Abstract
Phytochemical investigations on the chloroform and ethyl acetate soluble fractions of the roots of Ranunculus repens led to the isolation of methyl 3,4,5-trihydroxybenzoate 1, R(+)- 4-methoxydalbergione 2 and R(+)-dalbergiophenol 3. The structures of these compounds were established through spectral studies including 1D and 2D NMR experiments and by comparison with literature data. These compounds showed potent inhibitory activity against urease.
Introduction
Ranunculus repens Linn. belongs to family Ranunculaceae and is an herbaceous perennial that usually creeps along the ground, native to North Africa, the Middle East to China and Japan, and most of Europe. Plants of the genus are reported to have anemonins [Citation1], carotenes [Citation2], flavone glycosides [Citation3] and ranuncosides [Citation4]. Very little chemical work so far has been carried out on the species Ranunculus repens. In this report we describe the isolation of three phenolic compounds Citation5-7, reported for the first time from this species and their urease inhibiting activity.
Ureases (E.C 3.5.1.5) have been shown to be an important virulence determinant in the pathogenesis of many clinical conditions, which is detrimental for human and animal health as well as for agriculture. Urease is directly involved in the formation of infection stones and contributes to the pathogenesis of urolithiasis, pyelonephritis, ammonia and hepatic encephalopathy, hepatic coma, urinary catheter encrustation [Citation8,Citation9]. It is also known to be a major cause of pathologies induced by Helicobacter pylori (HP), which allows bacteria to survive at the low pH of the stomach during colonization and, therefore, plays an important role in the pathogenesis of gastric and peptic ulcer (including cancer) [Citation9]. In agriculture, high urease activity causes significant environmental and economic problems by releasing abnormally large amounts of ammonia into the atmosphere during urea fertilization. This further induces plant damage primarily by depriving them from their essential nutrient and secondly ammonia toxicity, which increase the pH of the soil [Citation10,Citation11]. Therefore strategies based on urease inhibition are now considered as first line treatment for infections caused by urease-producing bacteria.
Materials and methods
General techniques
Optical rotations were measured on a JASCO DIP-360 polarimeter. Melting points were determined on a Buchi melting point apparatus and are uncorrected. UV spectra were recorded on a Hitachi U-3200 spectrophotometer. IR spectra were recorded on a FTIR-8900 Shimadzu spectrometer. The 1H, 13C-NMR, HMQC, and HMBC spectra were recorded on Bruker spectrometers operating at 500, 300 MHz for 1H and 125, 75 MHz for 13C, respectively. The chemical shift values are reported in ppm (δ) units and the coupling constants (J) are in Hz. MS and HR-MS were respectively obtained on a JMS-HX-110 with a data system and on a JMS-DA 500 mass spectrometers. Aluminum sheets pre-coated with silica gel 60 F254 (20 × 20 cm, 0.2 mm thick; E-Merck) were used for TLC and silica gel (230–400 mesh) was used for column chromatography. Visualization of the TLC plates was carried out under UV at 254 and 366 nm and also by spraying with ceric sulphate reagent with heating.
Plant material
The roots of Ranunculus repens Linn. were collected from Nathia Gali, Hazara Hills and identified by Dr. Syed Iftikhar Hussain Shah, plant taxonomist, Faculty of Pharmacy, Gomal University D I Khan, Pakistan, where a voucher specimen has been deposited.
Extraction and isolation
The shade dried roots (10 kg) of Ranunculus repens was extracted three times, seven days on each occasion, with methanol. The combined methanol extract was evaporated in vacuo. The resulting residue (0.65 kg) was suspended in water and extracted successively with n-hexane, chloroform, and ethyl acetate. The chloroform soluble fraction (90 g) was subjected to column chromatography over silica gel eluting with hexane/ethyl acetate solvent system in increasing order of polarity. As a result fractions C1–C10 were obtained. The fraction which eluted with hexane/ethyl acetate (85:15) was subjected to repeated column chromatography eluting with hexane/ethyl acetate (88:12) to afford R(+)-dalbergiphenol 3 (10 mg) and R(+)-4-methoxydalbergione 2 (8 mg). The ethyl acetate soluble fraction (45 g) on repeated column chromatography using solvent system n-hexane/acetone (80:20) afforded compound 1 (40 mg).
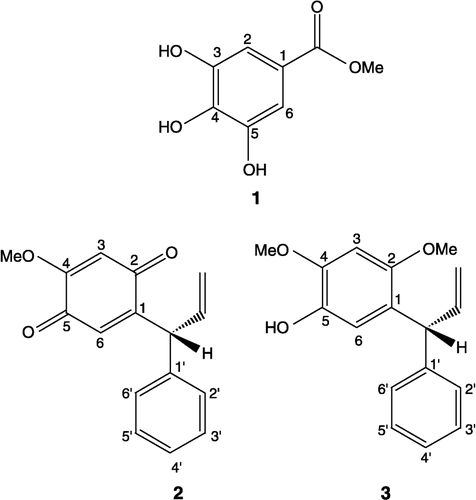
Methyl 3,4,5-trihydroxybenzoate (1)
Brown solid. M.p. 198–200°C. UV λmax (MeOH) (log ε) nm: 289 (3.82), 296 (3.86). IR (KBr) νmax cm− 1: 3370,1695, 1620, 1540, 1440, 1330, 1262, 1212, 1060, 1004, 866, 765 cm− 1. HREIMS: m/z 184.0371 (calcd. for C8H8O5; 184.0365). EIMS: m/z [rel. int.]: 184 [M+], 153 [100], 127 [6.5], 125 [25.9], 107 [6.5], 79 [30.1], 68 [5.9], 51 [14.8]. 1H NMR (CD3OD, 500 MHz): δ 7.12 (2H, s, H-2, 6), 3.88 (3H, s, OCH3).
13C NMR (CD3OD, 125 MHz): δ 138.5 (C-1), 145.7 (C-2, 6), 109.1 (C-3, 5), 124.1 (C-4), 166.5 (C-7), 52.2 (C-8).
R(+)-4-Methoxydalbergione (2)
Yellow crystals. M.p. 112-113°C. [α]D22+16.2 (CHCl3, c, 1.2). IR (KBr) νmax cm− 1: 1672, 1650, 1606, 1195, 1175, 992, 928, 904. HREIMS: m/z 254.0940 (calcd. for C16H14O3; 254.0936) 1H NMR (CDCl3, 300 MHz): δ 3.78 (OMe), 7.15- 7.32 (5H, H-2′ → 6′), 6.46 (1H, s, H-6), 5.89 (1H, s, H-3), 4.90 (1H, d, J = 6.58, Allylic), 5.0 (1H, d, J = 17.14, CH2trans), 5.2 (1H, d, J = 8.8, CH2cis), 6.1 (1H, m, vinylic). 13C NMR (CDCl3, 75 MHz): δ 186.2 (C-2), 182.3 (C-5), 158.4 (C-4), 151.02 (C-1), 139.3 (C-1′), 137.2 (vinylic), 131.5 (C-4′), 128.7 (C-3′,5′), 128.5 (C-2′,6′), 127.1 (C-6), 128.1 (CH2), 107.9 (C-3), 56.2 (OMe), 46.9 (allylic).
R(+)-Dalbergiphenol (3)
Light brown oil. UV λmax (MeOH) (log ε) nm: 208 (4.70), 231 (4.17), 296 (3.98). IR (KBr) νmax cm− 1: 3500, 3440, 2830, 1630, 1445, 910, 905. HREIMS: m/z 270.1248 (calcd. for C17H18O3; 270.1253). EIMS: m/z [rel.int.]: 270 [100], 269 [11], 256 [32], 255 [10], 253 [16], 165 [15], 115 [30], 91 [55], 77 [12], 69 [20]. [α] D22+31.9 (c, 0.64 CHCl3).1H NMR (CDCl3, 500 MHz): δ 3.74 (3H, s, OMe), 3.91 (3H, s, OMe), 4.90 (1H, dd, J = 17.0, 1.6, = CH2trans) 5.18 (1H, dd, J = 10.0, 1.6, = CH2cis), 6.23 (1H, ddd, J = 17.0, 10.0, 6.0, vinylic), 5.03 (1H, d, J = 6.0, allylic), 6.59 (1H, s, H-3), 6.83 (1H, s, H-6), 7.33 (5H, s, H-2′ → H-6′). 13C NMR (CDCl3, 125 MHz): δ 124.7 (C-1), 150.5 (C-2), 97.5 (C-3), 145.1 (C-4), 139.3 (C-5), 115.2 (C-6), 143.2 (C-1′), 127.9 (C-2′), 128.4 (C-3′), 125.8 (C-4′), 128.4 (C-5′), 127.9 (C-6′), 46.7 (allylic), 140.0 (vinylic), 115.7 ( = CH2), 56.0 (2-OMe), 56.9 (4-OMe).
Urease assay and inhibition
Reaction mixtures comprising 25 μl of enzyme (Jack bean and Bacillus pasteurii Urease) solution were incubated for 30 min with 5 μl test compounds (1, 2, 3) at 30°C for 15 min in 96-well plates and then 55 μl of buffers containing 100 mM urea were incubated for 15 min. At the end final urease activity was determined by measuring ammonia production using the indophenol method as described by Weatherburn [Citation11]. Briefly, 45 μl each of phenol reagent (1% w/v phenol and 0.005% w/v sodium nitroprusside) and 70 μl of alkali reagent (0.5% w/v NaOH and 0.1% active chloride NaOCl) were added to each well. The increasing absorbance at 630 nm was measured after 50 min, using a microplate reader (Molecular Device, USA). All reactions were performed in triplicate in a final volume of 200 μl. The results were processed by using SoftMax Pro software (Molecular Device, USA). All the assays were performed at pH 8.2 (0.01 M K2HPO43H2O, 1 mM EDTA and 0.01 M LiCl). Percentage inhibitions were calculated from the formula 100-[(ODtestwell/ODcontrol) × 100]. Thiourea was used as the standard inhibitor of urease.
Results and discussion
Urease is an enzyme that is present in many plants and in soil. It catalyzes the hydrolysis of urea to ammonium and carbamate ions, which decompose to carbon dioxide and ammonia. The active site contains two nickel (II) atoms which, as shown by X-ray analysis, are linked by a carbamate bridge; furthermore, two imidazole nitrogen atoms are bound to each nickel atom, and a carboxylate group and a water molecule fill the remaining coordination site of the metal ion. The coordination geometry of the first nickel atom is pseudo tetrahedral, while that of second is roughly trigonal bipyramidal [Citation8].
Compounds 1 and 3 displayed potent inhibitory potential while compound 2 () showed moderate to potent inhibition against the enzyme urease. From the results () it is clear that the compounds with more hydroxyl moieties are the most active inhibitors of urease; compound 1 with three hydroxyl groups is the most potent with an IC50 value of 22.5 μM, compound 3 with one hydroxyl and two methoxy moieties showed somewhat less inhibition than 1 although comparable to 1. Compound 2 with no free hydroxyl group showed the least inhibition in this series. This gradual increase in inhibition with the increasing number of hydroxyl groups may be apparently due to chelation of these phenolic moieties with nickel in the active site of the urease enzyme and as a result enhances the inhibitory potential of the compounds. Clearly there are many questions regarding the mode of action of these ligands that need to be answered and these studies will play an important role in the development of future generations of these inhibitors. Consequently we are synthesizing several derivatives of compounds 1–3 to be evaluated against urease in vitro through STD NMR and molecular dynamics simulation and kinetics studies to establish the detailed mechanism of inhibition by these compounds.
Table I. In vitro quantitative inhibition of urease by compounds 1-3.
Acknowledgements
The authors acknowledge the Higher Education Commission Islamabad, Pakistan for financial support.
References
- Ruijgrok HWL. Planta Med 1963; 11: 338
- Eschenmor W. Helv Chim Acta 1979; 62: 2534
- Litvineko V. Farm Zh (Kiev) 1969; 24: 77
- Tschesche R. Chim Ber 1972; 5: 290
- Muangnoicharoen N, Frahm AW. Phytochemistry 1982; 21: 767
- Chan SC, Chang YS, Kuo SC. Phytochemistry 1997; 46: 947
- Lajis NH, Khan MN. Indian J Chem 1994; 33B: 609
- Mobley HLT, Island MD, Hausinger RP. Microbiol Rev 1995; 59: 451
- Mobley HLT, Hausinger RP. Microbiol Rev 1989; 53: 85–100
- Bremner JM. Fert Res 421995; 321–329
- Weatherburn MW. Anal Chem 1967; 39: 971