through targeted gastrointestinal diagnostic follow-up.
While methylated BCAT1, IKZF1 and SEPT9 each show value in detecting colorectal, oesophageal, gastric and some pancreatic adenocarcinomas, they were not the most differentially methylated regions assessed within each of the analyzed datasets. The biomarker discovery process for SEPT9 commenced prior to the availability of methylation arrays (Lofton-Day et al. Citation2008). Similarly, BCAT1 and IKZF1 were shortlisted as candidate methylated biomarkers based on coverage within the Illumina HumanMethylation27 BeadChip (Mitchell et al. Citation2014), limiting the pool of available candidate biomarkers by more than 16× fewer CpG sites than those analyzed within the current study. As next-generation sequencing technologies have become more affordable, further advances in genome-wide methylation technologies have been made. As such, it is possible that new candidate biomarkers with greater sensitivity and specificity for gastrointestinal adenocarcinomas may be discovered and developed with more contemporary ‘omic’ approaches.
Similarly, methylated BCAT1, IKZF1 and SEPT9 were not identified as the most promising candidates for detection of cholangiocarcinoma. Methylated biomarkers with better differential capacity for cholangiocarcinoma have previously undergone early development in plasma ctDNA assays (Andresen et al. Citation2015) and could be considered for further development within an integrated methylated biomarker panel alongside BCAT1, IKZF1 and SEPT9.
While the methylated BCAT1/IKZF1 (COLVERA) and methylated SEPT9 (Epi proColon) ctDNA assays have similar biological rationale and test methodologies, Epi proColon remains the only ctDNA method approved by the United States FDA for CRC screening in individuals who have declined first-line screening tests (Shirley Citation2020). While methylated BCAT1/IKZF1 has been investigated for CRC screening (Symonds et al. Citation2016; Citation2018B), COLVERA was developed and used for recurrence monitoring and detecting minimal residual disease in CRC. The most commonly used tools for surveillance of gastrointestinal adenocarcinoma recurrence are low-sensitivity and low-specificity protein biomarker blood tests including CEA or CA 19-9, in combination with radiological scans. Given the high rate of BCAT1, IKZF1, and/or SEPT9 methylation in gastrointestinal adenocarcinomas, we consider further work up of these ctDNA assays justified. This could be either as a gastrointestinal cancer screening test option for individuals non-compliant for other screening methods, or as a ctDNA-based recurrence surveillance tool alongside diagnostic radiology.
Herein, we used in silico data to demonstrate hypermethylation of BCAT1, IKZF1, and SEPT9 in different gastrointestinal adenocarcinomas and rationalize further development of existing methylated ctDNA blood tests; COLVERA and Epi proColon, for pan-gastrointestinal adenocarcinomas. An important limitation of this study surrounds use of in silico methylation array data as a proxy for a methylation-specific qPCR ctDNA assays. First, we were unable to characterize methylation of each biomarker over the entire region detected within the COLVERA and Epi proColon assays, instead limiting observations to one to two probes located either internally (BCAT1, IKZF1) or immediately flanking (SEPT9) the regions detected by the corresponding COLVERA and Epi proColon ctDNA assays. Secondly, our positivity and selectivity analyses, beta regression, and ROC curves were performed using tissue, and not plasma cell-free DNA. Given that the vast majority of cell-free DNA come from haematopoietic cells, or sometimes tumour cells (particularly in late-stage cancers), a low positivity rate of BCAT1, IKZF1 and SEPT9 in cell-free DNA from most (cancer-free) individuals would be expected. Further, a tumour methylated for BCAT1, IKZF1 and/or SEPT9 may not be detectable in the blood ctDNA, nor shed into circulation, for a variety of reasons including tumour size, site, stage, and pathophysiological reasons. As such, results from tissue methylation arrays presented herein should be extrapolated to methylation-specific qPCR tests with caution. Regardless, our data provides valuable evidence to show that primary gastrointestinal adenocarcinomas are hypermethylated for BCAT1, IKZF1 and SEPT9 at a rate higher than other non-gastrointestinal cancers, rationalizing further development and application of methylated BCAT1, IKZF1 and SEPT9 qPCR-based assays for pan-gastrointestinal adenocarcinoma. This study is the first to directly assess methylation of BCAT1, IKZF1 and SEPT9 in pan-gastrointestinal adenocarcinoma tissue. BCAT1, IKZF1 and/or SEPT9 were frequently methylated in colorectal, oesophageal, gastric and some pancreatic adenocarcinomas, highlighting the value in adaption of these existing ctDNA assays for broader detection of gastrointestinal adenocarcinoma. The results presented warrant further prospective studies to investigate the accuracy of these blood-based biomarkers for detection of gastrointestinal cancers other than colorectal cancer.
Author contributions
Conceptualization: Ganessan Kichenadasse, Graeme P. Young, Erin L. Symonds, Jean M. Winter; Methodology: Geraldine Laven-Law, Jean M. Winter; Formal analysis and investigation: Geraldine Laven-Law; Writing - original draft preparation: Geraldine Laven-Law; Writing - review and editing: Ganessan Kichenadasse, Graeme P. Young, Erin L. Symonds, Jean M. Winter; Supervision: Erin L. Symonds, Jean M. Winter.
Ethics approval and consent to participate
All deidentified data used in this study obtained from a publicly accessible database. Ethical approval to conduct the observations within this study is not required.
Data availability
All data used in this study was publicly accessible. Analyzed data and code that supports the findings presented in this study are available from the corresponding author upon reasonable request.
Supplemental Material
Download MS Word (2.8 MB)Acknowledgements
The authors would like to thank sample donors and The Cancer Genome Atlas Research Network for access to study data.
Disclosure statement
G.P.Y. reports personal fees from Clinical Genomics Inc, NJ, USA. E.L.S. reports grants from Clinical Genomics outside of the submitted work. No disclosures were reported by the other authors.
References
- Andresen K, Boberg KM, Vedeld HM, Honne H, Jebsen P, Hektoen M, Wadsworth CA, Clausen OP, Lundin KE, Paulsen V, et al. 2015. Four DNA methylation biomarkers in biliary brush samples accurately identify the presence of cholangiocarcinoma. Hepatology 61(5):1651–1659. doi: 10.1002/hep.27707
- Arnold M, Abnet CC, Neale RE, Vignat J, Giovannucci EL, McGlynn KA, Bray F. 2020. Global burden of 5 major types of gastrointestinal cancer. Gastroenterology 159(1):335–349 e315. doi: 10.1053/j.gastro.2020.02.068
- Australian Institute of Health and Welfare. 2023. National Bowel Cancer Screening Program monitoring report 2023. AIHW, Canberra. CAN 154 112. https://www.aihw.gov.au/reports/cancer-screening/nbcsp-monitoring-2023/summary.
- Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al. 2014. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 6(224):224ra24. doi: 10.1126/scitranslmed.3007094
- Bhatia A, Shatanof RA, Bordoni B. 2023. Embryology, Gastrointestinal. StatPearls Publishing, Treasure Island, Florida.
- Chan MM, Winter JM, Pedersen SK, Roy AC, Watson DI, Hussey D, Young GP, Symonds EL. 2022. Assessment of methylated BCAT1 and IKZF1 circulating tumor DNA as a potential diagnostic and prognostic biomarker in esophagogastic cancers. JCO. 40(4_suppl):348–348. doi: 10.1200/JCO.2022.40.4_suppl.348
- Church TR, Wandell M, Lofton-Day C, Mongin SJ, Burger M, Payne SR, Castaños-Vélez E, Blumenstein BA, Rösch T, Osborn N, et al. 2014. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut 63(2):317–325., doi: 10.1136/gutjnl-2012-304149
- Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. 2016. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 44(8):e71–e71. doi: 10.1093/nar/gkv1507
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. 2010. Comparison of beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinf. 11(1):587. doi: 10.1186/1471-2105-11-587
- Engler J. 2022. Tidyheatmaps: heatmaps from Tidy Date. R package version 0.1.0. https://github.com/jbengler/tidyheatmaps.
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. 2013. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 49(2):359–367. doi: 10.1016/j.molcel.2012.10.016
- Harrold E, Latham A, Pemmaraju N, Lieu CH. 2023. Early-onset GI cancers: rising trends, genetic risks, novel strategies, and special considerations. Am Soc Clin Oncol Educ Book 43:e398068. doi: 10.1200/EDBK_398068
- Hu J, Hu B, Gui YC, Tan ZB, Xu JW. 2019. Diagnostic value and clinical significance of methylated sept9 for colorectal cancer: a meta-analysis. Med Sci Monit. 25:5813–5822. doi: 10.12659/MSM.915472
- Johnston AD, Ross JP, Ma C, Fung KYC, Locke WJ. 2023. Epigenetic liquid biopsies for minimal residual disease, what’s around the corner? Front Oncol. 13:1103797. doi: 10.3389/fonc.2023.1103797
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 32(Database issue):D493–496. doi: 10.1093/nar/gkh103
- Kuhn M, Vaughan D, Hvitfelt E, Posit Software P. 2023. yardstick: tidy characterizations of model performance. R package version 1.2.0. https://cran.r-project.org/web/packages/yardstick/index.html.
- Laird PW. 2003. The power and the promise of DNA methylation markers. Nat Rev Cancer 3(4):253–266. doi: 10.1038/nrc1045
- Lamb YN, Dhillon S. 2017. Epi proColon((R)) 2.0 CE: A blood-based screening test for colorectal cancer. Mol Diagn Ther. 21(2):225–232. doi: 10.1007/s40291-017-0259-y
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ. 2013. Software for computing and annotating genomic ranges. PLoS Comput Biol. 9(8):e1003118. doi: 10.1371/journal.pcbi.1003118
- Levenson VV. 2010. DNA methylation as a universal biomarker. Expert Rev Mol Diagn. 10(4):481–488. doi: 10.1586/erm.10.17
- Liu MC, Oxnard GR, Klein EA, Swanton C, Seiden MV, & Consortium, C. 2020. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol. 31(6):745–759. doi: 10.1016/j.annonc.2020.02.011
- Locke WJ, Guanzon D, Ma C, Liew YJ, Duesing KR, Fung KYC, Ross JP. 2019. DNA Methylation Cancer Biomarkers: Translation to the Clinic. Front Genet. 10:1150. doi: 10.3389/fgene.2019.01150
- Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M, et al. 2008. DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem. 54(2):414–423. doi: 10.1373/clinchem.2007.095992
- Maestranzi S, Przemioslo R, Mitchell H, Sherwood RA. 1998. The effect of benign and malignant liver disease on the tumour markers CA19-9 and CEA. Ann Clin Biochem. 35 (Pt 1)(1):99–103. doi: 10.1177/000456329803500113
- Massarweh NN, El-Serag HB. 2017. Epidemiology of hepatocellular carcinoma and intrahepatic cholangiocarcinoma. Cancer Control 24(3):1073274817729245. doi: 10.1177/1073274817729245
- Mitchell SM, Ho T, Brown GS, Baker RT, Thomas ML, McEvoy A, Xu ZZ, Ross JP, Lockett TJ, Young GP, et al. 2016. Evaluation of methylation biomarkers for detection of circulating tumor DNA and application to colorectal cancer. Genes (Basel) 7(12):125. doi: 10.3390/genes7120125
- Mitchell SM, Ross JP, Drew HR, Ho T, Brown GS, Saunders NF, Duesing KR, Buckley MJ, Dunne R, Beetson I, et al. 2014. A panel of genes methylated with high frequency in colorectal cancer. BMC Cancer 14(1):54. doi: 10.1186/1471-2407-14-54
- Moss J, Magenheim J, Neiman D, Zemmour H, Loyfer N, Korach A, Samet Y, Maoz M, Druid H, Arner P, et al. 2018. Comprehensive human cell-type methylation atlas reveals origins of circulating cell-free DNA in health and disease. Nat Commun. 9(1):5068. doi: 10.1038/s41467-018-07466-6
- Navarro M, Nicolas A, Ferrandez A, Lanas A. 2017. Colorectal cancer population screening programs worldwide in 2016: An update. World J Gastroenterol. 23(20):3632–3642. doi: 10.3748/wjg.v23.i20.3632
- NCI Genomic Data Commons. 2022. Bioinformatics pipeline: methylation analysis. In: GDC data user’s guide, vol. 35.0, pp. 56–57. https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/Expression_mRNA_Pipeline/.
- Paulovich AG, Whiteaker JR, Hoofnagle AN, Wang P. 2008. The interface between biomarker discovery and clinical validation: the tar pit of the protein biomarker pipeline. Proteomics Clin Appl. 2(10-11):1386–1402. doi: 10.1002/prca.200780174
- Pedersen SK, Baker RT, McEvoy A, Murray DH, Thomas M, Molloy PL, Mitchell S, Lockett T, Young GP, LaPointe LC. 2015. A two-gene blood test for methylated DNA sensitive for colorectal cancer. PLoS One. 10(4):e0125041. doi: 10.1371/journal.pone.0125041
- Pedersen SK, Symonds EL, Roy AC, Cornthwaite KJ, LaPointe LC, Young GP. 2023. Detection of methylated BCAT1 and IKZF1 after curative-intent treatment as a prognostic indicator for colorectal cancer recurrence. Cancer Med. 12(2):1319–1329. doi: 10.1002/cam4.5008
- Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R, Clark SJ, Molloy PL. 2015. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 8(1):6. doi: 10.1186/1756-8935-8-6
- Potter NT, Hurban P, White MN, Whitlock KD, Lofton-Day CE, Tetzner R, Koenig T, Quigley NB, Weiss G. 2014. Validation of a real-time PCR-based qualitative assay for the detection of methylated SEPT9 DNA in human plasma. Clin Chem. 60(9):1183–1191. doi: 10.1373/clinchem.2013.221044
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7):e47–e47. doi: 10.1093/nar/gkv007
- Robinson JT, Thorvaldsdottir H, Turner D, Mesirov JP. 2023. igv.js: an embeddable JavaScript implementation of the Integrative Genomics Viewer (IGV). Bioinformatics 39(1):btac830. doi: 10.1093/bioinformatics/btac830
- Sanz-Garcia E, Zhao E, Bratman SV, Siu LL. 2022. Monitoring and adapting cancer treatment using circulating tumor DNA kinetics: current research, opportunities, and challenges. Sci Adv. 8(4):eabi8618. doi: 10.1126/sciadv.abi8618
- Shirley M. 2020. Epi proColon((R)) for colorectal cancer screening: a profile of its use in the USA. Mol Diagn Ther. 24(4):497–503. doi: 10.1007/s40291-020-00473-8
- Shvetsov YB, Song MA, Cai Q, Tiirikainen M, Xiang YB, Shu XO, Yu H. 2015. Intraindividual variation and short-term temporal trend in DNA methylation of human blood. Cancer Epidemiol Biomarkers Prev. 24(3):490–497. doi: 10.1158/1055-9965.EPI-14-0853
- Sjoberg DD, Whiting K, Curry M, Lavery JA, Larmarange J. 2021. Reproducible summary tables with the gtsummary package. The R J. 13(1):570–580. doi: 10.32614/RJ-2021-053
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. 2021. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 71(3):209–249. doi: 10.3322/caac.21660
- Swanton C, Neal RD, Johnson PWM, Dur CC, Hamilton SA, Zhang N, Kumar H, Sasieni P. 2022. NHS-Galleri trial design: equitable study recruitment tactics for targeted population-level screening with a multi-cancer early detection (MCED) test. JCO. 40(16_suppl):TPS6606–TPS6606. doi: 10.1200/JCO.2022.40.16_suppl.TPS6606
- Symonds EL, Cock C, Meng R, Cole SR, Fraser RJL, Young GP. 2018B. Uptake of a colorectal cancer screening blood test in people with elevated risk for cancer who cannot or will not complete a faecal occult blood test. Eur J Cancer Prev. 27(5):425–432. doi: 10.1097/CEJ.0000000000000352
- Symonds EL, Pedersen SK, Baker RT, Murray DH, Gaur S, Cole SR, Gopalsamy G, Mangira D, LaPointe LC, Young GP. 2016. A blood test for methylated BCAT1 and IKZF1 vs. a fecal immunochemical test for detection of colorectal neoplasia. Clin Transl Gastroenterol. 7(1):e137. doi: 10.1038/ctg.2015.67
- Symonds EL, Yao B, Pedersen SK, Murray D, Young GP. 2018A. Specificity of methylated BCAT1 and IKZF1 for colorectal cancer. JCO 36(4_suppl):580–580. doi: 10.1200/JCO.2018.36.4_suppl.580
- Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, The Cancer Genome Atlas Research Network. 2013. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 45(10):1113–1120. doi: 10.1038/ng.2764
- Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, Grolemund G, Hayes A, Henry L, Hester J, et al. 2019. Welcome to the Tidyverse. JOSS. 4(43):1686. doi: 10.21105/joss.01686
- Winter JM, Sheehan-Hennessy L, Pedersen SK, Young GP, Symonds EL. 2022A. Detection of circulating DNA methylated BCAT1 and IKZF1 in pancreatic adenocarcinoma. JCO. 40(4_suppl):597–597. doi: 10.1200/JCO.2022.40.4_suppl.597
- Winter JM, Sheehan-Hennessy L, Yao B, Pedersen SK, Wassie MM, Eaton M, Chong M, Young GP, Symonds EL. 2022B. Detection of hypermethylated BCAT1 and IKZF1 DNA in blood and tissues of colorectal, breast and prostate cancer patients. Cancer Biomark. 34(3):493–503. doi: 10.3233/CBM-210399
- Wise J. 2022. A blood test for multiple cancers: game changer or overhyped? BMJ 378:o2279. doi: 10.1136/bmj.o2279
- World Cancer Research Fund International. 2020. Worldwide cancer data. https://www.wcrf.org/cancer-trends/worldwide-cancer-data/.
- Zaimi I, Pei D, Koestler DC, Marsit CJ, De Vivo I, Tworoger SS, Shields AE, Kelsey KT, Michaud DS. 2018. Variation in DNA methylation of human blood over a 1-year period using the Illumina MethylationEPIC array. Epigenetics 13(10-11):1056–1071. doi: 10.1080/15592294.2018.1530008
- Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. 2011. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics 6(5):623–629. doi: 10.4161/epi.6.5.15335

