Abstract
Context: H3K18ac is linked to gene expression and DNA damage. Nevertheless, whether H3K18ac participates in regulating Ras-ERK1/2-affected lung cancer cell phenotypes remains unclear.
Objective: We explored the effects of H3K18ac on Ras-ERK1/2-affected lung cancer cell phenotypes.
Material and methods: NCI-H2126 cells were transfected with, pEGFP-K-RasWT and pEGFP-K-RasG12V/T35S plasmids for 48 h, and transfection with pEGFP-N1 served as a blank control. Then H3K18ac and AKT and ERK1/2 pathways-associated factors were examined. Different amounts of the H3K18Q (0.5, 1, and 2 μg) plasmids and RasG12V/T35S were co-transfected into NCI-H2126 cells, cell viability, cell colonies and migration were analyzed for exploring the biological functions of H3K18ac in NCI-H2126 cells. The ERK1/2 pathway downstream factors were detected by RT-PCR and ChIP assays. The regulatory functions of SIRT7, GCN5 and MDM2 in Ras-ERK1/2-regulated H3K18ac expression were finally uncovered.
Results: RasG12V/T35S transfection decreased the expression of H3K18ac about 2.5 times compared with the pEGFP-N1 transfection group, and activated ERK1/2 and AKT pathways. Moreover, H3K18ac reduced cell viability, colonies, migration, and altered ERK1/2 downstream transcription in NCI-H2126 cells. Additionally, SIRT7 knockdown increased H3K18ac expression and repressed cell viability, migration and the percentage of cells in S phase by about 50% compared to the control group, as well as changed ERK1/2 downstream factor expression. Besides, Ras-ERK1/2 decreased H3K18ac was linked to MDM2-regulated GCN5 degradation.
Conclusion: These observations disclosed that Ras-ERK1/2 promoted the development of lung cancer via decreasing H3K18ac through MDM2-mediated GCN5 degradation. These findings might provide a new therapeutic strategy for lung cancer.
Introduction
Lung cancer is one of the most common and catastrophic cancers with high morbidity and mortality (Zhang et al. Citation2014), leading to about 1.8 million and 1.6 million deaths per year (Hirsch et al. Citation2017). Lung cancer diagnosed at an advanced stage often has a poor survival rate, and is a primary cause of cancer-related death (Hussain Citation2016). With the development of technology, a deeper understating of lung cancer has been obtained. However, there is still no effective therapeutic strategy for lung cancer due to the complex pathological mechanism and metastatic disease (Hensing et al. Citation2014). Nowadays, considerable attention has turned to epigenetics to look for potential mechanisms.
The Ras pathway has been discovered to be closely linked to the occurrence and development of human cancers, including lung cancer (Zhao et al. Citation2015; Baietti et al. Citation2016). K-RasG12 is a common signaling pathway, often studied as an oncogenic gene (Voisset et al. Citation2013). Previous literature reported that the main downstream factors of Ras signaling included extracellular regulated protein kinases (ERK)1/2, phosphatidylinositol 3′-kinase (PI3K), and Ras-like (Ral) 2 guanine nucleotide exchange factors (RalGEFs) (Yang et al. Citation2012; Cooper et al. Citation2013; Niu et al. Citation2015), which can be continuously activated by Ras. Activation of ERK1/2 has been shown to be associated with advanced and aggressive lung cancer (Vicent et al. Citation2004). Even though the Ras pathway is well-known to be linked to human cancer, the exact mechanism is still not well established.
In the field of epigenics, histone modification has become an important research topic (Lawrence et al. Citation2016). It is well known that eukaryotic DNA is wrapped by histone octamers, which is made of four different histones, including H2A, H2B, H3 and H4 (Shim et al. Citation2012). Interestingly, every histone has tails that can be modified, and thereafter to affect various kinds of biological processes, such as chromatin compaction, nucleosome dynamics and transcription (Lawrence et al. Citation2016). Importantly, many studies have shown that histone modification was involved in mediating tumor development, including lung cancer (Hattori and Ushijima Citation2014; Sundar and Rahman Citation2016). Histone H3 acetylation is often observed in multiple cancers because of the different acetyl points on lysine residues (Rodrigues et al. Citation2017). For instance, histone H3 lysine 23 acetylation (H3K23ac) is intimately related to poor prognosis in breast cancer (Ma et al. Citation2016). H3K18ac has been shown to be linked to several biological processes, such as mitosis and cellular senescence (Tasselli et al. Citation2016). Furthermore, previous studies disclosed that Sirtuin7 (SIRT7) and H3K18 acetylation were involved in mediating protein biosynthesis and maintaining tumor phenotypes (Martinez-Redondo et al. Citation2012). Therefore, we inferred that H3K18ac might also play important roles in lung cancer.
In our research, we explored the relationship between the Ras-ERK1/2 pathway and H3K18ac in lung cancer NCI-H2126 cells. Additionally, the functions of H3K18ac in cell viability, colonies, migration and the underlying mechanisms were probed in NCI-H2126 cells. The research might provide new insight for the treatment of lung cancer.
Materials and methods
Cell culture
NCI-H2126 cells (ATCC® CCL-256™) were obtained from American Type Culture Collection (ATCC, Rockville, USA). Cells were maintained at 37 °C in an incubator comprising 5% CO2 in Dulbecco's modified Eagle medium (DMEM, Gibco Laboratorties, Grand Island, NY) with 100 units/mL penicillin, 100 μg/mL streptomycin and 5% fetal bovine serum (FBS, Life Science, UT, USA).
Plasmid construction and siRNA
The pEGFP-N1, pEGFP-K-RasWT and pEGFP-K-RasG12V/T35S plasmids were transfected into NCI-H2126 cells. Meanwhile, transfection with specific gene with HA-tag was used for screening out the target factor through western blot. pEGFP-K-RasG12V/T35S plasmid was mutated by site-directed mutagenesis. SiRNAs, which were using interference RNA to silence the target RNA of mouse double minute 2 homolog (MDM2) or SIRT7 were produced by Shanghai GenePharma (Shanghai, China). These vectors were named as si-MDM2, si-SIRT1 and si-SIRT2, respectively. The plasmids of MDM2-His and MDM2-MU were acquired from Abace Biotechnology Co., Ltd. (Beijing, China). The pEGFP-H3K18Q construct was constructed through utilizing the TaKaRa MutanBEST Kit (TaKaRa, Shiga, Japan), as recommend by the manufacturer. Different amounts of the H3K18Q (0.5, 1, and 2 μg) were utilized for cell transfection in the next experiments.
Transfection
NCI-H2126 cells at a density of 5 × 105 per well were cultured in 6-well plates for 12 h. These cells were then transfected with the above-mentioned plasmids or siRNA through adopting Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After transfection for 48 h, RT-PCR and/or western blots were executed to determine the transfection efficiency.
Cell viability
MTT [3-(4, 5-dimethyl thiazol-2-yl)-2, 5-diphenyl tetrazolium bromide, Sigma-Aldrich, St. Louis, MO, USA] was utilized for the detection of cell viability. After cultivation for 48 h, 20 µL MTT (5 mg/mL) were added into each well, and then NCI-H2126 cells were cultivated with MTT solution for another 4 h at 37 °C. Afterward, 100 μL dimethyl sulfoxide (DMSO, Sigma-Aldrich) were added to lyse formazan crystal and then the absorbance was read at 570 nm utilizing a multiwell spectrophotometer (Emax; Molecular Devices, Sunnyvale, CA, USA).
Reverse transcription polymerase chain reaction (RT-PCR)
RT-PCR assay was performed according to a previous report (Bustin Citation2002). Total RNA was obtained and isolated from transfected NCI-H2126 cells through utilizing TRIzol reagent (Invitrogen). DNase-I-treated 1 μL total RNA was supplied for first-strand cDNA synthesis by M-MuLV reverse transcriptase (Fermentas, York, UK) and oligo-dT primers (Invitrogen). QuantiTect SYBR Green PCR Kit (Qiagen, Hilden, Germany) was used to amplify the target sequence. GAPDH served as an internal control for detecting RNA expression (triplicate experiments).
Soft-agar colony formation assay
Soft-agar formation assay was executed for the detection of colony formation ability (Horibata et al. Citation2015). NCI-H2126 cells were firstly suspended in the full culture medium with 0.35% low-melting agarose. Subsequently, these cells (1 × 103 cells/well) were transferred into solidified 0.6% agarose in six-well culture plates. After cultivation for 3-4 weeks, the number of the colonies was determined utilizing a microscope (40×, Olympus, Tokyo, Japan).
Transwell migration assay
After transfection with the correlative plasmids, cell migration was evaluated by exploiting a modified two-chamber migration Transwell (Corning Costa, NY, US) with a pore size of 8 μm. The 100 μL cell suspension (2 × 105 cells/mL) without serum was added to the upper compartment. Then, 600 μL culture medium with 10% FBS was added in the lower compartment of a 24-well Transwell. Subsequently, NCI-H2126 cells were cultivated for 24 h at 37 °C with humidified air and 5% CO2. After incubation, the un-migrated cells at the upper surface of the filter were removed with a cotton swab, meanwhile the migrated cells at the lower surface of the filter were fixed with methanol for 5 min and stained with 0.1% Giemsa (Sigma-Aldrich) for 15 min. These cells were counted by employing a microscope (Olympus). The OD-values of these migrated cells were finally determined utilizing a multiwell spectrophotometer (Emax; Molecular Devices) at 570 nm.
Western blot
Western blot was executed to detect protein expression. Total protein was obtained from transfected NCI-H2126 cells via utilizing RIPA lysis buffer (Cat. No:R0010, Solarbio, Beijing, China) supplemented with protease inhibitors (Thermo Fisher Scientific, Rockford, IL). The BCA™ Protein Assay Kit (Pierce, Appleton, WI, USA) was used for the detection of protein concentrations. The western blot system was established using a Bio-Rad Bis-Tris Gel system following the manufacturer’s instructions. Primary antibodies of anti-Histone H3 (acetyl K18) antibody (ab40888), anti-Histone H3 antibody (ab176842), anti-p-ERK1/2Thr202 antibody (ab223500), anti-ERK1/2 antibody (ab115799), anti-p-AKT antibody (ab38449), anti-AKT antibody (ab8805), anti-Ras antibody (ab52939), anti-HA antibody (ab20084), anti-GFP antibody (ab6556), anti-General control of nucleotidesynthesis 5 (GCN5) anti-body (ab217876), anti-His (ab83831), MDM2 (ab226939) and β-actin (ab16039, Abcam, Cambridge, UK) were prepared and diluted according to the product instruction. These primary antibodies were incubated with membrane and maintained at 4 °C overnight at recommended concentration. Afterward, the membranes were incubated with horseradish peroxidase (HRP) conjugated second antibody (ab205718, Abcam) for an extra 2 h at indoor temperature. Western Blotting Luminol Reagent (Santa Cruz Biotechnology, CA, USA) was performed to capture the signals and the intensity of the bands was quantified via adopting Image Lab™ Software (Bio-Rad, Shanghai, China).
Flow cytometric analysis of cell cycle distribution
NCI-H2126 cells were cultured until reaching 75–80% confluence, and then cells were washed by PBS to remove the non-adherent cells. After transfection with the related plasmids, these cells were washed with PBS again and were fixed with cold 70% ethanol. After that, cells were stained with 4, 6-diamidino-2-phenylindole (DAPI) (Partec, Germany) for 30 min keeping in darkness. Flow cytometry (Becton Dickinson, San Jose, CA, USA) was utilized for detecting cell cycle distribution. The percentage of cells in the G0/G1, S and G2/M phases of cell cycle was calculated.
Chromatin immunoprecipitation (ChIP)
NCI-H2126 cells were cultured and transfected, then cells were fixed in 1% formaldehyde for 10 min at room temperature, and subsequently rinsed twice with PBS, meanwhile lysed with SDS lysis buffer (Beyotime). Afterward, these above lysates were subjected to an ultrasonic bath for sonication (Bioruptor, Diagenode, Belgium). After centrifugation at 15,000 × g for 10 min, the supernatants were diluted with ChIP dilution buffer (Upstate Biotechnology, Lake Placid, NY, USA) and then were immunoprecipitated with anti-H3K18ac (2 μg, ab1191, Abcam) all night long at 4 °C. The normal anti-IgG antibody (2 μg, ab2410, Abcam) was regarded as a control of immunoprecipitation. The dynabeads were washed with in low-salt for 5 min at 4 °C, in the meantime were washed twice in 1 × TE (Upstate Biotechnology) for 2 min at room temperature. The DNA eluted from the beads according to previous literature (Schulz and Haussler Citation2014). After that, the purified DNA was used for PCR amplification at the CYR61, IGFBP3, WNT16B, NT5E, GDF15, CARD16 promoters.
Statistical analysis
All the data in this research were analyzed by Graphpad 6.0 statistical software (GraphPad, San Diego, CA, USA) and the data were presented as mean + SD. The statistical analyses were performed using the one-way ANOVA followed by Duncan post hoc multiple comparisons. *p < 0.05, **p < 0.01 and ***p < 0.001 were considered to be significant consequences.
Results
H3K18ac was reduced by Ras-ERK1/2 pathway
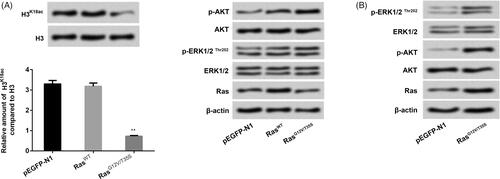
Ras/ERK pathway has been observed to be linked to lung cancer (Cheng et al. Citation2015). In the current study, NCI-H2126 cells were transfected with pEGFP-N1, pEGFP-K-RasWT and pEGFP-K-RasG12V/T35S plasmids. In , we discovered that transfection with RasG12V/T35S significantly reduced the expression level of H3K18ac about 2.5 times as compared with that transfection with pEGFP-N1 group (p < 0.01). Furthermore, we found that transfection with RasG12V/T35S activated p-ERK1/2 expression and activated p-AKT expression (), hinting that RasG12V/T35S could simultaneously activate AKT and ERK pathways. These observations indicated that H3K18ac expression was indeed specifically reduced by the Ras-ERK1/2 pathway.
Figure 1. H3K18ac expression was reduced by the Ras-ERK1/2 pathway. NCI-H2126 cells were transfected with pEGFP-N1, pEGFP-K-RasWT (RasWT), pEGFP-K-RasG12V/T35S (RasG12V/T35S) plasmids. (A and B) The expression levels of H3K18ac, Ras and the factors of AKT and ERK pathways were then measured by western blots. Data presented as mean + SD, ** p < 0.01 (n = 3).
H3K18ac participated in regulating the functions of Ras-ERK1/2 in lung cancer cell phenotypes
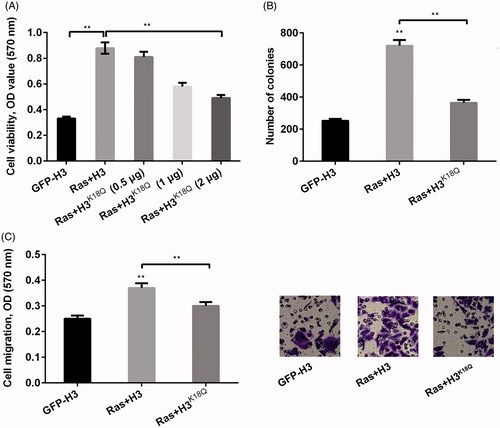
It is well-known that histone modification can influence cell growth and cell metastasis (Jiao et al. Citation2014). In the current study, we constructed H3K18Q plasmid to mimic the situation of the acetylation of H3K18 and the mimicked H3K18Q plasmid at different amounts of 0.5, 1 and 2 μg was co-transfected with RasG12V/T35S plasmid into NCI-H2126 cells. Thereafter, we examined the functions of acetylation of histone H3K18 in lung cancer cell viability, cell colony ability and cell migration. Results showed that Ras-ERK1/2 activation significantly increased cell viability about three times (p < 0.01), while transfection with H3K18Q reduced cell viability in a concentration-dependent manner (p < 0.01, ). This result suggested that Ras-ERK1/2 had the ability to increase cell viability, while H3K18Q explained a suppressive function in cell viability. In addition, the number of colonies (p < 0.01, ) and cell migration (p < 0.01, ) both revealed the similar trend by the Ras-ERK1/2 pathway, which was reversed by H3K18Q. Taken together, these results suggested that H3K18ac restrained Ras-ERK1/2-triggered acceleration of cell proliferation and migration in NCI-H2126 cells.
Figure 2. H3K18ac was involved in regulating the functions of Ras-ERK1/2 signaling pathway in lung cancer cell phenotypes. NCI-H2126 cells were transfected with pEGFP-N1, pEGFP-H3, pEGFPH-RasG12V/T35S, or pEGFP-H3K18Q (0.5, 1 and 2 μg) (indicated as GFP, H3, Ras and H3K18Q, respectively) plasmids. (A) Cell viability, (B) numbers of colonies and (C) cell migration were determined by MTT assay, soft agar formation, and Transwell assays, respectively. Data presented as mean + SD, **p < 0.01 (n = 3).
H3K18ac participated in mediating the downstream factors of the ERK1/2 pathway
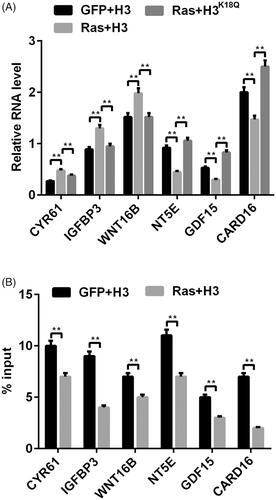
Ras-ERK1/2 pathway is a complex and exact regulation pathway, which is modulated by diverse downstream factors (Harada et al. Citation2015). We next probed the expression of these correlative important downstream factors of CYR61 (Xu et al. Citation2009), IGFBP3 (Guo et al. Citation2016), WNT16B (Johnson et al. Citation2013), NT5E (Wang et al. Citation2018), GDF15 (Li et al. Citation2016), CARD16 (Karasawa et al. Citation2015), which were all involved in mediating tumor cell growth and metastasis. We observed that, transfection with RasG12V/T35S obviously increased CYR61 (p < 0.01), IGFBP3 (p < 0.01) and WNT16B (p < 0.01) expression by less than 50%, while reducing NT5E (p < 0.01), GDF15 (p < 0.01) and CARD16 expression by less than 50% (p < 0.01, ). On the other hands, co-transfection with RasG12V/T35S and H3K18Q showed the opposite trend in the expression levels of these factors relative to transfection with the RasG12V/T35S group (p < 0.01, ). In addition, ChIP assay results revealed the reduced input levels of H3K18ac in these gene promoter regions (all p < 0.01, ). Taken together, the above-mentioned results corroborated that H3K18ac participated in modulating the transcription of the ERK1/2 downstream factors.
Figure 3. The transcription of Ras-ERK1/2-targeted genes was mediated by H3K18ac. (A) The mRNA levels of ERK1/2 downstream genes (CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16) were tested via RT-PCR after transfection with the corrective plasmids. (B) The transcription of these above-mentioned genes was detected through using ChIP after transfection with the related plasmids. Data presented as mean + SD, **p < 0.01 (n = 3).
Knockdown of SIRT7 mediated H3K18ac expression and recovered Ras-ERK1/2-affecting lung cell phenotypes
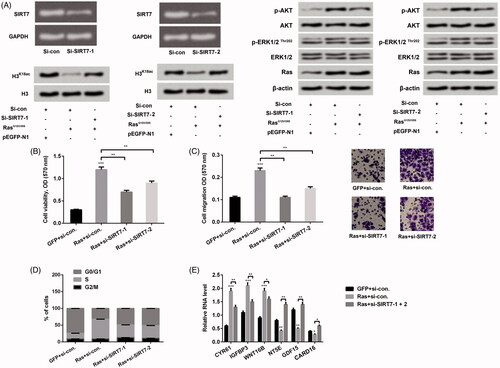
From a former report, we understand that SIRT7 is an NAD±-dependent H3K18ac deacetylase, which has potential functions in transforming the cancer cell state (Barber et al. Citation2012). Therefore, we next probed the impact of increasing H3K18ac triggered by SIRT7 silence on NCI-H2126 cell phenotypes. RT-PCR and western blot were both conducted to determine the transfection efficiency when NCI-H2126 cells were transfected with si-SIRT7-1 and si-SIRT7-2, respectively. Results in showed that transfection with si-SIRT7-1 and si-SIRT7-2 both decreased SIRT7 expression in mRNA level in NCI-H2126 cells. In addition, transfection with si-SIRT7-1/-2 reversed the declined expression of H3K18ac triggered by Ras-ERK1/2 in NCI-H2126 cells. Meanwhile, we observed that transfected with si-SIRT7-1 and si-SIRT7-2 both hindered activation of the ERK1/2 and AKT pathways. We then further detected the effects of transfection with si-SIRT7-1/2 on cell viability, cell migration, cell cycle stages and relative mRNA levels of ERK1/2 downstream factors. Results disclosed that co-transfection with RasG12V/T35S and si-SIRT7-1/2 decreased cell viability (p < 0.01), migration (p < 0.01) and the percentage of NCI-H2126 cells in S phase about 50% as compared with transfection with RasG12V/T35S alone (). Furthermore, co-transfection with RasG12V/T35S and si-SIRT7-1/2 also changed the mRNA expression of the downstream factors of ERK1/2 pathway as compared with transfection with RasG12V/T35S alone in NCI-H2126 cells (p < 0.05, p < 0.01 or p < 0.001, ). These investigations hinted that knockdown of SIRT7 reversed Ras-ERK1/2-evoked decrease of H3K18ac, meanwhile impeded the tumor-promoting functions of Ras-ERK1/2 in NCI-H2126 cells.
Figure 4. Knockdown of SIRT7 adjusted H3K18ac expression and mediated Ras-ERK1/2-affecting lung cell phenotypes. (A) After si-SIRT7-1 and si-SIRT7-2 plasmids transfection, the expression levels of SIRT7, Ras, AKT and ERK1/2 pathway associated factors were detected through RT-PCR and western blot. NCI-H2126 cells were co-transfected with pEGFP-K-RasG12V/T35S or pEGFP-N1 plasmids and SIRT7-specific siRNA or control siRNA as indicated (Ras, GFP, si-SIRT7-1, si-SIRT7-2, or si-con, respectively). (B) Cell viability, (C) cell migration, (D) cell cycle progression and (E) ERK1/2 downstream genes were examined by MTT assay, Transwell, flow cytometry and RT-PCR assays. Data presented as mean + SD, *p < 0.05, **p < 0.01, ***p < 0.001 (n = 3).
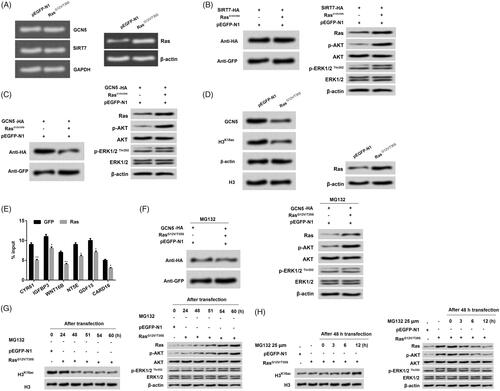
Activation of Ras-ERK1/2 triggered GCN5 degradation to reduce H3K18ac expression
GCN5 has an acetyl-lysine binding bromodomain module, which can regulate nucleosomal acetylation in vitro, as well as site-specificity of lysine acetylation on histone H3 (Cieniewicz et al. Citation2014). Further experiments were carried out to explore the exact mechanism of how GCN5 affected the acetylation of H3K18. Results from showed that no obvious difference was observed in the mRNA expression of GCN5 and SIRT7 in the pEGFP-N1 and RasG12V/T35S groups, which suggested that GCN5 expression level was unaffected by RasG12V/T35S at the transcription level. Afterwards, we found that the protein level of SIRT7 was not different (), while GCN5 protein level was significantly decreased in the group RasG12V/T35S relative to the pEGFP-N1 group (). Combined with the result from , we inferred that the expression of GCN5 regulated by RasG12V/T35S was at the translation level. Moreover, we observed that SIRT7 and GCN5 both activated the ERK1/2 and AKT pathways in NCI-H2126 cells. Interestingly, results in confirmed that GCN5 and H3K18ac expression had a similar trend in RasG12V/T35S group. Further results showed that RasG12V/T35S reduced the recruitment of GCN5 to the promoters of ERK1/2 downstream genes (CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16) (). Moreover, proteasome inhibitor MG132 administration made the changes of GCN5 expression by RasG12V/T35S disappear, which indicated high inhibition of GCN5 (). Afterwards, we found that without MG132 administration, the expression levels of H3K18ac were decreased with delayed transfection times (24, 48, 51, 54 and 60 h) in NCI-H2126 cells (). On the other hand, with the MG132 supplement, the decreased expression level of H3K18ac by RasG12V/T35S was abolished. Instead, the expression level of H3K18ac was increased after administration with MG132 at different time points (0, 3, 6 and 12 h) (). The similar phenomenon was presented in p-ERK1/2 and p-AKT expression in NCI-H2126 cells. Taken together, these findings indicated that RasG12V/T35S pathway affected the expression level of H3K18ac through the GCN5.
Figure 5. Ras-ERK1/2 activation degraded GCN5 to decline H3K18ac expression. (A) NCI-H2126 cells were transfected with pEGFP-K-RasG12V/T35S, pEGFP-N1, and GCN5-HA or SIRT7-HA plasmids. After transfection, the mRNA levels of GCN5 and SIRT7 were detected by RT-PCR. (B-D) The protein levels of GCN5, SIRT7, H3K18ac, Ras and AKT and ERK1/2 pathways associated factors were analyzed by western blot. (E) The recruitment of GCN5 to the ERK1/2 downstream genes was analyzed via ChIP analysis. (F) After treatment with MG132 (25 μM), the GCN5, Ras, and AKT and ERK1/2 pathways associated factors protein levels were analyzed by western blot with antibodies against HA and GFP. (G-H) After 24, 48, 51, 54, and 60 h transfection without MG132 treatment or 48 h transfection with MG132 treatment (25 μM, 0, 3, 6, and 12 h), the protein levels of H3K18ac, Ras and AKT and ERK1/2 pathways associated factors were analyzed by western blot. Data presented as mean + SD, *p < 0.05, **p < 0.01 (n = 3).
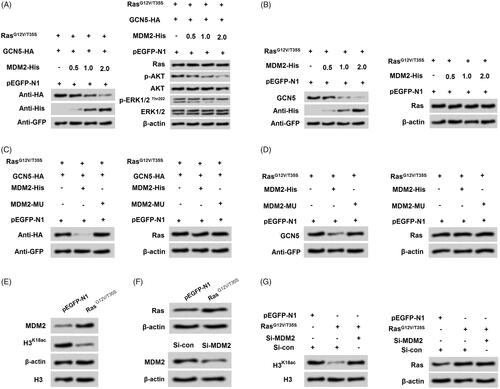
Activation of Ras-ERK1/2 degraded GCN5 via modulating MDM2
It is well validated that histone modification is mediated by MDM2 in various cells (Minsky and Oren Citation2004). Further experiments were performed to explore if the mechanism of MDM2 was involved in the regulation of GCN5 on H3K18ac expression. After co-transfection with RasG12V/T35S and GCN5 with tagged HA, we found that GCN5 expression was decreased with increasing amounts of MDM2 (0.5, 1 and 2 μg, ). The results indicated that there might be a negative relationship between MDM2 and GCN5. Likewise, the ascending p-AKT and p-ERK1/2 triggered by GCN5 were both reduced with increasing amounts of MDM2 (0.5, 1 and 2 μg). After transfection with MDM2-MU, the decreased GCN5 was notably recovered (), which further strongly suggested that there was a close negative relationship between MDM2 and GCN5. We thereafter determined the relationship between MDM2 and H3K18ac, and the results in showed that transfection with RasG12V/T35S inhibited H3K18ac while up-regulated MDM2 expression in NCI-H2126 cells. Then transfection with si-MDM2 to silence MDM2 (), and we found that transfection with si-MDM2 enhanced the expression of H3K18ac in NCI-H2126 cells (). These results disclosed that RasG12V/T35S triggered reduction of H3K18ac might be through MDM2-mediated GCN5 degradation in NCI-H2126 cells.
Figure 6. Ras-ERK1/2 pathway-induced degradation of GCN5 was mediated by MDM2. (A-B) NCI-H2126 cells were co-transfected with GCN5-HA, pEGFP-K-RasG12V/T35S, of pEGFP-N1 plasmids and increasing amounts of MDM2-His plasmid (0.5, 1, and 2 μg), the protein levels of GCN5, MDM2, Ras, and AKT and ERK1/2 pathways associated factors were examined by western blot. (C-D) After transfection with a mutation in MDM2 (MDM2-MU), GCN5 and Ras protein levels were detected by western blot. (E and F) After pEGFP-N1, pEGFP-K-RasG12V/T35S or si-MDM2 transfection, MDM2, H3K18ac and Ras protein levels were analyzed via western blot. (G) After co-transfection with pEGFP-K-RasG12V/T35S and MDM2-specific siRNA, H3K18ac and Ras protein levels were determined by western blot.
Discussion
Lung cancer is the leading cause of cancer-related deaths worldwide with the 5-year survival rate consistently less than 7% (Del Vescovo and Denti Citation2015; Semenova et al. Citation2015). Poor outcomes and high mortality motivated researchers to pay attention to explore the potential pathogenesis of lung cancer. In our study, we investigated the effects of H3K18ac and Ras-ERK1/2 pathway on NCI-H2126 cell phenotype. Results showed that Ras-ERK1/2 played a role as a switch for the acetylation of H3K18. Additionally, H3K18ac affected cell viability, cell colonies and cell migration as well as regulated the transcription of ERK1/2 downstream genes by regulating SIRT7 and GCN5. Outside of these, Ras-ERK1/2 decreased H3K18ac might be associated with MDM2-mediated GCN5 degradation.
Histone modification is mainly divided into acetylation, methylation, phosphorylation and ubiquitylation, which becomes a dominating reason causing aberrant gene damage and is also a hallmark of human cancer development (Lippman et al. Citation2005). The Ras pathway has been shown to be involved and play important roles in diverse human cancers, such as colorectal cancer (Rui et al. Citation2015) and colon cancer (Goel et al. Citation2015). Furthermore, activated Ras-ERK1/2 pathway often discovered in multiple human cancers (Samatar and Poulikakos Citation2014). In our study, we found that transfection with RasG12V/T35S decreased the accumulated level of H3K18ac, while activated ERK pathway, which suggested that Ras-ERK1/2 might play a vital role in regulation of H3K18ac expression and further influence other cell activities.
Cell metastasis is one main reason for lack of cancer control and ultimately causing death (Chaffer and Weinberg Citation2011). Cell viability, colony formation and migration are three important aspects, which can reveal cell growth and metastasis. Therefore, we investigated the functions of RasG12V/T35S and H3K18ac in cell viability, colonies and migration. Results demonstrated that Ras-ERK1/2 increased cell viability, colonies and migration, which was consistent with the previous report that inactivation of Ras-ERK1/2 suppressed lung tumor progression (Jiang et al. Citation2018). However, the promoting trends were inhibited by transfection with H3K18ac, which indicated that H3K18 might inhibit the tumorigenic impacts of Ras-ERK1/2 in lung cancer cells. Additionally, the down-stream genes of ERK1/2 (CYR61, IGFBP3, WNT16B, NT5E, GDF15 and CARD16) were further investigated to uncover the underlying mechanisms. We discovered that H3K18ac could modulate the transcription of these ERK1/2 downstream genes. These above-involved explorations further verify the involvement of H3K18ac in the development of lung cancer.
SIRT7 is a histone deacetylase of H3K18ac, which has been confirmed to influence gastric cancer cell growth and cell apoptosis (Zhang et al. Citation2015). According to this report, we inferred that SIRT7 might play a role in lung cancer cells through regulating H3K18ac expression. As expected, we discovered that SIRT7 silencing restored Ras-ERK1/2-decreased H3K18ac expression, as well as decreased cell viability, migration and the percentage of cells at S phase, meanwhile altered the downstream factors of ERK1/2 at the mRNA level. These results were consistent with a previous study that SIRT7 promoted ovarian cancer cell viability and SIRT7 knockdown induced cell apoptosis (Wang et al. Citation2015). These findings indicated that SIRT7 might be a pivotal adjustor in Ras-ERK1/2 and H3K18ac affected NCI-H2126 cells. What’s more interesting, we discovered that SIRT7 silence restrained the activations of ERK1/2 and AKT pathways. According to this, we speculated that the functions of SIRT7 silence in NCI-H2126 cells might be associated with the regulation of ERK1/2 and AKT pathways. Further research is still needed to confirm this speculation.
GCN5 is a histone H3 acetylase, which has been reported to be involved in the progression of H3 acetylation (Burgess et al. Citation1999). Interesting research disclosed that GCN5 degradation was linked to the acetylation of H3K56 (Li et al. Citation2012). Therefore, we hypothesized that GCN5 might be also involved in the progress of acetylation of H3K18. Interestingly, we found that GCN5 functions at the translational level. Further research uncovered that activation of Ras-ERK1/2 induced GCN5 degradation to decrease H3K18ac in NCI-H2126 cells. MDM2 is frequently observed in human cancers and also related to histone acetylation (Nihira et al. Citation2017). In our study, the correlative result demonstrated that Ras-ERK1/2 activation degraded GCN5 through up-regulation of MDM2. Taken together, the whole cascade process might be Ras-ERK1/2 activation evoked the declined H3K18ac expression through MDM2-regulated GCN5 degradation in NCI-H2126 cells.
To our best knowledge, the relationship between SIRT7 and GCN5 has not been reported in existing research. In our research, we discovered that SIRT7 silence recovered Ras-ERK1/2-decreased H3K18ac expression, and restrained the carcinogenesis of Ras-ERK1/2 in NCI-H2126 cells. Moreover, Ras-ERK1/2 decreased H3K18ac was linked to GCN5 degradation. Therefore, we speculated that SIRT7 might affect GCN5 degradation, thereby influencing Ras-ERK1/2-regulated H3K18ac expression, as well as affecting lung cancer cell phenotypes. However, further research is still needed for confirmation.
Conclusions
These results demonstrated that Ras-ERK1/2 activation regulated lung cancer cell biological processes via suppression of H3K18ac. The processes might be modulated by SIRT7 and MDM2-GCN5 axis. This research might provide novel insight in the relationship between histone modification and the treatment of lung cancer. Nevertheless, the relationship between SIRT7 and GCN5 is still not shown in this research. Additionally, the wide impact of Ras-ERK1/2-H3K18ac axis in lung cancer is worthy for further investigation.
Acknowledgement
Thanks for Linyi Central Hospital who hosted the research.
Disclosure statement
The authors declare no conflict of interest.
References
- Baietti MF, Simicek M, Abbasi Asbagh L, Radaelli E, Lievens S, Crowther J, Steklov M, Aushev VN, Martinez Garcia D, Tavernier J, et al. 2016. OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol Med. 8(3):288–303.
- Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. 2012. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 487(7405):114–118.
- Burgess SM, Ajimura M, Kleckner N. 1999. GCN5-dependent histone H3 acetylation and RPD3-dependent histone H4 deacetylation have distinct, opposing effects on IME2 transcription, during meiosis and during vegetative growth, in budding yeast. Proc Natl Acad Sci USA. 96(12):6835–6840.
- Bustin SA. 2002. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 29(1):23–39.
- Chaffer CL, Weinberg RA. 2011. A perspective on cancer cell metastasis. Science. 331(6024):1559–1564.
- Cheng D, Zhao L, Xu Y, Ou R, Li G, Yang H, Li W. 2015. K-Ras promotes the non-small lung cancer cells survival by cooperating with sirtuin 1 and p27 under ROS stimulation. Tumour Biol. 36(9):7221–7232.
- Cieniewicz AM, Moreland L, Ringel AE, Mackintosh SG, Raman A, Gilbert TM, Wolberger C, Tackett AJ, Taverna SD. 2014. The bromodomain of Gcn5 regulates site specificity of lysine acetylation on histone H3. Mol Cell Proteomics. 13(11):2896–2910.
- Cooper JM, Bodemann BO, White MA. 2013. The RalGEF/Ral pathway: evaluating an intervention opportunity for Ras cancers. Enzymes. 34:137–156.
- Del Vescovo V, Denti MA. 2015. microRNA and lung cancer. Adv Exp Med Biol. 889:153–177.
- Goel S, Huang J, Klampfer L. 2015. K-Ras, intestinal homeostasis and colon cancer. Curr Clin Pharmacol. 10(1):73–81.
- Guo L, Costanzo-Garvey DL, Smith DR, Zavorka ME, Venable-Kang M, MacDonald RG, Lewis RE. 2016. Cell non-autonomous regulation of hepatic IGF-1 and neonatal growth by Kinase Suppressor of Ras 2 (KSR2). Sci Rep. 6:32093.
- Harada H, Omi M, Sato T, Nakamura H. 2015. Pea3 determines the isthmus region at the downstream of Fgf8-Ras-ERK signaling pathway. Dev Growth Differ. 57(9):657–666.
- Hattori N, Ushijima T. 2014. Compendium of aberrant DNA methylation and histone modifications in cancer. Biochem Biophys Res Commun. 455(1–2):3–9.
- Hensing T, Chawla A, Batra R, Salgia R. 2014. A personalized treatment for lung cancer: molecular pathways, targeted therapies, and genomic characterization. Adv Exp Med Biol. 799:85–117.
- Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ, Jr., Wu YL, Paz-Ares L. 2017. Lung cancer: current therapies and new targeted treatments. Lancet. 389(10066):299–311.
- Horibata S, Vo TV, Subramanian V, Thompson PR, Coonrod SA. 2015. Utilization of the soft agar colony formation assay to identify inhibitors of tumorigenicity in breast cancer cells. J Vis Exp. 20:e52727.
- Hussain S. 2016. Nanomedicine for treatment of lung cancer. Adv Exp Med Biol. 890:137–147.
- Jiang W, Wei K, Pan C, Li H, Cao J, Han X, Tang Y, Zhu S, Yuan W, He Y, et al. 2018. MicroRNA-1258 suppresses tumour progression via GRB2/Ras/Erk pathway in non-small-cell lung cancer. Cell Prolif. 51(6):e12502.
- Jiao F, Hu H, Yuan C, Jin Z, Guo Z, Wang L, Wang L. 2014. Histone deacetylase 3 promotes pancreatic cancer cell proliferation, invasion and increases drug-resistance through histone modification of P27, P53 and Bax. Int J Oncol. 45(4):1523–1530.
- Johnson LM, Price DK, Figg WD. 2013. Treatment-induced secretion of WNT16B promotes tumor growth and acquired resistance to chemotherapy: implications for potential use of inhibitors in cancer treatment. Cancer Biol Ther. 14(2):90–91.
- Karasawa T, Kawashima A, Usui F, Kimura H, Shirasuna K, Inoue Y, Komada T, Kobayashi M, Mizushina Y, Sagara J, et al. 2015. Oligomerized CARD16 promotes caspase-1 assembly and IL-1β processing. FEBS Open Bio. 5:348–356.
- Lawrence M, Daujat S, Schneider R. 2016. Lateral thinking: how histone modifications regulate gene expression. Trends Genet. 32(1):42–56.
- Li C, Wang J, Kong J, Tang J, Wu Y, Xu E, Zhang H, Lai M. 2016. GDF15 promotes EMT and metastasis in colorectal cancer. Oncotarget. 7(1):860–872.
- Li Y, Jaramillo-Lambert AN, Yang Y, Williams R, Lee NH, Zhu W. 2012. And-1 is required for the stability of histone acetyltransferase Gcn5. Oncogene. 31(5):643–652.
- Lippman Z, Gendrel AV, Colot V, Martienssen R. 2005. Profiling DNA methylation patterns using genomic tiling microarrays. Nat Methods. 2(3):219–224.
- Ma L, Yuan L, An J, Barton MC, Zhang Q, Liu Z. 2016. Histone H3 lysine 23 acetylation is associated with oncogene TRIM24 expression and a poor prognosis in breast cancer. Tumour Biol. 37(11):14803–14812.
- Martinez-Redondo P, Santos-Barriopedro I, Vaquero A. 2012. A big step for SIRT7, one giant leap for Sirtuins… in cancer. Cancer Cell. 21(6):719–721.
- Minsky N, Oren M. 2004. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 16(4):631–639.
- Nihira NT, Ogura K, Shimizu K, North BJ, Zhang J, Gao D, Inuzuka H, Wei W. 2017. Acetylation-dependent regulation of MDM2 E3 ligase activity dictates its oncogenic function. Sci Signal. 10(466):eaai8026.
- Niu Z, Zheng L, Wu S, Mu H, Ma F, Song W, Zhu H, Wu J, He X, Hua J. 2015. Ras/ERK1/2 pathway regulates the self-renewal of dairy goat spermatogonia stem cells. Reproduction. 149(5):445–452.
- Rodrigues LM, Uribe-Lewis S, Madhu B, Honess DJ, Stubbs M, Griffiths JR. 2017. The action of β-hydroxybutyrate on the growth, metabolism and global histone H3 acetylation of spontaneous mouse mammary tumours: evidence of a β-hydroxybutyrate paradox. Cancer Metab. 5:4.
- Rui Y, Wang C, Zhou Z, Zhong X, Yu Y. 2015. K-Ras mutation and prognosis of colorectal cancer: a meta-analysis. Hepato-gastroenterology. 62(137):19–24.
- Samatar AA, Poulikakos PI. 2014. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat Rev Drug Discov. 13(12):928–942.
- Schulz S, Haussler S. 2014. Chromatin immunoprecipitation for ChIP-chip and ChIP-seq. Methods Mol Biol. 1149:591–605.
- Semenova EA, Nagel R, Berns A. 2015. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 29(14):1447–1462.
- Shim Y, Duan MR, Chen X, Smerdon MJ, Min JH. 2012. Polycistronic coexpression and nondenaturing purification of histone octamers. Anal Biochem. 427(2):190–192.
- Sundar IK, Rahman I. 2016. Gene expression profiling of epigenetic chromatin modification enzymes and histone marks by cigarette smoke: implications for COPD and lung cancer. Am J Physiol Lung Cell Mol Physiol. 311:1245–1258.
- Tasselli L, Xi Y, Zheng W, Tennen RI, Odrowaz Z, Simeoni F, Li W, Chua KF. 2016. SIRT6 deacetylates H3K18ac at pericentric chromatin to prevent mitotic errors and cellular senescence. Nat Struct Mol Biol. 23(5):434–440.
- Vicent S, López-Picazo JM, Toledo G, Lozano MD, Torre W, Garcia-Corchón C, Quero C, Soria J-C, Martín-Algarra S, Manzano RG, Montuenga LM. 2004. ERK1/2 is activated in non-small-cell lung cancer and associated with advanced tumours. Br J Cancer. 90(5):1047–1052.
- Voisset E, Oeztuerk-Winder F, Ruiz EJ, Ventura JJ. 2013. p38α Negatively regulates survival and malignant selection of transformed bronchioalveolar stem cells. PloS One. 8(11):e78911.
- Wang HL, Lu RQ, Xie SH, Zheng H, Wen XM, Gao X, Guo L. 2015. SIRT7 exhibits oncogenic potential in human ovarian cancer cells. Asian Pac J Cancer Prev. 16(8):3573–3577.
- Wang N, Xiang X, Chen K, Liu P, Zhu A. 2018. Targeting of NT5E by miR-30b and miR-340 attenuates proliferation, invasion and migration of gallbladder carcinoma. Biochimie. 146:56–67.
- Xu W, Chou CL, Israel DD, Hutchinson AJ, Regan JW. 2009. PGF(2alpha) stimulates FP prostanoid receptor mediated crosstalk between Ras/Raf signaling and Tcf transcriptional activation. Biochem Biophys Res Commun. 381(4):625–629.
- Yang HW, Shin MG, Lee S, Kim JR, Park WS, Cho KH, Meyer T, Heo WD. 2012. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol Cell. 47(2):281–290.
- Zhang S, Chen P, Huang Z, Hu X, Chen M, Hu S, Hu Y, Cai T. 2015. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 5:9787.
- Zhang Y, Yang Q, Wang S. 2014. MicroRNAs: a new key in lung cancer. Cancer Chemother Pharmacol. 74(6):1105–1111.
- Zhao YY, Yu L, Liu BL, He XJ, Zhang BY. 2015. Downregulation of P-gp, Ras and p-ERK1/2 contributes to the arsenic trioxide-induced reduction in drug resistance towards doxorubicin in gastric cancer cell lines. Mol Med Rep. 12(5):7335–7343.
