Abstract
Iron(III) have been combined to well known quinolones (ciprofloxacin) and some Schiff bases with the help of coordination approach. Characterization of these compounds have been done using elemental analysis, magnetic measurements, thermogravimetric analysis, IR, UV-VIS, 1H NMR and 13C NMR spectral investigation. Analytical studies suggest that the iron(III)-quinolone complexes assume a six-coordinated dimeric distorted octahedral geometry. All the compounds show a good antibacterial activity against broad range of bacteria like Bacillus cereus, Staphylococcus aureus, Escherichia coli, Bacillus subtilis, Salmonella typhi and Serratia marcescens, whereas no significant inhibition towards growth of fungal strains like Aspergillus Niger, Aspergillus flavus and Lasiodiplodia theobromae. Analyses of all these compounds show effective sperm herring DNA inhibition.
Introduction
Iron is the most abundant element found in the surrounding environment and plays crucial role for the survival of terrestrial organisms. Iron also participates in biochemical processes like ribonucleic reduction, energy production, photo synthesis, nitrogen reduction, oxygen transport, and oxygenation [Citation3]. Ligands are designed and functionalized in order to provide tissue specificity for the declivity of metal toxicity and improvement of pharmacological properties. Several iron(III) complexes have been known as contrast agent for magnetic resonance imaging(MRI)[Citation4]. Some Cu and Fe complexes have been known for catalytic, antitumor, hypoxic selective cyatotoxins, and antimicrobial activity Citation5-9. Iron and its complexes both are known for its DNA damaging property Citation10-12. In continuation of our earlier work we have synthesized eight quinolone blended iron(III) compounds with eight different neutral bidentate ligands, all the compounds have been screened for antibacterial activity, and their DNA binding property [Citation1,Citation2].
Experimental
Materials
All the chemicals used were of analytical grade. Ferric nitrate, m-chloro aniline, cyclohexanone, o-phenylenediamine, benzaldehyde, ethylenediamine, p-anisaldehyde, benzil, were purchased from the E. Merck (India) Limited, Mumbai. Ciprofloxacin hydrochloride was purchased form Bayer AG (Wuppertal, Germany). Thiophene-o-carboxaldehyde and 2,2′-bipyridylamine, 2-aminopyridine, and 1,8-diaminonaphthalene were purchased from Eastgate, White Lund, Morecambe, Lancaster, England. Luria broth and agar-agar ware purchased from SRL, India. Sperm herring DNA, sucrose, bromophenol blue, xylene cyanol FF, agarose, acetic acid and EDTA were purchased from Sigma Chemical Co., India.
Chemistry
Synthesis of schiff bases
The A2–A8 Schiff bases were synthesized according to reported processes [Citation1].
Synthesis of Fe(III)-complexes
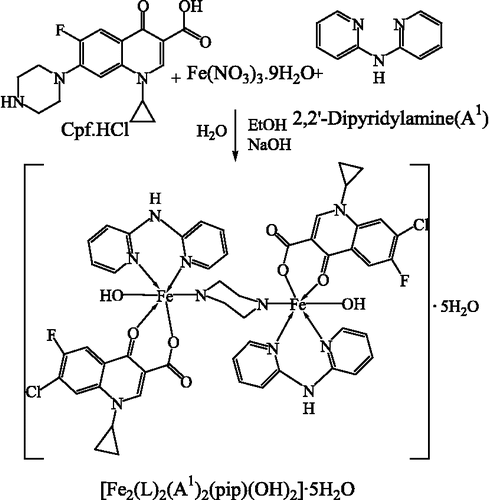
A general preparative method for Fe(III) complexes is shown in .
[Fe2(cip)2(bipym)2(pip)(oh)2]·5h2o (I)
A methanolic solution of Fe(NO3)2·9H2O (4.04 g, 10 mmol) was added to bipym (1.71 g, 10 mmol) in ethanol (100 mL). Followed by addition of formerly primed solution of Cpf·HCl (3.67 g, 10 mmol) in water; the pH was adjusted to 5.0 ∼ 6.0 pH with dilute HNO3 or NaOH solution. The resulting reddish brown solution was refluxed for 7 h, and then heated on steam bath to concentrate for 4–5 h. The reaction mixture was kept for overnight at room temperature. A fine colored product was obtained. Obtained product was washed with ether and dried over vacuum desiccator. Yield: 69%, m.p.: 345°C, Found %: C, 49.18, H, 4.38, N, 11.41; Fe, 9.07. C50H54Cl2F2Fe2N10O13 (1223.62) requires %: C, 49.08, H, 4.45, N, 11.45; Fe, 9.13%.
[Fe2(cip)2(bap)2(pip)(oh)2]·5h2o (Ii)
Prepared from bap (1.82 g, 10 mmol) was used instead of bipym (1.71 g, 10 mmol). Yield: 65%, m.p.: 130°C, Found %: C, 51.97, H, 4.58, N, 8.91; Fe, 9.06. C54H56Cl2F2Fe2N8O13 (1245.66) requires %: C, 52.07, H, 4.53, N, 9.00; Fe, 8.97.
[Fe2(cip)2(tca)2(pip)(oh)2]·5h2o (Iii)
Prepared from tca (1.87 g, 10 mmol). Yield: 62%, m.p.: 355°C, Found %: C, 49.85, H, 4.23, N, 6.70; Fe, 8.95. C52H54Cl2F2Fe2N6O13S2 (1255.74) requires %: C, 49.74, H, 4.33, N, 6.69; Fe, 8.89.
[Fe2(cip)2(bendan)2(pip)(oh)2]·5h2o (Iv)
Prepared from bendan (3.34 g, 10 mmol). Yield: 59%, m.p.: >360°C, Found %: C, 60.50, H, 4.56, N, 7.19; Fe, 7.20. C78H72Cl2F2Fe2N8O13 (1550.05) requires %: C, 60.44, H, 4.68, N, 7.23; Fe, 7.21.
[Fe2(cip)2(benen)2(pip)(oh)2]·5h2o (V)
Prepared from benen (2.36 g, 10 mmol). Yield: 58%, m.p.: 210°C, Found %: C, 54.93, H, 5.10, N, 8.25; Fe, 8.30. C62H68Cl2F2Fe2N8O13 (1353.84) requires %: C, 55.00, H, 5.06, N, 8.28; Fe, 8.25.
[Fe2(cip)2(bmbbd)2(pip)(oh)2]·5h2o (Vi)
Prepared from bmbbd (3.44 g, 10 mmol). Yield: 48%, m.p.: 235°C, Found %: C, 56.65, H, 4.90, N, 7.20; Fe, 7.04. C74H76Cl2F2Fe2N8O17 (1570.03) requires %: C, 56.61, H, 4.88, N, 7.14; Fe, 7.11.
[Fe2(cip)2(bcpded)2(pip)(oh)2]·5h2o (Vii)
Prepared from bcpded (4.29 g, 10 mmol). Yield: 45%, m.p.: 350°C, Found %: C, 56.60, H, 4.17, N, 6.37; Fe, 6.48. C82H72Cl6F2Fe2N8O13 (1739.89) requires %: C, 56.61, H, 4.17, N, 6.44; Fe, 6.42.
[Fe2(cip)2(dcbd)2(pip)(oh)2]·5h2o (Viii)
Prepared from dcbd (2.68 g, 10 mmol). Yield: 46%, m.p.: >360°C, Found %: C, 55.91, H, 6.00, N, 7.84; Fe, 7.90. C66H84Cl2F2Fe2N8O13 (1418.01) requires %: C, 55.90, H, 5.97, N, 7.90; Fe, 7.88.
Other methods
The organic solvents were purified by recommended methods [Citation13]. The metal contents of the complexes were analyzed by EDTA titration [Citation14] after decomposing the organic matter with a mixture of HClO4, H2SO4, and HNO3 (1:1.5:2.5). The magnetic moments were measured by Gouy's method using mercury tetrathiocyanatocobaltate(II) as the calibrant (χg = 16.44 × 10− 6 cgs units at 20°C). The diamagnetic correction was made using Pascal's constant [Citation15].
Spectral investigation
The 1H NMR and 13C NMR was recorded on Bruker Avance (400 MHz). The diffuse reflectance spectra of the complexes were recorded in the range 1700–350 nm (as MgO discs) on a Beckman DK-2A spectrophotometer. Infrared spectra were recorded on an FT-IR Shimadzu spectrophotometer as KBr pellets in the range 4000–400 cm–1. Carbon, hydrogen, nitrogen, and/or sulfur elemental analyses were performed with a model 240 Perkin Elmer elemental analyzer. Thermogravimetric analyses study were obtained with a model 5000/2960 SDTA, TA instrument (USA).
Preparation of stock solution
A stock solution of 2.5 ppm was prepared by dissolving 0.25 mg of each complex in 5% DMSO solution.
Biological studies
Determination of MIC value
The biocidal test was screened by minimal inhibitory concentration (MIC). MIC was determined with the help of progressive double dilution method [Citation16] in liquid media containing 1ppm to 50ppm of the compound being tested. All the compounds were more effective with MIC value at 2.5ppm ≈ 2.5μg/mL. The biocidal activity of the ofloxacin, levofloxacin, flucanozole, ligands, Fe(NO3)2·9H2O, and its complexes were analyzed against various gram-negative and gram-positive bacterial cultures of Staphylococcus aureus, Bacillus subtilis, Bacillus cereus, Salmonella typhi, Escherichia coli and Serratia marcescens using the Agar-plate technique [Citation17].
Preparation of agar plates
The media was made up by dissolving bacteriological agar (20 g.) and luria broth (20 g.) in 1 L distilled water. The mixture was autoclave for 15 min at 120°C and then dispended into sterilized petri dishes, allowed to solidify, and then used for inoculation.
Procedure of inoculation
The target microorganism cultures were prepared separately in 15 mL of liquid LB medium for activation. Inoculation was done with the help of micropipette with sterilized tips; 100 μL of activated strain is placed onto the surface of an agar plate, and spread evenly over the surface by means of a sterile, bent glass rod. Then two well having diameter 10 mm is done with the help of sterilized borer in each plate.
Application of discs
Sterilized stock solutions (2.5 μg/mL) were used for the application in the well of previously inoculated agar plates. When the discs were applied, they were incubated at 37°C for 24 h. The control experiment was performed and then the zone of inhibition was measured (in mm) around the disc and the results are represented in . All experiments were performed in triplicate and ofloxacin, levofloxacin, and flucanozole were used as a standard drug. The growth was compared with solvent as the control and is expressed as zone of inhibition.
Absorption titration
DNA binding affinity study was performed on Shimadzu UV-VIS spectrophotometer. Absorption titration of compounds were done by keeping fixed amount of iron compounds (where compound: I = 12.23, II = 12.45, III = 12.55, IV = 15.50, V = 13.53, VI = 15.70, VII = 17.39, VIII = 14.18 μg), variable amount of DNA i.e.0 to 7 μg and maintaining total volume of 20 mL using phosphate buffer, pH 7.2. Compound-DNA solutions were employed to record absorption spectra.
Gel analyses and quantification
The inspection of super coiled pBR322 have been done in TAE [tris(hydroxymethyl)methylamine(T), acetic acid(A) and EDTA(E)] buffer pH 8.0. Pattern of inspection designed as DNA alone (control), DNA in presence of ligands and DNA in presence of Fe(III)-complexes. Nuclease activity experiments have been accomplished by mixing pBR322 (50 μM) in TE [40 mM Trisacetate(T) and 1 mM EDTA(E)] buffer (pH 8.0), and ligand or Fe(III)-complexes (50 μM). Reaction mixture was incubated at room temperature for 1 h. then it was diluted with 6 × loading buffer (40% sucrose, 0.02% bromophenol blue and 0.02% xylene cyanol FF) and loaded on 0.8% agarose gel. Electrophoresis was carried out on constant voltage (100V) in submarine electrophoresis unit (Genei, Banglore, India). Gel was stained with ethidium bromide. The same experimental conditions were maintained in control assays. The gels were viewed on UV transilluminator, images captured with an attached camera and estimated using AlphaDigiDoc™ RT. Version V.4.1.0 PC-Image software.
Results and discussion
Chemistry
The ligands A2–A8 were isolated by reacting aldehyde/ketone with amine in ethanol as reported in earlier work of our laboratory [Citation11]. The synthesized ligands were analyzed using elemental analyses, IR, 1H and 13C NMR spectroscopy. The iron(III) complexes of ciprofloxacin were synthesized by reacting ferric nitrate and variable ligands A1–A8 in a 1:1:1 ratio. Iron complexes assume six coordinated octahedral geometry coordinating with N–N/N–S of neutral bidentate ligands, two oxygen of cip.(LH = 7-Chloro-1-cyclopropyl-6-fluoro-4-oxo-1,4-dihydro-quinoline-3- carboxylic acid), nitrogen of piperazine ring and oxygen of hydroxyl group. The iron complexes are dimeric in nature. In continuation of earlier work on Cu(II) and Fe(II) complexes [Citation1,Citation2] we have synthesized eight dimeric Fe(III) complexes which follow the proposed probable reaction scheme .
The synthesized dimeric compounds are characterized using IR spectra, electronic spectra, magnetic measurements and thermogravimetric analyses. The thermal decomposition suggests five water molecule of crystallization and stepwise decomposition of complexes. The elemental analyses were in good agreement with the proposed 1:1:1; Fe(III):Cip:An formulation of all complexes.
IR spectra
The IR spectral bands for stretching and bending vibrations are listed in . The sharp band in ciprofloxacin at 3520 cm− 1 [Citation18] is due to hydrogen bonding; which is contributed to ionic resonance structure and peak observed because of free hydroxyl stretching vibration. This band completely absent in the spectra of metal complexes indicating deprotonation of carboxylic proton. The broad absorption band obtained at 1624 cm− 1 and 1384 cm− 1 in ciprofloxacin is observed at ∼1579 cm− 1 and ∼1367 cm− 1 for ν (COO)asy and ν (COO)sym in complexes respectively, in present case separation frequency Δν>200 cm− 1 (Δν = ν COOasy - ν COOsym) suggesting the unidentate binding of carboxylato group Citation19-22. The ν (C = O) stretching vibration band appears at 1708 cm− 1 in the spectra of ciprofloxacin, in the complexes this band shifted towards lower energy at about 1630 cm− 1; suggesting that coordination occurs through the carbonyl oxygen atom [Citation19]. In the investigated compound the ν(C = N) of 2, 2′-bipyridylamine appears at 1580 cm− 1. This band shifted to higher frequency at 1612 cm− 1 [Citation23] in complexes indicates the bidentate N–N coordination of the ligand. Similarly for benzylidene-2-amino pyridine two strong band at 1619 and 1593 cm− 1 assigned to ν(C = N) stretching vibration of azomethine and pyridine ring respectively. On complexation these bands are shifted to 1664 cm− 1 and 1619 cm− 1 suggesting the bidentate N–N participation in coordination [Citation24,Citation25]. The ν (C = N) peak for synthesized Schiff bases A3–A8 observed at 1601–1629 cm− 1. On complexatation of ligand these bands are shifted to 1550–1610 cm− 1 indicating the N–S or N–N bidentate coordination of ligand Citation26-30. Absorption at about 3420 cm− 1 for the ν OH frequency indicates the coordinated hydroxo anion to iron, and is further supported by shoulder at about 340 nm in UV-vis spectra [Citation31]. These data are further supported by ν (M–O) [Citation32], and ν (M–N) [Citation33] appear at 500 ∼ 515 cm− 1, and 535 ∼ 545 cm− 1 respectively. In case of [Fe2(Cip)2(tca)2(pip)(OH)]·5H2O new band observed at 420 cm− 1 which can be assigned to ν (M–S) [Citation27,Citation34] mode.
Table I. Infrared spectral data of complexes.
Electronic spectra and magnetic measurements
Electronic spectral data and magnetic moments are summarized in . The diffuse reflectance spectra of diiron complexes [Fe2(L)2(An)2(pip)(OH)2] ·5H2O have been taken in solid state. The electronic spectra of the complexes exhibit three absorption bands at about ∼19000, ∼23000, and ∼25500 cm− 1, which may be assigned to transitions 6A1g → 4T1 g, 6A1g → 4T2 g, 6A1g → 4A1 g, 4Eg, respectively [Citation35] and suggests octahedral geometry. The magnetic moment of all compounds obtained in between 5.71–6.09 B.M. is good agreement for six-coordinated dinuclear iron(III) system and consistent with the presence of a five-unpaired electrons [Citation36].
Table II. Electronic spectral data of complexes.
Thermogravimetric analysis
The thermogravimetric analysis suggest that initially there is weight loss occurring in the 50–130°C temperature range for all Fe(III)-complexes is attributed to as a loss of the water of crystallization. In the second step weight loss during 140–180°C corresponding to two hydroxyl (OH) molecules. In case of third step weight loss during 180–220°C corresponding to piperazine (pip) molecule, followed by liberation of (L) in between 220–490°C. At last decomposition of An occurs in temperature range 510–710°C, and remaining weight is in good agreement with oxide of iron.
Biology
Biocidal activity
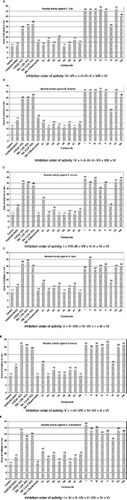
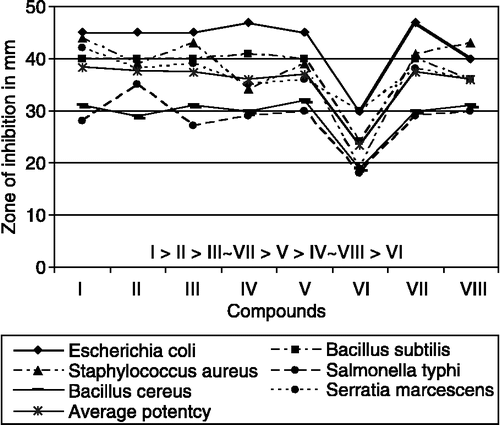
Comparative analysis show higher biocidal activity of the Fe(III)-complexes than free ligands, metal salt and the control (DMSO). The Fe(III)-complexes exhibit higher activities except complex VI against S. aureus, B. subtilis, B. cereus, E. coli, S. marcescens and moderate activity against S. typhi as compared to the standard drugs ofloxacin, levofloxacin, and flucanozole. Comparative study of biocidal activity shown in . Average potency of biocidal activity is determined and order of activity has been found out. The compound-VI has lowest activity while, compound-I has highest activity may be due to the compound-I have highest number of N atoms in its geometry. Order of average potency to inhibit the microorganism is I > II > III ∼ VII > V > IV ∼ VIII> VI. All the iron(III) complexes exhibit marked augment in biocidal activity may be due to the effect of the metal ion on the normal cell process [Citation37]. A possible mode for increase in biocidal activity may be considered in light of Overtone's concept [Citation38] and Tweedy's chelation theory [Citation39].
DNA binding
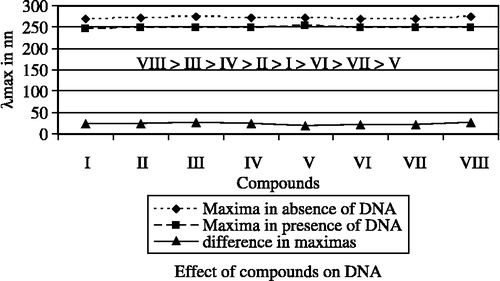
Absorption titration is comprehensively used and renowned to decide the binding of the complexes with DNA helix. Complexes bound to DNA through interaction results in red shift (bathochromism) and blue shift (hypochromism) due to interaction between chromophores and the base pair of DNA helix. The level of hypochromism is generally consistent with the strength of intercalative interaction Citation40-42. The data of the DNA binding with complexes are represented in . The maxima at about 271 nm is observed in spectrum of Fe(III)-complexes in absence of DNA, which is decreases as the amount of DNA increases and observed at about 249 nm in presence of 7 μgm DNA. The absorption spectra of the complex [Fe2(Cip)2(bipym)2(pip)(OH)2].5H2O is shown in . Similarly in case of variable ligands (A1 to A8), Fe(NO3)2.9H2O, and ciprofloxacin maxima observed at about 260 nm in absence of DNA and in presence of 7 μgm DNA maxima observed about 245 nm. All data leads to suggest that in presence of 7 μgm of DNA (maxima at about 245 nm) whole complex dissociate, and free Fe(III), constant ligand(Cip), and variable ligands(An) bind with DNA. The effect of compounds in presence of DNA and in absence of DNA have been studied on the basis of difference in λmax of compounds and presented in . Order of effect is VIII > III > IV > II > I > VI > VII > V, and suggesting that compound- VIII has highest binding property while; compound-V has lowest binding property.
Table III. DNA binding with complexes data.
Electrophoretic behavior of complexes-DNA systems
The effect of the binding of complexes on supercoiled (SC) pBR322 was determined by its ability to make it bulky by binding with reactive sites of pBR322 DNA and produce changes in its conformation from supercoiled(SC) to nicked open circular(OC) form. When pBR322 is subjected to electrophoresis, the fastest migration is observed for SC. If one strand is cleaved due to binding with reactive species, the SC form is converted in OC form. shows the electrophoretic process of all eight Fe(III)-complexes after incubation for 1 h. and the comparison of same experiments have been carried out with Fe(III) and ligands. Fe(III)-complexes exhibit nuclease higher activity than that of Fe(III).
Lane 1: pBR322 (control); lane 2: pBR322 + I; lane 3: pBR322+ II; lane 4: pBR322+ III; lane 5: pBR322+ IV; lane 6: pBR322 + V; lane 7: pBR322+ VI; lane 8: pBR322+ VII; lane 9: pBR322+ VIII
The more cleavage activity of complexes respect to the ligands is clearly evidenced from the figure and data (). It is observed that SC migrates faster than OC. SC smudge on the gel while OC stay behind in well. It may have two reasons, (I) OC become bulky having high molecular weight due to intercalation of compounds. (II) OC requires more time to run on gel than SC. From the experiment we can conclude that the conversion of SC to OC is higher in presence of complexes than that of in presence of free ligands and Fe(III).
Table IV. Cleavage of pBR322 DNA.
Notes
References
- Pansuriya PB, Patel MN, Dhandhukia P, Thakkar V. J Enz Inhib Med Chem, [In press-DOI: 10.1080/14756860701228988]
- Pansuriya PB, Patel MN. Enz Inhib Med Chem, [In press-DOI: 10.1080/147566360701384039]
- Theil EC. Iron biomineralization, RB Frankel. Plenum Press. 1990
- Schwert DD, Davies JA, Richardson N. Non gadolinium based MRI contrast agents. Topics in current chemistry, W Krause. Springer, New York 2001; 221: 165–200
- Torre MH, Gambino D, Araujo J, Cerecetto H, González M, Azqueta ML, Cerain AL, Vega AM, Abram U, Costa-Filho AJ. Eur J Med Chem 2005; 40: 473–480
- Rechardson DR, Bernhardt PV. J Biol Inorg Chem 1999; 4: 166–273
- Brookhart M, Small BL. Macromolecules 1999; 32: 2120–2130
- Flavia N, Angela L, Giancarlo M, Carlo P, Vincenzo P, Genevieve C, Pierrette B, Daniel M. J Biol Inorg Chem 1998; 3: 671–681
- Patel KN, Patel KM, Patel MN, Patel CR, Patel NH. Synth React Inorg Met-Org Chem 2000; 30(8)1617–1627
- Pedreño E, López-Contreras AJ, Cremades A, Peñaflel R. J Bioinorg Chem 2005; 99: 2074–2080
- Routier S, Vezin H, Lamour E, Bernier JL, Cattean JP, Bailly C. Nucleic Acid Research 1999; 27: 4160–4166
- Catherinr H, Marguerite P, Michael R, Heinz G, Stéphanie S, Bernard M. J Biol Inorg Chem 2001; 6: 14–22
- , Furniss BS, Hannaford AJ, Smith PWG, Tatchell AR. 5th ed. Vogel's textbook of Practical Organic Chemistry 2004.
- Vogel AI. A textbook of quantitative inorganic analysis. ELBS and Longman, London 1989
- Weiss A, Witte H. Magnetochemie. Verlag Chemie, Weinheim 1973
- Dimitra KD, Demertzis MA, Filiou E, Pantazaki AA, Yadav PN, Miller JR, Zheng Y, Kyriakidis DA. Biometals 2003; 16: 411–418
- Pelczar MJ, Chan ECS, Krieg NR. Microbiology5th Ed. 1993; 23: 488
- Silverstein RM, Webster FX. Spectrometric identification of organic compounds6th Ed. John Wiley & Sons, Inc. 2004
- Turel I, Leban I, Bukovec N. J Inorg Biochem 1999; 241–245
- Chohan ZH, Suparan CT, Scozzafava A. J Enz Inhib Med Chem 2005; 20: 303–307
- Deacon GB, Philips RJ. Coord Chem Rev 1980; 23: 227–250
- Nakamoto K. Infrared and raman spectra of inorganic and coordination compounds4th ed. A Wiley Interscience Publication. 1986
- Panchal PK, Pansuriya PB, Patel MN. J Enz Inhib Med Chem 2006; 21(2)203–209
- Mandal SS, Ghorai PC, Ray S, Saha HK. J Ind Chem Soc 1995; 72: 807
- Gudasi KB, Goudar TR. Synt React Inorg Met-Org Chem 2000; 30(10)1859–1869
- Panchal PK, Pansuriya PB, Patel MN. J Enz Inhib Med Chem 2006; 21(4)671–681
- Raman N, Kulandaisamy A, Jayasubramananian K. Polish J Chem 2002; 76: 1085–1094
- Parekh HM, Panchal PK, Pansuriya PB, Patel MN. Polish J Chem 2006; 80: 989–992
- Singh K, Barwa MS, Tyagi P. Eur J Med Chem 2006; 41: 147–153
- Sharma S, Athar F, Maurya MR, Azam A. Eur J Med Chem 2005; 40: 1414–1419
- Bénisvy L, Halut S, Donnadieu B, Tuchagues JP, Chottard JC, Li Y. Inorg Chem 2006; 45: 2403–2405
- Sönmer M, Berber İ, Akbaş A. J Med Chem 2006; 41: 101–105
- Chandra S, Gupta N, Gupta LK. Synth React Inorg Met-Org Chem 2004; 34(5)919–927
- Sargent AL, Titus EP, Riordan CG. Inorg Chem 1996; 35(21)7095
- Lever ABP. Electronic spectra of dn ions. Inorganic electronic specroscopy2nd Ed. 1984
- Cotton FA, Wilkinson G. The element of first transition series. Advanced inorganic chemistry3rd Ed. 1992
- Dharmaraj N, Viswanathamurthi P, Natarajan K. Trans Met Chem 2001; 26: 105, (b) Chohan ZH, Supuran CT, Rauf A. Metal-Based Drugs. 2002; 8: 287
- Chohan ZH, Khan KM, Supuran CT. Appl Organomet Chem 2004; 18: 305, (b) Anjaneyula Y, Rao RP. Synt React Inorg Met-Org Chem 1986;16:257
- El-Metwaly NM. Trans Met Chem 2007; 32: 88–94, (b) Tweedy BG. Phytopathology 1964;55:910
- Barton JK, Danishefsky AT, Goldberg JM. J Am Chem Soc 1984; 106: 2172
- Kelly TM, Tossi AB, McConnell DJ, Strekas TC. Nucl Acid Res 1985; 13: 6017
- Tysoe SA, Margan RJ, Baker AD, Strekas TC. J Phy Chem 1993; 97: 1707–1711

![Figure 3 Absorption titration curve of complex [Fe2(L)2(A1)2(pip)(OH)2]·5H2O.](/cms/asset/2b015f62-2d60-419a-9718-906488482945/ienz_a_247344_f0005_b.gif)