Abstract
Context: The bulb of Allium sativum Linn (Alliaceae) has numerous medicinal values. Though the petroleum ether extract of the bulb has shown to exhibit antimycobacterial activity, the phytochemical(s) responsible for this inhibitory activity is not known.
Objective: To characterize the bioactive compounds in the petroleum ether extract of Allium sativum (garlic) that inhibit the growth of Mycobacterium tuberculosis H37Ra.
Materials and methods: Bioactivity-guided fractionation was employed to isolate the bioactive compounds. Antimycobacterial activity was evaluated by well-diffusion method and microplate alamar blue assay (MABA). Infrared spectroscopy, mass spectrometry and nuclear magnetic resonance spectroscopy were used to characterize the bioactive compounds. Autodock was used to obtain information on molecular recognition, and molecular dynamics simulation was performed using GROMACS.
Results: The bioactive compounds that inhibited the growth of M. tuberculosis H37Ra were found to be lauric acid (LA) and myristic acid (MA). The minimal inhibitory concentration of LA and MA was found to be 22.2 and 66.7 μg/mL, respectively. In silico analysis revealed that these fatty acids could bind at the cleft between the N-terminal and C-terminal lobes of the cytosolic domain of serine/threonine protein kinase B (PknB).
Discussion and conclusion: The inhibition activity was dependent on the alkyl chain length of the fatty acid, and the amino acid residues involved in binding to fatty acid was found to be conserved across the Pkn family of proteins. The study indicates the possibility of using fatty acid derivatives, involving Pkn family of proteins, to inhibit the signal transduction processes in M. tuberculosis.
Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis is one of the most prolific infectious disease with high mortality worldwide (Dye et al. Citation1999). In 2013, 8.6 million people were estimated to develop TB, of whom 1.3 million died (including 360,000 deaths among HIV-positive) [Global Tuberculosis Report, World Health Organization (WHO), Citation2013]. Annual reports indicate that the number of multidrug-resistant (MDR) cases is on the rise (Global Tuberculosis Report, WHO, Citation2014). Therefore, it is necessary to look for alternate strategies to combat this infectious bacterium.
Medicinal plants are being used worldwide by traditional healers to treat various ailments, including infectious diseases. Literature indicates that the primary sources of 65–75% medicines used today are plants (Newton et al. Citation2000). Extracts of various common herbal plants have been shown to exhibit antimycobacterial activity. Gebreyohannes and Gebreyohannes (Citation2013) recently reviewed the medicinal values of Allium sativum Linn (Alliaceae) and documented that the extracts of this common plant have potential pharmaceutical application for the treatment of heart diseases, hypertension, cancer, diabetics, inflammation, atherosclerosis, hyperlipidaemia, viral and microbial infections.
Allicin from A. sativum inhibits the growth of M. tuberculosis in vitro (Schauenberg & Paris Citation1990; Gupta et al. Citation2011), but loses its beneficial properties within an hour as it rapidly gets oxidized to diallyl sulfides, diallyl disulfides, diallyl trisulfides, vinyl dithiins and ajoenes. Geraniol, from the same plant but at higher concentration, also inhibits the growth of M. tuberculosis (Dini et al. Citation2011). The in vivo antimycobacterial activity of garlic oil was studied in tubercle bacilli-exposed guinea pigs and it was comparable with the acivity of first-line drugs such as streptomycin and rifampicin (Jain Citation1998). The pharmacological activity of garlic and the phytochemicals therein are being studied for the past three decades, of which the organosulphur compounds, especially allicin is one among the most-studied compounds (Mikaili et al. Citation2013). Apart from the antimicrobial activity, the selected sulphur compounds from garlic have found to induce apoptosis by suppressing NF-kB signaling through destabilization of TRAF6 in primary effusion lymphoma (Shigemi et al. Citation2016); attenuates oxidation stress, inflammation and endothelial dysfunction in hypercholesterolemic rabbits (El-Sheakh et al. Citation2016) and protects against ischemic stroke in a rat model (Zhang et al. Citation2015).
We have shown, recently, that the petroleum ether extract of the bulb of A. sativum inhibits the growth of M. tuberculosis H37Ra. Since the initial analysis revealed that the phytochemical responsible for the inhibition activity could be different from allicin and geraniol (Rajiniraja & Jayaraman Citation2014), the present study aimed to isolate and characterize the active compounds from the bulbs of A. sativum using various analytical techniques and to arrive at a possible mechanism of action through molecular modelling and simulations.
Materials and methods
Chemicals
Petroleum ether, chloroform-AR, rifampicin were purchased from SDF Chem. Ltd (Gujarat, India); Silica gel 230–400 mesh from HiMedia (Mumbai, India); Alamar Blue, 96-well micro titre plate, Loewenstein-Jensen medium base from Fluka-63237 (Bangalore, India); Middlebrook 7H9 broth base from AccumixTM (Bangalore, India); glycerol, lauric acid (LA), myristic acid, palmitic acid and stearic acid were purchased from Sigma-Aldrich (Bangalore, India).
Preparation of petroleum ether extract
Allium sativum (garlic) bulbs were purchased from the local market (Vellore, Tamil Nadu, India) on September 2011. The bulbs were authenticated by Dr. K. Madhava Chetty and a specimen (Voucher no. 2011) was deposited at the herbarium of Sri Venkateswara University, Tirupati, India.
The bulb was peeled, washed with double-distilled water, chopped into small pieces, air dried in shade at room temperature (25 °C) for two weeks and then ground into fine paste. The paste was stored in sterile bag at 4 °C until use (Harborne Citation1973).
The non-polar components in the above paste were extracted (40 °C, 9 h) with petroleum ether (250 mL) using Soxhlet apparatus. The extract was then evaporated to dry using rotary evaporator (40 °C) and was stored in sterile bottle (covered with aluminum foil) as reported earlier (Chaudhar Citation1996; Rajiniraja & Jayaraman Citation2014).
Experimental design
Column chromatography
The petroleum ether extract (0.5 g) of A. sativum was subjected to column chromatography (1.5 x 54 cm; 15 g of silica gel 230–400 mesh) and the extracts were eluted using a step gradient of petroleum ether and chloroform (10:0, 9.5:0.5, 9:1, 8.5:1.5, 8:2, 7.5:2.5, 7:3, 6.5:3.5, 6:4, 0:10 each 1 L). Fractions were transferred into tightly capped sterile glass bottles and stored at 4 °C until further use (Patra et al. Citation2012).
High-performance liquid chromatography
Column chromatographic fractions that exhibited antimycobacterial activity were subjected to HPLC (LC-8A prominence and Rheodyne type injector, Shimadzu Corporation, Japan). Phenomenex Luna (250 mm x 10 mm) column was used and elution was done in a reverse-phase mode at ambient temperature. The elution was performed in isocratic mode with acetonitrile:water (60:40), flow rate of 8.6 mL/min and detection wavelength of 220 nm. The fractions were collected, evaporated to dryness and stored (at 4 °C) in a sterile glass bottle.
Culture medium and organism growth
Mycobacterium tuberculosis H37Ra (MTCC 300) was purchased from IMTECH, Chandigarh (India). The organism was sub-cultured by suspending 6.2g of the Loewenstein–Jensen medium base in 100 mL water containing 2 mL glycerol and autoclaved at 15 lbs for 15 min. Egg emulsion (50 mL) was mixed with the base to obtain uniform mixture. The medium was aliquoted (10 mL) in sterile screw capped bottles and was made to coagulate by heating it at 85–90 °C for 45 min (NCIM). After solidification, M. tuberculosis was streaked (one loop full) and incubated at room temperature for 4 weeks.
The pure culture was isolated from the slant (LJ medium) and inoculated into 5 mL Middlebrook 7H9 broth base (Middlebrook & Cohn Citation1958) supplemented with 10% (v/v) albumin dextrose catalase (AccumixTM) and 0.2% (v/v) glycerol (Sigma-Aldrich). The inoculated media was maintained at 37 °C for 72 h on a shaker until an OD600 of 0.6–0.8 was reached (equivalent to 3.2 × 107 cfu/mL).
Mycobacterium tuberculosis assay
In order to obtain the qualitative information on the antimycobacterial activity of the extracts agar well diffusion assay was performed (Parish & Stroker Citation1998, Molina-Salinas et al. Citation2007, Kouznetsov & Sánchez Citation2010; Rajiniraja & Jayaraman Citation2014). In all these experiments rifampicin (5 μg/mL) served as a positive control.
Determination of minimum inhibitory concentration
MIC was determined by microplate Alamar blue assay (MABA) in a 96-well micro titre plate. To 180 μL of freshly cultured medium, 20 μL of the sample (2–20 μg) was added. Rifampicin (5 μg) was used as the positive control. The plate was incubated at 37 °C for 48 h and 10 μL of Alamar blue was served into the wells (total volume of 210 μL). The MIC was defined as the lowest concentration of the extract in which the change in colour from blue to pink was not observed (Collins & Franzblau Citation1997; Prescott et al. Citation1999).
EI-MS, IR and NMR spectroscopy analysis
Electron impact-mass spectrometry (EI-MS) was carried out using JEOL GCMATE II GC-MS (Tokyo, Japan) with EI mode of ionization. The Fourier transform-infrared (FT-IR) spectra were recorded using a Perkin–Elmer instrument in the range of 400–4000 cm−1 in thallium bromide. Nuclear magnetic resonance (NMR) spectra were acquired on Bruker Accent multi nuclei probe spectrophotometer at 400 MHz (1H, 13C). Chemical shifts (δ ppm) were referenced with respect to tetra methyl silane (TMS). The spectra (1H, 13C, DEPT, COSY and HSQC) were recorded in CDCl3 (Lanos et al. Citation2012).
Docking of fatty acids to PknB and PstP
The three-dimensional structures of serine/threonine protein kinase B (PknB) (PDB ID: 1MRU) and phosphatase (PDB ID: 2CM1) were retrieved from the Protein Data Bank. Since the structure of PknB (1MRU) lacked the region spanning from Ile163 to Thr179, the conformation of the missing region was predicted using Phyre2 (Protein fold recognition server: http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index). The full-length protein was used for docking studies.
Docking of PknB and protein serine/threonine phosphatase (PstP) with the ligand was performed by Autodock (4.0) using the Lamarckian genetic algorithm (Hu & Shelver Citation2003). The active site was defined using AutoGrid. The grid size was set to 90 × 90 × 90 points with a grid spacing of 0.375 Å, centered on the binding site of the protein molecules (Dario & Roberto Citation2009). The grid box included the entire protein molecule to ensure enough space for the ligand translational and rotational walk. Step sizes of 1 Å for translation and 50° for rotation were chosen and the maximum number of energy evaluations was set to 54,00,000. One hundred runs were performed for each ligand. For each of the 100 independent runs, the genetic algorithm was set to generate 27 000 operations on a single population of 200 individuals. Default values were used for crossover, mutation and elitism operator weights (0.80, 0.02, and 1.00, respectively). Docked conformations of 100 runs were clustered on the basis of their root mean square deviation (RMSD) tolerance of the ligand conformation. Consequently, the best docked conformation was selected and visualized using Pymol 0.99 (New York, NY).
Molecular dynamics simulation
Molecular dynamics (MD) simulation of the free and ligand-bound protein was carried out with the GROMACS (Groningen, Netherland) 4.5.3 package using GROMOS96 force field (Berendsen et al., Citation1995; Schuler et al., Citation2001). The conformation of the protein–ligand complex with lowest binding energy (most negative) was used for MD simulation. The topology parameters for the proteins were created by using the GROMACS program. The topology parameters of the ligand were built using the Dundee PRODRG server (Scotland, UK) (Schuttelkopf & Van Aalten, Citation2004). The entire scheme was subjected to MD production run at a temperature of 300 K and a pressure of 1 bar for 5000 ps.
Results
Bioactivity-guided fractionation
The petroleum ether extract of A. sativum was recently shown to exhibit antimicrobial property against selected microorganisms, including M. tuberculosis H37Ra (Rajiniraja & Jayaraman Citation2014). In continuation of our efforts to investigate the nature of the active component, we embarked on purifying the petroleum ether extract of A. sativum by chromatographic techniques. The fractions collected were pooled (P1–P11) based on the Rf value and the homogeneity observed on the TLC plates. The fraction P9 exhibited a significant antimycobacterial activity (Supplementary Figure 1). The yield (19.3% DW of the petroleum ether extract) of this component was apparently the highest ().
Table 1. Chromatographic fractions of petroleum ether extract of A. sativum Linn (Alliaceae).
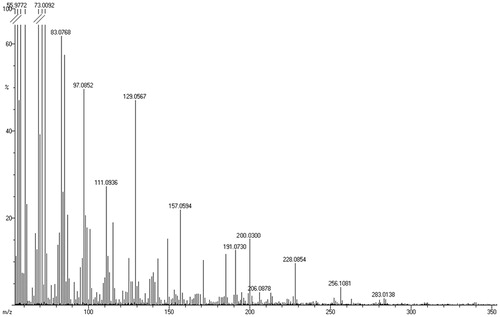
The EI-MS obtained, for P9, in the positive-ion mode displayed peaks at m/z 200, 228, 256 and 283 () with their relative intensity (%) as 15, 10, 4, and 2, respectively. The difference of 28 in the m/z value between the subsequent peaks is suggestive of the presence of long chain aliphatic compounds.
TLC and HPLC analysis indicated that the active fraction, P9, contained more than one component (Supplementary Figures 2–4) and therefore subsequent fractionation was performed with semi-preparative reverse-phase HPLC (Supplementary Figure 5). The one-dimensional (1D) proton NMR of the subfractions that displayed antimycobacterial activity (F3–F5), were not identical but had similar peak patterns. This indicated that the phytochemicals present in these subfractions could be of similar chemical nature. In addition, the spectral range of the chemical shifts (0.75–2.50 ppm) indicated that these compounds could be aliphatic, which corroborate with the mass spectral data. summarizes the steps involved in the purification of the active component. The overall yield and purification fold of the bioactive component (subfraction F3) was found to be 31.0 mg (31.95% DW with respect to P9) and 43, respectively.
Table 2. Purification process of the petroleum ether extract of A. sativum with information on yield, specific inhibition activity and purification fold.
Determination of the minimal inhibitory concentration (MIC) of the subfractions was performed, as per MABA (Supplementary Figure 6). The MIC of the subfractions F3 and F4 were found to be 22.2 and 66.7 μg/mL (), which are lower than the value reported for allicin and comparable with geraniol (Delaha & Garagusi Citation1985; Bueno-Sánchez et al. Citation2009).
Table 3. The EI-Mass, 1H-NMR and antimycobacterial activity by plate assay and MABA of the subfractions F3, F4 and F5.
IR, mass spectrometric and NMR spectroscopic analysis
The bioactive component in subfraction (F3) was found to be colourless and odourless solid substance. The FT-IR spectrum () showed absorptions for aliphatic C–H stretches (at 2911 and 2845 cm−1) and one carbonyl stretch at 1690 cm−1. Such spectral characteristics indicated that the compound could be aliphatic carbonyl/carboxyl compound. EI-MS displayed peaks at m/z 200 with the relative intensity of 38% (). The molecular ion is clearly abundant, and there are ions representing fragmentations of methylene groups.
Figure 2. (a) IR spectrum, (b) EI-MS, (c) COSY (expanded spectrum), (d) HSQC (expanded spectrum) in CDCl3 for the bioactive subfraction (F3).
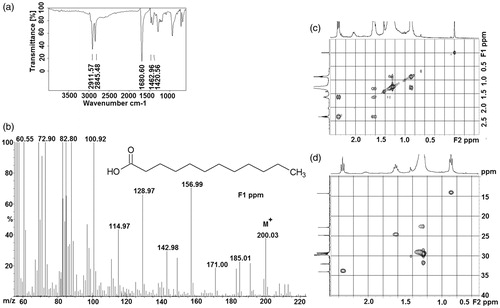
In order to get further insight into the nature of the active compound (in F3 fraction), various one-dimensional (1D) and two-dimensional (2D) experiments were performed. The resonances in 1H (0.75–2.5 ppm) and 13C-NMR (20–40 ppm) (Supplementary Figures 7) confirmed the presence of a long chain aliphatic group in the active compound. Presence of carbonyl was inferred from the signal at 180 ppm in 13C-NMR. The scalar connectivities obtained from the 2D experiments, COSY () and HSQC (), confirmed that the active molecule in the F3 fraction to be LA. The chemical shifts of 1H and 13C values are given in Supplementary Table 1. Similarly, the bioactive compound in the subfractions F4 and F5 were identified to be myristic acid (MA) and palmitic acid, respectively (Supplementary Figures 8).
Docking of lauric acid to PknB and PstP
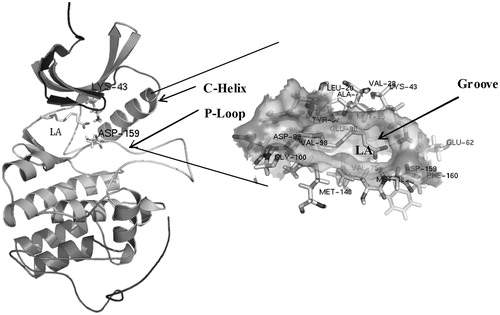
The list of membrane proteins, common in both virulent (H37Rv) and avirulent (H37Ra) strains of M. tuberculosis, for which the three-dimensional structures are known is given in (Hiwa et al. Citation2011). The superimposed structure of the modelled full-length protein on the original truncated protein of PknB (1MRU) is given in .
Figure 3. Superposition of the modeled cytosolic domain of PknB on the loop-deficient structure available at the PDB (PDB ID: 1MRU). The modeled loop region, present in between the N-terminal and C-terminal lobe is indicated.
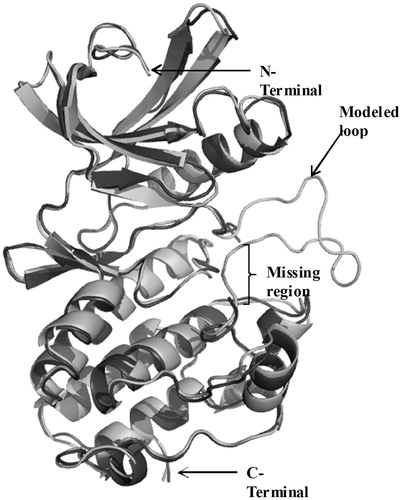
Table 4. The protein IDs, gene name and PDB codes for 12 membrane proteins are shown.
In order to derive unbiased information on the possible ligand binding site in PknB and PstP, blind docking procedure was adapted. One-hundred structures were generated and a root mean square (RMS) tolerance of 4 Å was set to form a cluster. In each cluster, structures were ranked by their binding energy. The best docked conformation was selected based on the lowest binding energy and with higher number of conformations in a given cluster (Rohit et al. Citation2009). It was observed that PknB displayed efficient binding (binding energy of −5.14 kcal/mol) with LA ().
Table 5. Results of lauric acid docking in PknB modeled structure (modeled 1MRU) and PstP (pdb code 2CM1) using Autodock 4.0 software.
Moreover, cluster analysis revealed that 56% of PknB–LA complex have the lowest binding energy compared to 13% for PstP–LA complexes. Additionally, identical binding regions were observed in both the clusters representing PknB–LA complex. These inferences indicate that LA could have greater preference for binding to PknB than PstP-LA.
Binding site residues
The frequency of occurrence of a given amino acid in the LA binding site of PknB is given in . It could be seen that the amino acid residues Val28, Lys43, Met148, Met158 and Asp159 are involved in all the 71 conformations. Moreover, the ligand binding site in PknB was predicted using 3DLigandSite server (http://www.sbg.bio.ic.ac.uk/3dligandsite/) (Wass et al. Citation2010). It was observed that the amino acids Leu20, Gly21, Phe22, Gly23, Gly24, Met25, Ser26, Val28, Ala41, Lys43, Met95, Glu96, Tyr97, Val98, Thr102, Ala145, Asn146, Met148, Met158 and Asp159 were presented in the predicted binding site. This site was identical to the binding region of KT5720 reported earlier (Carl et al. Citation2008).
Table 6. Binding site residues traced from the ligand with the distance of 4 Å using Swissprot deep viewer.
depicts the binding mode of PknB with LA. Amino acid residues Lys43 and Asp159 in the activation loop of PknB were found to interact with the carboxylic group of LA.
Effect of chain length
In order to have a comprehensive understanding on the chain length dependency on the bi-molecular interaction, molecular docking was performed with other long-chain fatty acids. The results indicated that both lauric and MA interact with Lys43 and Asp159. However, palmitic acid was found to interact only with Asp159 (Supplementary Figures 9) possibly because of the strain conferred by the increased length of the alkyl chain and therefore could fail to fit into the binding cavity of PknB. Clustering analysis indicated that the number of conformations with lower binding energy decreases with increase in alkyl chain length, thus reflecting on its decreasing binding efficiency (Supplementary Table 2). However, the mean binding energy of lauric and MA was comparable.
Stability of the complex
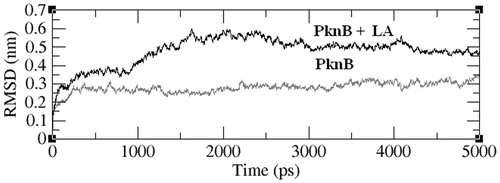
The stability of PknB–LA complex was evaluated by performing molecular dynamic simulations. The RMSD of the simulated structures with respect to the initial structure is given in . It could be seen that the RMSDs of both PknB and PknB-LA are stable after a simulation time period of 1500 ps, thus reflecting on their stability. However, the conformation of the amino acid residues in the ligand binding site seems to change upon binding to LA.
Discussion
LA, as approved by FDA, is a permitted food additive for human consumption as long as its concentration does not change the physical, chemical and other technical effect of the food. In pharmaceutical applications, LA attachment enhances the bioavailability of certain drugs (Kravchenko et al. Citation2003). It also has the ability to control mastitis, an infection caused by Staphylococcus aureus (Kelsey et al. Citation2006). Antimycobacterial assay as well as molecular docking analysis emphasizes that LA is very effective in inhibiting the growth of M. tuberculosis. Studies on bioactivity of the molecules using agar well diffusion assay is of limited value (Collins & Franzblau Citation1997) and, in certain instances, may not reflect the real biological activity of the molecules. Diffusion of the molecules in the agar medium is the major rate limiting step and therefore cannot be used for any quantitative analysis. With this in view, in the present study, the antimycobacterial effect of the long chain fatty acids was analyzed both by agar well diffusion assays and by MABA. It should be noted that in both the assays, the inhibition activity of these fatty acids is indirectly proportional to the chain length, with LA being the most potent molecule to display this property. However, it should be mentioned that Kondo and Kanai (Citation1977) and Saravanakumar et al. (2008) indicated greater antimycobacterial activity for myristic acid. Direct correlation of the carbon chain length was also observed previously with regard to the cytotoxicity (with respect to leukemic cell lines) of these fatty acids (Lima et al., Citation2002). In addition to LA, MA extracted from other sources has been shown to possess antimycobacterial activity (Saravanakumar et al. Citation2008).
Among the long chain fatty acids used in the present study, the antimicrobial activity of MA is well documented (Miller et al. Citation1977; Skrivanova et al. Citation2006; Huang et al. Citation2011). It is present in the catalytic subunit of cyclic AMP-dependent protein kinase. MA can form N-myristylated protein by the catalytic enzyme N-myristyl transferase (Aitken et al. Citation1982; Carr et al. Citation1982). N-myristoylation is a co-translational modification involved in protein–protein interactions as well as in anchoring polypeptides to phospholipid bilayers by covalently attached to its N-terminus for proper protein localization or biological function; however, its role in targeting proteins to specific subcellular compartments has not been clearly defined (Peitzseh & MeLaughlin Citation1993; Michael et al. Citation1996). MA has also been indicated that myristoylated reductase localizes to mitochondria and endoplasmic reticulum by different mechanisms, and reveals a novel role for MA in protein targeting (Borgese et al. Citation1996). However, sufficient information on the exact mode of action of other long-chain fatty acids is not available. Also, the involvement of other fatty acids in the post-translational/co-translational modification of proteins is unknown. Supportively, a few of these fatty acids have been shown to interfere in the signal transduction process of the selected prokaryotic organisms and mammalian cells, thus affecting the growth and proliferation of the cell (Tommy et al. Citation1992; Tyagi & Deepak Citation2004; Yogesh et al. Citation2014).
In M. tuberculosis, the protein kinase family of proteins (PknA-PknG) have been shown to play crucial role in the bacterial survival or growth and therefore considered as one of the factor contributing to virulence. The membrane-bound members of Pkn family share greater homology in their sequence and therefore their domain organization. The cytosolic domain of this group of proteins possesses Ser/Thr kinase activity. Based on the information on the inhibition of signal transduction process and the importance of Pkn family in M. tuberculosis, we embarked on investigating the possible binding mode of these selected alkyl fatty acids to Pkn family of proteins in M. tuberculosis. Sequence alignment of PknB with the other members of this group indicates conservation of the two amino acids, Lys43 and Asp159, which are involved in electrostatic interaction with the fatty acid. Also, the enlarged binding site contains Leu20, Gly21, Val28, Ala41, Glu62, Val73, Val75, Glu96, Tyr97, Val98, Met148 and Met158, thus making hydrophobic contact with the alkyl chain. Though blind docking was used to dock the ligand, approximately 80% of the binding site residues correlate with predicted binding site residues (according to 3DLigandSite). Residues Lys140-Ile146 comprises the activation loop of PknB, which functions to coordinate with either Mg2+ or Mn2+ ion (Young et al., Citation2003). The molecular modelling also indicates that both the length of the alkyl chain and the carbonyl group plays vital role in the molecular recognition.
Conclusions
The present study, for the first time, authenticates the antimycobacterial property of long-chain fatty acids, especially LA and MA extracted from A. sativum. It is proposed that these molecules could possibly interfere with the signal transduction process of M. tuberculosis (H37Ra) by binding to PknB and its variants. However, more detailed analysis need to be carried out to have an in-depth understanding on the in vivo significance of this bi-molecular interaction.
Acknowledgements
We acknowledge SIF, VIT University, Vellore (India) for extending the NMR, IR facility. We thank SAIF (IITM) for EI-MS. We also thank Mr. Vincent Sagayaraj from St Joseph College, Trichy, for their help in the semi-preparative HPLC. The authors thank the management of VIT University for providing the necessary infrastructure facility to carry out this work.
Disclosure statement
The authors report no declarations of interest.
References
- Aitken A, Cohen P, Santikarn S, Williams DH, Calder AG, Smith A, Klee CB. 1982. Identification of the NH2-terminal blocking group of calcineurin B as myristic acid. FEBS Lett. 150:314–318.
- Berendsen HJC, Van der Spoel D, Van Drunen R. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun. 91:43–56.
- Borgese N, Aggujaro D, Carrera P, Pietrini G, Bassetti M. 1996. A role for N-myristoylation in protein targeting: NADH-cytochrome b5 reductase requires myristic acid for association with outer mitochondrial but not ER membranes. J Cell Biol. 135:1501–1513.
- Bueno-Sánchez JG, Martínez-Morales JR, Stashenko EE, Ribón W. 2009. Anti-tubercular activity of eleven aromatic and medicinal plants occurring in Colombia. Biomedica. 29:51–60.
- Carl M, Anthony TI, Tom A. 2008. Auto-activation mechanism of the Mycobacterium tuberculosis PknB receptor Ser/Thr kinase. EMBO J. 27:3186–3197.
- Carr SA, Biemann K, Shoji S, Parmelee DC, Titani K. 1982. n-Tetradecanoyl is the NH2-terminal blocking group of the catalytic subunit of cyclic AMP-dependent protein kinase from bovine cardiac muscle. Proc Natl Acad Sci USA. 79:6128–6131.
- Chaudhar RD. 1996. Herbal drug industry. New Delhi: Eastern Publishers.
- Collins LA, Franzblau SG. 1997. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 41:1004–1009.
- Dario G, Roberto S. 2009. Improving accuracy and efficiency of blind protein-ligand docking by focusing on predicted binding sites. Proteins. 74:417–424.
- Delaha EC, Garagusi VF. 1985. Inhibition of mycobacterial by garlic extract (Allium sativum). Antimicrob Agents Chemother. 27:485–486.
- Dini C, Fabbri A, Geraci A. 2011. The potential role of garlic (Allium sativum) against the multi-drug resistant tuberculosis pandemic: a review. Ann Ist Super Sanita. 47:465–473.
- Dye C, Scheele S, Dolin P, Pathania V, Raviglione MC. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 282:677–686.
- El-Sheakh AR, Ghoneim HA, Suddek GM, Ammar ESM. 2016. Attenuation of oxidative stress, inflammation, and endothelial dysfunction in hypercholesterolemic rabbits by allicin. Can J Physiol Pharmaco. 94:216–224.
- Gebreyohannes G, Gebreyohannes M. 2013. Medicinal values of garlic: a review. Int J Med Sci. 5:401–408.
- Global tuberculosis report. 2013. Report global tuberculosis control: surveillance, planning, financing. World Health Organization: Geneva, Switzerland; [cited 2015 April 20]. Available from http://apps.who.int/iris/bitstream/10665/91355/1/9789241564656_eng.pdf.
- Global tuberculosis report. 2014. Drug-resistant TB surveillance & response, World Health Organization; [cited 2015 April 20]. Available from http://apps.who.int/iris/bitstream/10665/137094/1/9789241564809_eng.pdf.
- Gupta R, Thakur B, Singh P, Singh HB, Sharma VD, Katoch VM, Chauhan SVS. 2011. Anti-tuberculosis activity of selected medicinal plants against multi-drug resistant Mycobacterium tuberculosis isolates. Ind J Med Res. 131:809–813.
- Harborne JB. 1973. Phytochemical methods. London: Chapman and Hall Ltd, 49–188.
- Hiwa M, Gustavo ADS, Sharad P, Tina S, Harald GW. 2011. Comparison of membrane proteins of Mycobacterium tuberculosis H37Rv and H37Ra strains. BMC Microbiol. 11:18.
- Hu X, Shelver WH. 2003. Docking studies of matrix metalloproteinase inhibitors: zinc parameter optimization to improve the binding free energy prediction. J Mol Graph Model. 22:115–126.
- Huang CB, Altimova Y, Myers TM, Ebersole JL. 2011. Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch Oral Biol. 56:650–654.
- Jain RC. 1998. Anti tubercular activity of garlic oil. Indian J Pathol Microbiol. 41:131.
- Kelsey JA, Bayles KW, Shafii B, McGuire MA. 2006. Fatty acids and monoacylglycerols inhibit growth of Staphylococcus aureus. Lipids. 41:951–961.
- Kondo E, Kanai K. 1977. The relationship between the chemical structure of fatty acids and their mycobactericidal activity. Jpn J Med Sci Biol.30:171–178.
- Kouznetsov VV, Sánchez JGB. 2010. Antimycobacterial susceptibility testing methods for natural products research. Braz J Microbiol. 41:270–277.
- Kravchenko IA, Golovenko NY, Larionov VB, Aleksandrova AI, Ovcharenko NV. 2003. Effect of lauric acid on transdermal penetration of phenazepam in vivo. Bull Exp Biol Med. 136:579–581.
- Lanos GG, Araujo LM, Jiménez IA, Moujir LM, Bazzocchi IL. 2012. Withaferin A-related steroids from Withania aristata exhibit potent antiproliferative activity by inducing apoptosis in human tumor cells. Eur J Med Chem. 54:499–511.
- Lima TM, Kanunfre CC, Pompeia C, Verlengia R, Curi R. 2002. Ranking the toxicity of fatty acids on Jurkat and Raji cells by flow cytometric analysis. Toxicol In Vitro. 16:741–747.
- Michael TK, Howard B, Frank R. 1996. Contributions of Myristoylation to calcineurin structure/function. J Biol Chem. 271:26517–26521.
- Middlebrook G, Cohn ML. 1958. Bacteriology of tuberculosis: laboratory methods. Am J Public Health Nations Health. 48:844–853.
- Mikaili P, Maadirad S, Moloudizargari M, Aghajanshakeri S, Sarahroodi S. 2013. Therapeutic uses and pharmacological properties of garlic, shallot, and their biologically active compounds. Iran J Basic Med Sci. 16:1031–1048.
- Miller RD, Brown K, Morse SA. 1977. Inhibitory action of fatty acids on the growth of Neisseria gonorrhoeae. Infect Immun.17:303–312.
- Molina-Salinas GM, Pérez-López A, Becerril-Montes P, Salazar-Aranda R, Said-Fernández S, Waksman de Torres N. 2007. Evaluation of the flora of northern Mexico for in vitro antimicrobial and antituberculosis activity. J Ethnopharmacol. 109:435–441.
- Newton SM, Lau C, Wright CW. 2000. A review of antimycobacterial natural products. Phytother Res. 14:303–322.
- Parish T, Stroker NG. 1998. Mycobacteria protocols: methods in molecular biology. Totowa (NJ): Humana Press.
- Patra JK, Gouda S, Sahoo SK, Thatoi HN. 2012. Chromatography separation, 1H NMR analysis and bioautography screening of methanol extract of Excoecaria agallocha L. from Bhitarkanika, Orissa, India. Asian Pac J Trop Biomed. 2:S50–S56.
- Peitzseh RM, MeLaughlin S. 1993. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristoylated proteins. Biochem. 32:10436–10443.
- Prescott ML, Harley J, Donald P, Klein A. 1999. Microbiology. 2nd ed. TX: Brown Publisher. p. 325.
- Rajiniraja M, Jayaraman G. 2014. Bioautography guided screening of selected Indian medicinal plants reveals potent antimycobacterial activity of Allium sativum extracts – implication of non sulfur compounds in inhibition. Int J Pharm Pharm Sci. 6:671–676.
- Rohit T, Kiran M, Ryan P, Chenglong L, Werner T. 2009. Carborane clusters in computational drug design: a comparative docking evaluation using Autodock, Flexx, Glide and Surflex. J Chem Inf Model. 49:1581–1589.
- Saravanakumar DEM, Folb PI, Campbell BW, Smith P. 2008. Antimycobacterial activity of the red alga Polysiphonia virgata. Pharm Biol. 46:254–260.
- Schauenberg P, Paris F. 1990. Guide to medicinal plants. First paperback edition, Paris: Lutterworth Press.
- Schuler LD, Daura X, Van Gunsteren WF. 2001. An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase. J Comput Chem. 22:1205–1218.
- Schuttelkopf AW, Van Aalten DMF. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D. 60:1355–1363.
- Shigemi Z, Furukawa Y, Hosokawa K, Minami S, Matsuhiro J, Nakata S, Watanabe T, Kagawa H, Nakagawa K, Takeda H, et al. 2016. Diallyl trisulfide induces apoptosis by suppressing NF-κB signaling through destabilization of TRAF6 in primary effusion lymphoma. Int J Oncol. 48:293–304.
- Skrivanova E, Marounek M, Benda V, Brezin P. 2006. Susceptibility of Escherichia coli, Salmonella sp. and Clostridium perfringens to organic acids and monolaurin. Vet Med – Czech. 51:81–88.
- Tommy N, Tomas M, Tiina P, Leif CA. 1992. Modulation of calcium fluxes in Jurkat T cells by myristic acid inhibition is independent of membrane potential and intracellular pH. Biochem J. 283:113–118.
- Tyagi JS, Deepak S. 2004. Signal transduction systems of mycobacteria with special reference to M. tuberculosis. Curr Sci. 86:93–102.
- Wass MN, Kelley LA, Sternberg MJ. 2010. 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res. 38:469–473.
- Yogesh C, Sandeep U, Shazia K, Sathya Narayanan N, Francesca F, Vinay Kumar N. 2014. Protein Kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J Biol Chem. 289:13858–13875.
- Young TA, Delagoutee B, Endrizzi JA, Falcik AM, Alber T. 2003. Structure of Mycobacterium tuberculosis PknB supports a universal activation mechanism for Ser/Thr protein kinases. Nat Struct Biol.10:168–174.
- Zhang B, Li F, Zhao W, Li J, Li Q, Wang W. 2015. Protective effects of allicin against ischemic stroke in a rat model of middle cerebral artery occlusion. Mol Med Rep. 12:3734–3738.