Abstract
In mammals, xanthine oxidase (E.C. 1.17.3.2) catalyzes the hydroxylation of a wide variety of heterocyclic substrates such as purines, pyrimidines, and pterins, in addition to aldehydes [Citation] as all-trans-retinaldehyde Citation. Here, we show that buttermilk xanthine oxidase was capable to oxidizing all-trans-retinol (t-ROL) to all-trans-retinaldehyde (t-RAL) that was successively oxidized to all-trans-retinoic acid (t-RA). A rise in the enzyme activity, when t-ROL-CRBP complex was assayed, with respect to the free t-ROL, was observed. Furthermore, treatment of the enzyme with Na2S and glutathione resulted in a significant increment in catalytic activity toward t-ROL and t-RAL, due to the reconstitution of the native structural organization of the molybdenum centre of molybdopterin cofactor of the desulfo form of xanthine oxidase.
Introduction
Xanthine oxidoreductase (XOR) a member of molybdenum hydroxylases, found from bacteria to humans, catalyzes the hydroxylation of a wide variety of heterocyclic substrates such as purines, pyrimidines, and pterins, in addition to aldehydes as all-trans- and 9-cis-retinaldehyde, which are oxidised to the respective acids Citation1-5. The enzyme can occur naturally in two inactive forms, the demolybdo-XOR which lacks Mo and desulfo-XOR, in which Mo = S, essential for catalytic activity, is replaced by Mo = O [Citation6]. The active enzyme can exist in one of two interconvertible forms, xanthine dehydrogenase (XDH; E.C. 1.17.1.4) and xanthine oxidase (XO) [Citation7,Citation8]. Both forms will reduce O2, although XO is more efficient in this respect, leading to formation of reactive oxygen species (ROS) [Citation9]. These ROS can interact, particularly in the presence of iron, to generate a range of cytotoxic agents, including hydroxyl radicals. XOR, detected on the outer surface of cultured endothelial and epithelial cells [Citation10], has been implicated in the pathogenesis of ischemia-reperfusion injury Citation11-12. It has recently been shown to catalyse the anaerobic reduction of inorganic nitrite to nitric oxide (NO) [Citation13]. Under aerobic conditions, oxygen reduced by XOR to superoxide, can react rapidly with NO to give peroxynitrite [Citation14] that has bactericidal properties [Citation15]. Bio-activation of ethanol to acetaldehyde and the contemporaneous free radical production in rat breast tissue [Citation16] has been also demonstrated. In the present work, experiments that show that xanthine oxidase can operate the oxidation of all-trans-retinol (t-ROL) and enzyme assays that demonstrate its sequential oxidation to all-trans-retinaldehyde (t-RAL) and to all-trans-retinoic acid (t-RA) are also reported.
Materials and methods
Chemicals
All chemicals were from Sigma-Aldrich (Milan, Italy) except sucrose, ammonium and sodium acetate that were from Merck Group (Milan, Italy).
XO preparation
Desulfo-enzyme was re-sulphurated by a slight modification of the method described by Wahl and Rajagopalan [Citation17] as follows. 50 μL (65 μg) of bovine buttermilk xanthine oxidase (Sigma X-4500 grade III; 1–2 units/mg protein) was dissolved in 2 mL of 50 mM Tris-HCl pH 7.4, containing 1 mM EDTA (0.64 mg × mL− 1). After stirring the sample was dialyzed against the same buffer (5 L) for 2 h to remove ammonium sulfate and sodium salicylate. Enzyme solution transferred in vacutainer was made anaerobic by repeated alternate evacuations and flushing with nitrogen using syringe needles inserted into the septa. 1 M sodium sulfide solution was added to the enzyme preparation to a final concentration of about 10 mM. After 10 min incubation at 37°C, glutathione at a final concentration of 5 mM was added and sample incubated for 1 h at 4°C. After filtration through a 0.22-μm membrane the enzyme preparation was applied to a Centricon Ultracel YM-50, centrifuged at 5000 × g for 10 min and concentrated to about 0.5 mL. After dilution to 1 mL with 50 mM Tris-HCl pH 8.8 the enzyme was spectrophotometrically analyzed and an A280/A450 ratio of 4.3 and an A450/A550 ratio of 3.9 were evaluated, with respect the corresponding values reported in the literature for the enzyme purified from milk, which are 5–6 and 2.8, respectively Citation18-20. The oxidase activity was determined by measuring the rate of oxidation of xanthine to uric acid spectrophotometrically at 295 nm in a Beckman DU8 spectrophotometer, using an absorption coefficient of 9.6 mM− 1 cm− 1 [Citation21]. Assays were performed at 25°C in air-saturated Tris-HCl buffer, pH 8.8, containing 100 μM xanthine.
CRBP-enriched fraction preparation
Homogenate from normal human mammary epithelial cells (HMECs), obtained from Clonetics (San Diego, CA), was prepared by cavitation of a cell suspension (about 30 × 106 cells in 3 mL of 0.25 M sucrose, 20 mM Tris-HCl pH 7.0, 0.4 mM EDTA) in a pre cooled cell disruption bomb (Parr Instrument Company) at a nitrogen pressure of 600 Psi for 10 min. The homogenate was centrifuged at 110,000 × g for 60 min in a L8-M Beckman ultracentrifuge and 3 mL of supernatant corresponding to about 24 × 106 cells were gel filtered on Shodex KW 804 column injecting 100 μL of cytosol aliquots (74 μg protein) to obtain separation of CRBPs. Proteins were eluted with 50 mM Tris-HCl pH 7.4 pumped at a flow rate of 1 mL/min. Protein peaks were monitored measuring their fluorescence (ex. 350 nm/ em. 480 nm) and absorbance at 280 nm and at 350 nm to identify retinoid binding protein peaks. Column eluates positive by fluorimetry were collected, pooled and concentrated to a third of their initial volume (about 1 mL) with a Centriprep YM-10 membrane. To assess the presence of CRBP and CRABP in this fraction, the sample was analyzed on a DEAE 5PW column [Citation22] monitoring the absorbance of protein peaks at 284 and 350 nm and evaluating their A350/A284 ratio [Citation23] (data not shown). In particular, HPLC analysis of cellular retinoid binding proteins (CRBP(s)) were performed equilibrating the column with 20 mM sodium acetate, pH 5.8 and eluting it with a linear gradient of NaCl from 0–250 mM in the same buffer following the indications reported by Bailey and Siu [Citation22]. After the anionic exchange chromatography analysis, to achieve a higher grade of purification, the sample was re-chromatographed on Protein-Pak I-60. CRBP and CRABP fractions, detected by absorbance and fluorescence, were collected, pooled, and concentrated on a Centripep YM-10 membrane to one millilitre and saved at − 80°C. The apo-CRBP(s) fraction was prepared by extraction twice with hexane (2:1 v/v); residual hexane was removed keeping the apo-protein preparation under nitrogen. Protein was determined using a Sigma protein assay reagent.
Radio-chromatography of retinoid binding proteins
Qualitative and quantitative analysis of the CRBP(s)-enriched fraction was performed by assaying it with the [3H]-retinoids (t-ROL or t-RA) and the binding evaluated by performing an HPLC analysis on Protein Pak I 60. To determine the binding activity of the partially purified retinoid binding proteins, 100 μL of sample (20.18 μg of total proteins) were mixed with 200 pmoles of [3H]-t-RA or [3H]-t-ROL (both at 55–85 Ci/mmoles) in 5 μL of ethanol, and incubated at 4°C for 2 h. Non-specific binding of labelled retinoids to proteins was corrected by adding 20 μL (40 nmoles) of cold t-ROL or t-RA to the samples and incubating at 4°C for 2 h. 100 μL of samples were analyzed on a Protein Pack I-60, as already described above, and retinoid binding proteins detected by their absorbance at 280 nm and radioactivity in a mixture of HPLC eluent-scintillator cocktail (Ultima-Flo M), in a ratio 1:6, in a 500 μL flow cell, at a spiked rate of 6 mL/min. Routine data integration was achieved by the Flo-One/beta F1B IC program and computed in net cpm, after correction for both residence time and background subtraction.
Retinoid solution and analysis
Individual stock solutions of t-RAL, t-RA and t-ROL, handled under indirect yellow light to minimize photo-oxidation, were prepared in absolute ethanol (1 mg/mL), in amber vials, and stored under inert gas at − 80°C. Dilutions were performed immediately prior to use. For calibration curves the stock solutions were diluted with ammonium acetate 10 mM pH 5.8 and acetonitrile (final ratio 35/65 v/v) to obtain a mixture of working standards whose concentration was in the range of the substrates added and products expected in enzyme assays. For enzyme assay, retinoid was diluted with 0.12 mM Tween 80 in Tris-HCl 50 mM, pH 8.8 solution just before the preparation of the enzyme assay medium, and kept at 4°C.
Enzyme assays
Enzyme assays with hypoxanthine, ethanol and t-ROL on native PAGE
Xanthine oxidase zymogram was prepared by loading the enzyme protein onto a non-denaturing 7.5% polyacrylamide gel, following the indications reported in reference [Citation24]. A current of 20–25 mA was passed through the gel for 60 min. The gel was incubated in the dark at 37°C in 50 mL of a staining solution containing 50 mM Tris-HCl, pH 8.8, 1 mM t-ROL dissolved in a minimal volume of acetone, 17.5 mg of nitro blue tetrazolium and 1 mg of phenazine methosulfate, as indicated in reference [Citation25]. The substrate concentrations used for ethanol or hypoxanthine assay were 1 mM and 0.1 mM respectively.
Enzyme assays with free t-ROL or t-RAL
27.6 μL of enzyme preparation was incubated with 0.25–5 μM t-ROL or t-RAL in 500 μL of 50 mM Tris-HCl, pH 8.8 containing 0.012 mM Tween 80. After 10 min of incubation at 37°C, the reaction was stopped by dipping the sample into a cold bath. After this, the mixture was acidified by adding 20 μL of 0.5 M acetic acid, and the retinoids were extracted by adding 3 mL of hexane and shaking for 1 min in the dark. The hexane phase was dried under nitrogen and stored at − 20°C. To evaluate xanthine oxidase activity on t-ROL or t-RAL, samples were re-suspended in 60 μL of ammonium acetate/acetonitrile solution (35:65 v/v) and one third of it was injected into the HPLC system and analyzed as reported by Frolik et al. [Citation26].
Enzyme assays with liganded t-ROL
A few minutes before the assay 10 μl of t-ROL (1 mg/mL in absolute ethanol) were diluted to 10 mL with 0.12 mM Tween 80 Tris-HCl 50 mM, pH 8.8 and after stirring, 171 μL (600 pmol) of this were added to 1 mL of apo-retinoid binding proteins enriched fractions (about 60 pmol of CRBP and 200 pmol of CRABP) and kept at 4°C. The molar concentration of CRBP and t-ROL in the mixture was 51.2 nM and 512 nM, respectively, with a CRBP/t-ROL ratio of about 0.1. 2.76 μL of XO solution (about 5 pmoles of enzyme) were incubated for 10 min at 37°C after addition of the t-ROL/CRBP solution (50–450 μL) in a final volume of 500 μL. At the end of inculation, the mixture was acidified by adding 20 μL of acetic acid solution (0.5 M), and the retinoids extracted by adding 3 mL of hexane and shaking for 1 min in the dark. The hexane phase was dried under nitrogen and stored at − 20°C.
Kinetic data processing
Leatherbarrow Eritacus Software made a provisional estimate of the kinetic constants with that aid of the Grafit program. The appropriate velocity equations describing the kinetic behavior [Citation27] were verified
Statistical analysis
Statistical significance of the data was evaluated using Student's t-test (Snedecor and Cochran, [Citation28]) and probability values below 0.05 (P < 0.05) were considered significant. Results are expressed as mean ± S.D. for the indicated set of experiments.
Results and discussion
Consistent with our previous results, where we showed the ability of milk xanthine oxidase to synthesize all-trans- and 9-cis-RA from the respective retinaldehydes [5], in the present work we report data that demonstrates that xanthine oxidase is capable of oxidising alcohols such as ethanol and t-ROL.
All experiments were performed using the Sigma buttermilk xanthine oxidase dialyzed in the presence of Na2S, and successively treated with glutathione (see: XO preparation in Materials and Methods). This treatment was performed to obtain an increment in the concentration of active enzyme due to the reconstitution of the native structural organization of the molybdenum centre of the molybdopterin in the xanthine oxidase desulfo form [19].
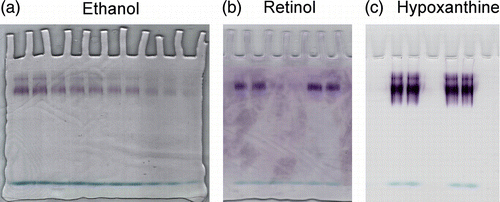
Detection of the XO activity on hypoxanthine, ethanol and t-ROL on native PAGE
Xanthine oxidase stained onto an electropherogram with 1 mM ethanol () showed a linear catalytic response in the range of 2.16–1.08 pmoles (about 130 nM–65 nM of enzyme in the loading wells). The ability of xanthine oxidase to oxidize ethanol suggested to us assaying the enzyme with t-ROL (). At the two different concentrations used (1.08–2.16 pmol), XO was capable of oxidizing t-ROL during incubation for one hour. Enzyme detection with hypoxanthine was also performed to evaluate the xanthine oxidase basal activity ().
Figure 1 Assays of xanthine oxidase on electrophoretic gels with ethanol, retinol and hypoxanthine via PMS/tetrazolium stain. Zymograms of xanthine oxidase acting on (a) 1 mM ethanol, (b) 1 mM t-retinol and (c) 0.1 mM hypoxanthine are shown. Staining solution composition varied only for the kind of substrate used. Development times were 10 min for hypoxanthine and 1 h for t-retinol and ethanol. The enzyme concentrations assayed with ethanol were decreasing from 2.16 to 1.08 pmoles and those assayed with t-retinol or hypoxanthine were 1.08 and 2.16 pmoles. One representative of three independent experiments is shown.
t-ROL oxidation was initially performed by using free retinoids
To analyze the products of t-ROL oxidation, xanthine oxidase was assayed in test-tubes. Preliminary assays were performed with 250 nM t-RAL in 0.012 mM Tween 80 and the results obtained were in accord with those reported in our previous work [Citation5]. The evaluation of xanthine oxidase activity on t-ROL was performed at a concentration range between 500 nM–5 μM (see: Assays with free and liganded t-ROL or t-RAL in Materials and Methods), and at substrate concentrations less than 2 μM it was not possible to see any enzymatic oxidation of t-ROL. On the contrary, when xanthine oxidase was assayed with a concentration of t-ROL higher than 2 μM it was possible to detect not only t-RAL but also t-RA as a product of aldehyde oxidation. Synthesis of t-RA as final product of the t-ROL oxidation was a very interesting phenomenon but the very low activity showed by xanthine oxidase toward free t-ROL () needed a more exhaustive examination.
Table I. Kcat values in the conversion of trans-retinol to trans-retinaldehyde and to trans-retinoic acid catalyzed by xanthine oxidase.
holo-CRBP is the substrate for xanthine oxidoreductase
Presuming that the low catalytic activity of xanthine oxidase toward t-ROL might be due to its insufficient dispersion in the water/detergent phase we have evaluated whether the addition of retinoid binding proteins, in the incubation medium, could be useful to the oxidative process by improving the interactions of t-ROL with the enzyme. Considering that most retinoids present in cells and plasma are bound to CRBP or RBP, we firstly purified CRBPs from human mammary epithelial cells and then successively assayed the catalytic activity of xanthine oxidase toward t-ROL in the presence of them or human RBP.
Purification of cellular retinoids binding proteins from HMEC
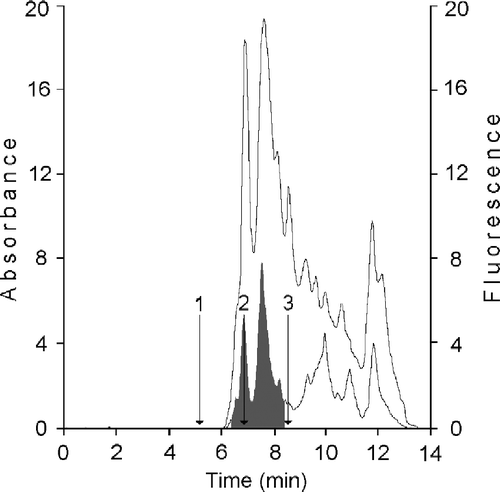
We have initially purified from HMEC cytosol a fraction containing the specific binding proteins CRBPs by gel permeation chromatography on KW 804 column. The elution profile reported in shows a peak, positive on fluorimetry and containing the cellular retinoid binding proteins (CRBP(s)) (grey area; RT 11.47 ± 0.3), where proteins with a mass between 14 and 20 kDa are separated. The further purification of the fraction containing CRBPs was obtained on Protein Pak I-60, which provides excellent separation in the molecular weight range of 1,000–20,000 for native globular proteins. Their elution profiles are shown in . Moreover, to assess the presence of CRBPs in this fraction, the sample was analyzed on a DEAE 5PW column [Citation22] by monitoring their absorbance at 284 and 350 nm and evaluating the A350/A284 ratio [Citation23] (data not shown). After the anionic exchange chromatography analysis, the fraction was assayed with the [3H]-retinoids (t-ROL or t-RA) and the binding evaluated by performing a HPLC analysis on Protein Pak I 60 (data not shown). The amounts of retinol binding proteins and retinoic acid binding proteins available in the sample were determined by measuring the specific binding activity assayed versus the two [3H]-retinoids. In 100 μL of the analyzed sample containing 20.18 μg of total protein, there were in all about 8.39 ng of CRBPs (about 5.6 pmoles) and 28.6 ng of CRABPs (19.6 pmoles) with a purification factor of about 11-fold. This fraction was used to optimize t-ROL oxidation performed by xanthine oxidase.
Figure 2 Purification of cellular binding proteins from cytosol of human mammary epithelial cells. 100 μL cytosol (0.8 × 106 cells - 74 μg protein) were gel filtered on a KW 804 column and eluted at 1 mL min− 1 with 50 mM Tris HCl pH 7.4 containing 1 mM glutathione. Elution peaks were monitored by absorbance at 280 nm (top trace) and fluorescence (bottom trace). Iterative analyses were carried out and fraction peaks were collected from the area containing cellular retinoid binding proteins (RT 11.47 ± 0.3) (grey peaks). Positions of standards used as molecular weight markers were: 1) bovine erythrocyte carbonic anhydrase (29.3 kDa); 2) horse heart cytochrome C (12.4 kDa).
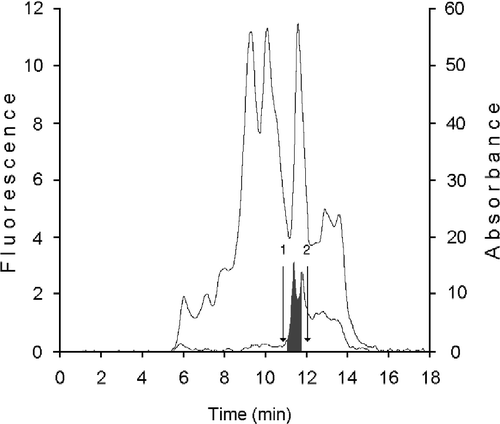
Figure 3 HPLC analysis of cellular retinoid binding proteins from human mammary epithelial cells. CRBP(s) fraction from KW 804 chromatography was analyzed on Protein Pak I-60 column. Bottom trace corresponded to ultraviolet detection at 280 nm; top trace is fluorescence reading (ex. 350 nm; em. 470 nm). CRBPs and CRABPs were separated in the RT interval of 6.4–8.3 min (grey area). Retention times were determined for (1) bovine erythrocytes carbonic anhydrase, 29.3 kDa, (2) lactalbumin, 14.7 kDa, and (3) horse heart cytochrome C, 12.4 kDa.
Retinol oxidation was performed using all-trans-retinol-CRBPs or -RBP complexes
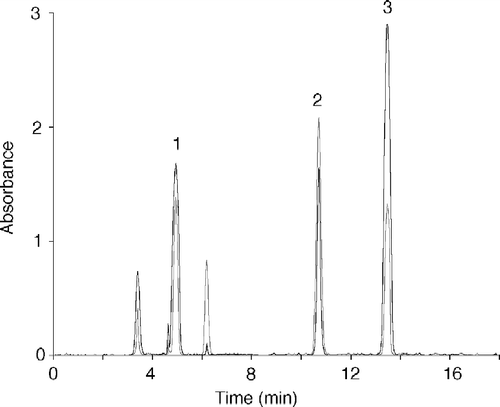
The XO-catalyzed oxidation of t-ROL in the presence of variable concentrations of RBP or CRBPs was followed by observing the rate of t-RAL and t-RA production. While it was not possible to see any activity toward t-ROL-RBP in the concentration range of 0.2–1 μM, a significant catalytic activity (Kcat = 255 min− 1) was determined with t-ROL-CR-BP (holo-CRBP) (50–460 nM) with respect to that of free t-ROL (Kcat = 0.266 min− 1) (). It is probable that the higher catalytic activity of the enzyme toward holo-CRBP is due to the true substrate availability, which will result in a higher t-RAL production, and its build up will initiate t-RA synthesis. The products of such catalytic behaviour by xanthine oxidase are shown in the chromatographic pattern in , where the peaks of t-RA and t-RAL, and that of t-ROL are well represented.
Figure 4 Chromatographic analysis of retinoids produced by xanthine oxidase assayed with t-ROL-CRBP. The chromatographic pattern reported is related to an assay performed with 5 nM xanthine oxidase, 256 nM t-ROL, 25.6 nM CRBP and 250 nM CRABP. Chromatographic elution was performed with acetonitrile-10 mM ammonium acetate (65:35 v/v) at a flow of 1 mL min− 1 and retinoids were monitored at 325 nm (grey profile) and 350 nm (dark profile). Peaks are as follows: 1, all trans-retinoic acid; 2, all trans-retinol; 3, all trans-retinaldehyde.
The possible influence exerted by free t-ROL on XO activity was evaluated performing assays with free t-RAL (0.20 μM) in the presence of variable concentrations of free t-ROL (1–5 μM). The presence of 1 μM t-ROL in the incubation medium resulted in a reduction of Kcat for t-RAL of over 90% () showing that t-retinol in the free state is not the appropriate substrate for xanthine oxidase.
Conclusion
The observation that milk xanthine oxidase (XO), an enzyme of broad specificity, is capable of converting t-ROL bound to CRBP to t-RAL and t-RAL to t-RA should be taken into due consideration, not only because the enzyme catalyzes the oxidation of an alcohol to the respective acid, but also because the alcohol involved is t-ROL and the produced acid is t-RA. In fact, the importance is well known of the various biological roles and effects of retinoic acid, such as the control of cell proliferation, differentiation and morphogenesis, epithelium protection and prevention effects in carcinogenesis Citation29-31. In particular, the results obtained stimulated us to attempt to define the role of the native form of xanthine oxidase, xanthine dehydrogenase, in the metabolic pathway that proceeds from t-ROL to t-RA biosynthesis.
Acknowledgements
We wish to thank Prof. Luigi Castagnetta and Dr. Giuseppe Carruba for providing generous access to the Laboratory of Experimental Oncology Unit, Department of Clinical Oncology, ARNAS-Civico, Palermo. We also thank Dr. Letizia Cocciadiferro for help with the cell culture and Dr. Grazia M. Granata for radio chromatography analysis. This work was supported by a MURST grant.
References
- Bray RC. The enzymeThird ed., PD Boyer. Academic Press, New York 1975; 299–419
- Futterman S. Enzymatic oxidation of vitamin A aldehyde to vitamin A acid. J Biol Chem 1962; 237: 677–680
- Olson JA. The metabolism of vitamin A. Pharmacol Rev 1967; 19: 559–596
- Lee MO, Manthey CL, Sladek NE. Identification of mouse-liver aldehyde dehydrogenases that catalyze the oxidation of retinaldehyde to retinoic acid. Biochem Pharmacol 1991; 42: 1279–1285
- Taibi G, Paganini A, Gueli MC, Ampola A, Nicotra CMA. Xanthine oxidase catalyzes the synthesis of retinoic acid. J Enz Inhib 2001; 16: 275–285
- Harrison R. Milk xanthine oxidase: Properties and physiological roles. Int Dairy J 2006; 16: 546–554
- Enroth C, Eger BT, Okamoto K, Nishino T, Nishino T, Pai EF. Crystal structure of bovine milk xanthine dehydrogenase and xanthine oxidase: Structure-based mechanism of conversion. Proc Natl Acad Sci USA 2000; 97: 10723–10728
- Mc Manaman JL, Bain DL. Structural and conformational analysis of the oxidase to dehydrogenase conversion of xanthine oxidoreductase. J Biol Chem 2002; 277: 21261–21268
- Hille R, Nishino T. Xanthine oxidase and xanthine dehydrogenase. FASEB J 1995; 9: 995–1003
- Rouquette M, Page S, Bryant R, Benboubetra M, Stevens CR, Blake DR. Xanthine oxidoreductase is asymmetrically localized on the outer surface of human endothelial and epithelial cells in culture. FEBS Lett 1998; 426: 397–401
- Granger DN, Rutili G, McCord JM. Role of superoxide in feline intestinal ischemia. Gastroenterol 1981; 81: 22–29
- McCord JM. Oxygen-derived free radicals in post-ischemic tissue injury. New Eng J Med 1985; 312: 159–163
- Godber BLJ, Doel JJ, Sapkota GP, Blake DR, Stevens CR, Eisenthal R, Harrison R. Reduction of nitrite to nitric oxide catalysed by xanthine oxidoreductase. J Biol Chem 2000; 275: 7757–7763
- Godber BLJ, Doel JJ, Durgan J, Eisenthal R, Harrison R. A new route in peroxynitrite: A role of xanthine oxidoreductase. FEBS Letters 2000; 475: 93–96
- Brunelli L, Crow JP, Beckman JS. The comparative toxicity of nitric oxide and peroxynitrite to escherichia coli. Arch Biochem Biophys 1995; 316: 327–334
- Castro GD, Delgado de Layño AMA, Costantini MH, Castro JA. Cytosolic xanthine oxidoreductase mediated bioactivation of ethanol to acetaldehyde and free radicals in rat breast tissue. Its potential role in alcohol-promoted mammary cancer. Toxicology 2001; 160: 11–18
- Wahl RC, Rajagopalan KV. Evidence for the inorganic nature of the cyanolyzable sulfur of molybdenum hydroxylase. J Biol Chem 1982; 257: 1354–1359
- Olson JS, Ballou DP, Palmer G, Massey V. The reaction of xanthine oxidase with molecular oxygen. J Biol Chem 1974; 249: 4350–4362
- Massey V, Brumby PE, Komai H, Palmer G. Studies on milk xanthine oxidase. Some spectral and kinetic properties. J Biol Chem 1969; 244: 1682–1691
- Kim JH, Hille R. Reductive half-reaction of xanthine oxidase with xanthine. Observation of a spectral intermediate attributable to the molybdenum center in the reaction of enzyme with xanthine. J Biol Chem 1993; 268: 44–51
- Avis PG, Bergel F, Bray RC. Cellular constituents. The chemistry of xanthine oxidase. Part III. Estimations of the co-factors and the catalytic activities of enzyme fractions from cow's milk. J Chem Soc 1956; 1219–1226
- Bailey JS, Siu CH. Purification and partial characterization of a novel binding protein for retinoic acid from neonatal rat. J Biol Chem 1988; 263: 9326–9332
- Chytil F, Ong DE. Cellular retinoid-binding proteins, MB Sporn, AB Roberts, DS Goodman. Academic Press, Inc, Orlando, Florida 1984; 2: 89–123
- Walker JM. The protein protocolos handbook. Humana Press Inc, Totowa NJ 1996; 51–54, Chapter 10 pp.
- Manchenko G. Handbook of detection of enzyme on electrophoretic gels. CRC Press. 1994
- Frolik CA, Tavela TE, Peck GL, Sporn MB. High-pressure liquid chromatographic determination of 13-cis-retinoic acid and all-trans-retinoic acid in human plasma. Anal Biochem 1978; 86: 743–750
- Segel IH. Enzyme kinetics. Wiley Interscience, New York 1975
- Snedecor G, Cochran W. Statistical methods. Iowa State University Press, Ames, IA 1967; 120–134
- Sporn MB, Roberts AB. Role of retinoids in differentiation and carcinogenesis. Cancer Res 1983; 43: 3034–3040
- Chytil F. Retinoic acid, biochemistry, pharmacology, toxicology and therapeutic use. Pharmacol Rev 1984; 36: 93S–100S
- Lotan R, Pieniazek J, Gorge MD, Jettern AM. Identification of a new squamous cell differentiation marker and its suppression by retinoids. J Cell Physiol 1992; 151: 94–102
