Abstract
Matrix metalloproteinases (MMPs) play an important role in many physiological and pathological processes. To assay the activities of MMPs is important in diagnosis and therapy of the MMPs associated diseases, such as neoplastic, rheumatic and cardiovascular diseases. Several assay systems have been developed, which include bioassay, zymography assay, immunoassay, fluorimetric assay, radio isotopic assay, phage-displayed assay, multiple-enzyme/multiple-reagent assay and activity-based profiling assay. The principle, application, advantage and disadvantage of these assays have been reviewed in this article.
Keywords::
Introduction
The matrix metalloproteinases (MMPs) are zinc-binding endopeptidases that collectively degrade all components of the extracellular matrix. Overall, the MMP family consists of more than 25 enzymes, which include collagenases (MMP-1, 8, 13, 18), gelatinases (MMP-2, 9), stromelysins (MMP-3, 10, 11), matrilysins (MMP-7, 26), membrane type MMPs (MMP-14, 15, 16, 17, 24, 25) and others. They are important in normal physiological functions such as angiogenesis, wound healing, mammary gland and postpartum uterus involution, and cervical dilatation, but excess MMP activities are correlated with various diseases, such as tumor, arthritis, periodontal diseases, liver cirrhosis, atherosclerosis, and multiple sclerosis Citation1-4.
The activities of these enzymes are well regulated by endogenous tissue inhibitors of metalloproteinases (TIMPs). The synthetic inhibitors of MMPs, such as peptidomimetic compounds, nonpeptidic compounds, tetracycline derivative, biphosphonate are also of interest for the treatment of cancer and arthritis. Many MMP synthetic inhibitors have been studied using preclinical studies in vivo models. Although preclinical studies were so compelling to encourage several clinical trials, the past years have seen a consistent number of disappointments and limited success [Citation5]. Most preclinical studies have focused on the role of MMPs in the early stages (progression and metastases) of cancer, in which MMP inhibition seems to have its greatest effect. Unfortunately, clinical trials of MMP inhibitors were conducted almost uniformly in patients with advanced, metastatic disease, and all have failed to show any beneficial effect on patients.
The failure of these inhibitors in clinical trials has stimulated the methodology research in assay of MMPs and their inhibitors in tissue samples and body fluids. In 2005, Lombard et al [Citation6] published a very important review about MMPs activities assay, in which the assay methods were classified mainly by substrates, such as proteins, synthetic linear peptides and synthetic “mini-collagens”. In order to summarize the further development of MMPs assay methods since 2005, such as in vivo zymography and activity-based profiling assay, we delivered this review. Moreover, the assay methods of MMPs and their inhibitors were classified by the assay techniques, which were outlined below: bioassay, zymography assay, immunoassay, fluorimetric assay, radio isotopic assay, phage-displayed assay, multiple-enzyme/multiple-reagent assay, and activity-based profiling assay. The principle, application, advantage and disadvantage of these assay methods were described in this review.
Bioassay
Most of previous assays for MMPs rely on the biological activity of the enzyme to degrade natural substrates, and are so-called “bioassay”.
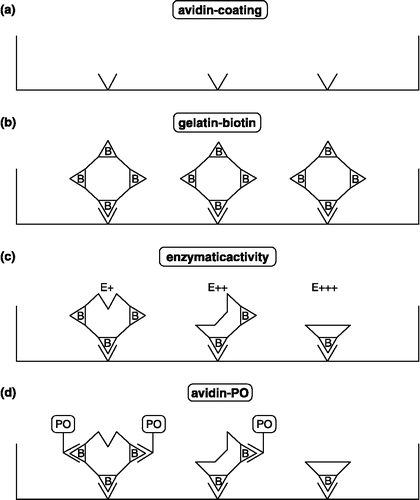
Assays using biotinylated gelatin (BG)
Gelatin is a substrate for MMPs such as MMP-2 and MMP-9. Gelatin was biotinylated to increase the sensitivity of the detection system. Biotinylated gelatin (BG) coated on microtiter plates provides a useful substrate for detecting gelatinase activity. Individual wells (illustration) of a microtiter plate were coated with avidin to which biotinylated gelatin substrate was bound. Enzymatic activity of gelatinases liberated gelatin fragments and decreased the number of residual biotin proportionally. The latter was specetrophotometrically quantified by binding of streptavidin-peroxidase and development of peroxidase activity (PO), using peroxide and tetra methyl benzidine as a color reagent () [Citation7,Citation8].
This 2 h assay permits detection of MMP-2 activity in concentrations as low as 0.16 ng/mL and can be used for monitoring MMP activity in tumor-derived samples and for evaluation of efficiencies of synthetic MMP inhibitors [Citation9].
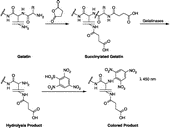
Assays using succinylated gelatin
The assay is based on the use of succinylated gelatin as substrate and measurement of primary amines exposed by hydrolysis of the substrate by gelatinases. The exposed primary amines are detected by reacting with 2,4,6-trinitrobenzene sulfonic acid (TNBSA) that produces a quantifiable color reaction with a λmax at 450 nm, which is directly proportional to the gelatinase activity (). Succinylated gelatin was primarily digested by MMP-2 and -9. The detection limit for catalytically active MMP-9 was 12.5 ng (1.0 nM) and that for MMP-2 was 20 ng (2 nM), using a 30 min incubation [Citation10].
The assay can be used to detect gelatinolytic activity in purified or crude samples, and as a high throughput screen for enzyme inhibitors of MMP-2 and -9, in particular.
Zymography assay
Zymography is an accurate and sensitive quantitative technique for the evaluation of matrix metalloproteinases or their inhibitors. This technique is a variation on acrylamide gel electrophoresis. The gel used for zymography, commonly referred to as a zymogram, contains a protease substrate (usually gelatin) incorporated directly into the gel during polymerization. Following electrophoresis of the sample containing a protease, SDS is removed from the gel by exchange in Triton X-100. This allows the gelatinases in the sample to renature and autoactivate. Its activity is revealed by an absence of protein staining in the region where the substrate has been digested.
Gelatin zymography
Gelatin zymography is the common method for examining gelatinases in cells and media samples. This is an SDS-PAGE-based method performed with copolymerized gelatin. This method allows the relative amount of each gelatinase to be determined in small samples of tissue ( < 10 mg) [Citation11] and can be used qualitatively for the analysis of gelatinase species present [Citation12,Citation13]. For collagenases 1 and 3, adding heparin could enhance the detection on gelatin zymograms at least 5-fold [Citation14].
There are controversial data on the effects of different anticoagulants as pre-analytical determinants of plasma MMP activities by gelatin zymography. In order to solve this problem, Gerlach et al Citation15-17 examined whether different anticoagulants affect plasma MMP-2 and MMP-9 activities using gelatin zymography. Their results suggest that similar MMPs activities are found in plasma samples separated from blood collected into EDTA, citrate, or heparin tubes. Further studies found that the circulating levels of MMP-9 should be assessed in citrate or heparin plasma samples. Serum samples, however, should not be used to assess MMPs activities for two main reasons: because artificially higher MMP-9 levels are found, independently of how rapidly the serum is separated from blood, and because they do not correlate with the MMP-9 levels found in plasma samples.
Despite its numerous applications [Citation18], the utility of gelatin zymography is limited by poor inter-laboratory comparison due to lack of a readily available MMP Mr referenced standard. Toward the goal of defining a readily available MMP Mr standard, Makowski et al [Citation19] investigated the feasibility of using gelatinase standards from human capillary blood for calibrating gelatin zymograms. The standards are easily obtained and well characterized and should thus facilitate inter-laboratory comparison and standardization.
Casein zymography
For stromelysins 1 and 2 (MMP-3 and MMP-10, respectively) and matrilysin (MMP-7), casein zymography is more suitable. However, casein zymography has a detection limit at least two orders of magnitude less sensitive than gelatin zymography; moreover, due to its relatively low molecular weight (23 kDa), casein migrates during the electrophoresis, resulting after staining in the existence of two clearly defined zones in the gel [Citation20]. The anodic migration of the substrate turns out to be especially problematic in the case of proteinases which are located near the casein migration boundary. This is the case of the latent and activated forms of matrilysin (29 and 20 kDa, respectively). Pre-running casein-embedded gels before the loading and electrophoresis of the samples can improve the definition of the caseinolytic band, increase the method sensitivity, and avoid the problem of substrate migration observed in the classical casein zymography [Citation21].
Collagen zymography
Unlike the gelatinases (MMP-2 and -9) and matrilysin (MMP-7), collagenases (MMP-1 and -13) are difficult to detect at low levels in conventional casein or gelatin zymography. Gogly et al [Citation22] describe the use of casting native collagen type I in SDS–polyacrylamide gel (collagen zymography) for the determination of interstitial collagenase.
This method proved to be very sensitive: 0.1 pg of APMA (p-aminophenylmercuric acetate)-activated procollagenase could be detected, and specific levels of active gelatinase or stromelysin lower than 5 ng were not detected under their experimental conditions [Citation23].
In situ zymography
In situ zymography is based on the same principles as gel-based zymography. Either a photographic emulsion containing gelatin or a fluorescence-labeled proteinaceous macromolecular substrate is brought into contact with a tissue section or cell preparation. After incubation, enzymatic activity is revealed as white spots in a dark background or as black spots in a fluorescent background [Citation24].
This technique is extremely useful for the detection and localization of MMP-2, 7, 9 in situ as has been demonstrated in several unrelated tissues Citation25-27.
In situ zymography preserves the fine morphological details of the tissue and can complement the study of enzyme expression by other microscopic techniques, such as immunocytochemistry [Citation28]. However, researchers must be aware of potential variations in zymographic analysis may be influenced by physical tissue parameters in addition to suspected gelatinase activity [Citation29].
Reverse zymography
Reverse zymography is an electrophoretic technique used to detect the presence of MMP inhibitors rather than MMPs. This technique allows concurrent semi-quantitative analysis of inhibitor levels and molecular weight. The technique has its origin in a fibrinogen–agarose plate assay for protease inhibitory activity in which a digesting agent was added to grooves after electrophoresis and allowed to diffuse into the gel [Citation30]. The original method had very crude sensitivity and resolving power by current standards, but was modified and improved by directly incorporating a proteinase source into the acrylamide-substrate gels. Recent methods have focused on the detection of MMP inhibitory activity using conditioned media as a good source of MMPs [Citation31].
To minimize the number of variables and make reverse zymography more consistent, sensitive, and quantitative, Oliver et al [Citation30] describe a novel system for reverse zymography using purified recombinant human gelatinase A or gelatinase B in place of conditioned media. This system can detect TIMPs 1, 2, and 3 simultaneously, but with differential sensitivities for TIMPs 1 and 2.
However, this technique is still problematic. For example, it is very hard to predict the proper incubation time for a sample containing an unknown concentration of TIMPs because the gel staining procedure terminates the reaction. As a result, multiple gel samples are typically required to obtain the best reaction time.
Real-time zymography
Hattori et al [Citation32] utilized a fluorescein-isothiocyanate (FITC, a more popular and readily available fluorescent dye)-labeled denatured collagen as the substrate to develop novel zymographic and reverse zymographic methods for detecting MMPs and TIMPs, respectively. Using a trans illuminator, the results can be observed visually without stopping the enzymatic reaction. For this reason, they have named these methods real-time zymography and real-time reverse zymography.
They further expanded this technique by incorporating two substrates with distinct fluorophores (FITC-labeled gelatin and Texas-red-labeled casein) into a single polyacrylamide gel as substrates. The separate detection of the gelatin and casein degradation could be assessed on the same gel using two types of optical filters. With this fluorescent detection system, MMP-3 could be distinguished from MMP-2 and MMP-9 and activities could be measured concurrently and in real time [Citation33].
Using this dual detection system, the time and reagent can be saved, and quantitative analysis of MMPs is possible. Higher sensitivity is achieved with a lower amount of substrate as compared with conventional methods.
In vivo zymography
Bremer and Crawford [Citation34,Citation35] developed a novel approach to assay MMP activity in vivo, which was named in vivo zymography. This technique is based on the injection and in situ degradation of heavily fluoresceinated native collagens. These substrates are only weakly fluorescent in their native state due to intra-molecular quenching. However, when proteolytically degraded, the quenching is relieved, resulting in an increase in fluorescent signal, which is proportional to the degree of collagen degradation.
Using this technique, Crawford et al [Citation34] characterized patterns of MMP activity in the zebrafish embryo, and Bremer et al [Citation35] sensed or imaged MMP-2 activity directly in intact tumors in nude mice. This technique could also be used to image the effect of MMP inhibition directly within hours after initiation of treatment using the potent MMP inhibitor, prinomastat (AG3340). This technology will enable the detailed analysis of MMPs and their inhibitors directly in vivo in intact tumor environments.
Immunoassay
Immunoassays have been developed for MMPs, TIMP and MMP-TIMP complex. Selection of antibodies of defined specificity enabled measurement of both the pro and active forms of the metalloproteinase. Free TIMP was quantified by the selection of a monoclonal antibody which did not recognize TIMP when complexed with metalloproteinases. Detection of enzyme-inhibitor complexes was achieved by capturing the TIMP component of the complex and revealing the metalloenzyme using specific antibodies.
Immuno capture assay
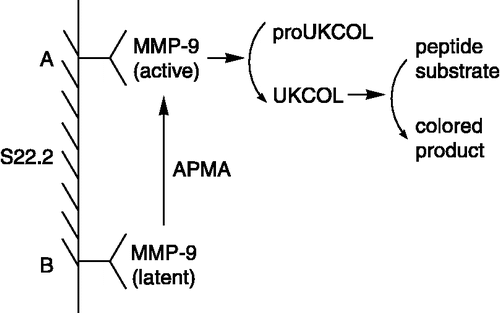
The immuno capture assay, also named biotrak assay [Citation36], was first described by Verheijen et al [Citation37]. MMPs was captured and immobilized by specific antibody, and Prior activation of the latent MMPs by APMA produced the active MMPs, which was necessary to perform this assay. Using protein engineering, a modified pro-urokinase (proUKCOL), as a substrate, was made in which the activation sequence, normally recognized by plasmin (ProArgPheLysIleIleGlyGly), was replaced by a sequence that is specifically recognized by MMPs (ArgProLeuGlyIlelleGlyGly). The active urokinase (UKCOL) resulting from the activation of proUKCOL by active MMPs can be measured directly using a chromogenic peptide substrate for UKCOL. The assay has been made specific for MMP-9 using an MMP-9 specific monoclonal antibody S22.2 () [Citation38,Citation39]. The detection limit for MMP-9 was below 15 pM, corresponding to 3. 75 × 10− 15 mol per assay.
By using other antibodies, they then adapted the assay to specific assays for the MMP-2 [Citation40]. Using this technique, Yoshida et al [Citation41] determined the molecular mechanism of SI-27, an anti-MMP agent.
This method was not time-consuming, and yet was quantitative for each subtype of MMP, so it may become a new standard that avoids the deficiencies of zymography and collagenolysis. This assay can easily be used for high throughput screening of compounds or samples.
Western blotting
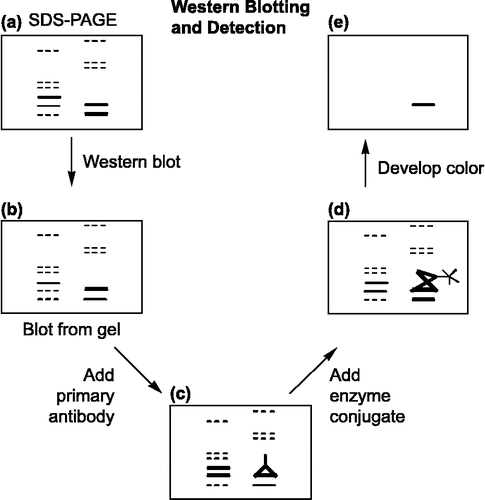
Western blotting (protein blotting or immunoblotting) is a powerful and important procedure for the immunodetection of MMPs post-electrophoresis [Citation42]. Western blotting follows five procedures (): (A) Unstained SDS–PAGE gel prior to western blot. (B) Exact replica of SDS–PAGE gel obtained as a blot following western transfer. (C) Primary anti-MMPs antibody binding to a specific band on the blot. (D) Secondary antibody conjugated to alkaline phosphatase or horse radish peroxidase (along with soluble substrates) binding to primary antibody. (E) Color development of specific band [Citation43].
Western Blotting offers the following specific advantages: (a) wet membranes are pliable and easy to handle, (b) the proteins immobilized on the membrane are readily and equally accessible to different ligands, (c) multiple replicas of a gel is possible, (d) prolonged storage of transferred patterns, prior to use, becomes possible and (e) the same protein transfer can be used for multiple successive analyses [Citation44].
However, western Blotting requires knowledge of the MMPs native substrate and the availability of anti-MMPs antibodies. And what is more, it is expensive and time-consuming.
Sandwich enzyme immunoassay
Obata et al [Citation45] developed a one-step sandwich enzyme immunoassay (EIA) for MMP-3 (stromelysin-1). The assay system used two simultaneous immunoreactions using a solid phase monoclonal antibody and a horseradish peroxidase-labeled monoclonal antibody (Fab’). The sensitivity of the assay system was 20 μg/mL and linearity was obtained between 31 and 500 μg/mL. The EIA system was capable of measuring both precursor and active forms of MMP-3 as well as the forms of MMP-3 complexed with tissue inhibitors of metalloproteinases. They further established the one-step sandwich enzyme immunoassay (EIA) for MMP-1 [Citation46], MMP-2 [Citation47], TIMP-2 [Citation48], MMP-7 [Citation49], MMP-8 [Citation50], MMP-20 [Citation51]. () Compared to the two-step sandwich enzyme immunoassay system [Citation52], this system is more sensitive and has advantages of simplicity and rapidity. This system can be applied to determine the levels of MMPs and proMMPs in human plasma from normal subjects and patients with cancer.
Table I. The sensitivity and linearity of sandwich enzyme immunoassay for various MMP subtypes.
Enzyme-linked immunosorbent assay (ELISA)
Enzyme-linked immunosorbent assays (ELISA) have recently been developed for the quantification of MMPs [Citation53,Citation54]. Clark et al [Citation55] has raised and characterized monoclonal antibodies against purified human fibroblast collagenase. One of these antibodies has been used in combination with a polyclonal anticollagenase antibody in a double antibody sandwich ELISA to measure collagenase. Two combinations were applicable to the immunoassay: (i) a monoclonal capture antibody with a polyclonal detecting antibody; (ii) two monoclonal antibodies. The assay measures total collagenase, i.e. pro- and active enzyme as well as collagenase in complex with TIMP Citation56-58. This assay has also been used to quantify stromelysin [Citation59,Citation60], and type II collagen [Citation61]. These commercially available ELISAs have good sensitivity.
Maliszewska et al [Citation62] further developed an ultra-sensitive two-side ELISA which allows for the first time to measure reliably MMP-9 concentrations in human cerebrospinal fluid.
Using ELISA, Jung et al [Citation63,Citation64] studied the pre-analytical pitfalls of blood sampling to measure true circulating MMP-9, TIMP-1 and TIMP-2. The results showed that serum does not seem to be an appropriate sample for determining circulating MMP-9 and TIMP-1 whereas the TIMP-2 determination is interfered by heparin. In order to avoid pre-analytical misinterpretations, citrate plasma has been suggested to be the sample of choice for measuring circulating MMP-2 and MMP-9. Their results were consistent with the above mentioned report from Gerlach Citation15-17 using gelatin zymography.
Urinary type II collagen neoepitope assay (uTIINE assay)
uTIINE assay is a technique that utilizes urinary type II collagen neoepitope to measure MMP activity. When an antibody is made that specifically recognizes a cleavage site that includes either the free amine or the carboxyl terminus in a peptide sequence, it is referred to as a neoepitope antibody. Poole et al [Citation65] applied neoepitope antibodies to the sequence GPQG-COOH, a principal cleavage site in type II collagen, detecting type II collagen fragments in synovial fluid from rheumatoid and osteoarthritic patients. Another neoepitope antibody to a new terminus GPP(OH)GPQG-COOH was generated for collagenase 1, 2, and/or 3 cleavage of type II collagen. Collagen fragments were detected in urine when an upstream anti-collagen II-specific antibody appeared. The urinary TIINE activity correlated well with osteoarthritis (OA) disease activity, reflecting the importance of collagenase activity in the OA disease process. In addition, quantitative assays of type II collagen neoepitopes may be useful markers of cartilage degeneration and joint disease in humans [Citation66]. However, these assays are cumbersome and do not discern a particular collagenase or the relative contribution of the various collagenases.
Fluorimetric assay
Fluorogenic substrates provide a particularly convenient enzyme assay method, as they can be monitored continuously and utilized at reasonably low concentration ranges. There are three types of fluorogenic substrates: (i) fluorescein isothiocyanate(FITC)-labeled proteins; (ii) intra-molecular fluorescence energy transfer substrates (IFETS); and (iii) fluorogenic triple-helical peptide (fTHP).
Fluorescein isothiocyanate (FITC)-labeled proteins
Intact fluorescein isothiocyanate (FITC)-labeled proteins are internally quenched due to the close proximity of the labels, so they have relatively low background fluorescence at excitation and emission wavelengths of 495 and 525 nm, respectively. Degradation of these substrates leads to exposure of covalently linked fluorescein isothiocyanate molecules and to a concomitant increase in relative fluorescence at these wavelengths. The increase in relative fluorescence is proportional to the degree of protein degradation. This phenomenon provides the basis for a sensitive assay for MMP activity [Citation67]. There is no requirement for the removal of undegraded substrate from the assay mixture prior to the measurement of fluorescence. Assays can be performed in 96-well micro-titer trays, enabling a large number of samples and their respective controls to be processed simultaneously and repeated determinations of fluorescence values may be made on the same assay Citation68-70.
FITC-proteins exhibit several advantages such as their low cost, stability, easy preparation and waste disposal. However, they are time-consuming due to the procedures required to separate bound from unbound marker.
Intra-molecular fluorescence energy transfer substrates (IFETS)
The substrates consist of peptide chains constructed to match the specificity of the particular enzyme and to bear a suitable chromophore at each side of the cleavable bond [Citation71]. One of the chromophores is a fluorophore (the fluorescence donor) and the other is a light-absorbing group (quencher, the fluorescence acceptor). The intact peptide has low intrinsic fluorescence because of the close proximity of the donor and quencher. When a proteinase cleaves the substrate, the liberated fluorophore is no longer quenched efficiently. The rate of fluorescence increase is a measure of enzyme activity. Quenching is almost unaffected by the nature of the intervening amino acids, so that with appropriate sequences some degree of specificity for the individual MMPs can be achieved Citation72-74.
Standard methods for performing such assays employ commercial 96-well micro-titer plates and compatible detection devices, with assay volumes of 0.05 to 0.2 mL and times of 30 to 60 minutes, which make this assay suitable for use in high-throughput screening of MMP inhibitors [Citation75].
In contrast to the known fluorogenic substrates that are used widely in the scientific and medical research, IFETS allow control of the enzymatic cleavage at any point of the peptide chain and thus permit simultaneous studies of enzymes of different specificity () Citation76-84. However, IFETS is limited by poor solubility, quenching and high background readings [Citation85].
Table II. The fluorophore, quencher, cleavage site and enzyme of various substrates*.
Fluorogenic triple-helical peptide (fTHP)
Fluorogenic triple-helical peptide (fTHP) substrates mimic the behavior of the native collagen substrate and may be useful for the investigation of collagenase triple-helical activity. Based on this idea, chemically synthesized triple-helical substrates have been used to study the mechanisms of MMP action on collagenous substrates [Citation86,Citation87].
Fluorogenic triple-helical peptide (fTHP) substrates could be assembled by either (a) covalent branching or (b) the “peptide-amphiphile” method (non-covalent self-assembly of lipophilic molecules) () [Citation88].
Radio isotopic assay
Method based on the degradation of radio-labeled (such as 14C or 3H) natural substrates has been used extensively for the detection of MMP activity, especially MMP-2 and -9 Citation89-92. The method is performed in plastic plates of 96 flat-bottom micro-wells in which labeled substrates are dried to a film from thermally reconstituted fibrils. The enzyme samples were added to the wells and were incubated, and the fluid was removed for scintillation counting of the release of labeled degradation products [Citation93,Citation94].
However, the method requires expensive devices for registering the released marker, as well as isotope handling, and generates radioactive waste disposal problems [Citation95,Citation96].
Phage-displayed assay
Recently two biological systems using phage display have been developed to quickly search and decode a large peptide library for substrate optimization. The first system uses monovalent substrate phage display to study substrate specificity of membrane type-1 MMPs [Citation97,Citation98]. The second system uses polyvalent substrate phage display to more quickly optimize substrates of stromelysin, matrilysin and human collagenase 3 Citation99-102.
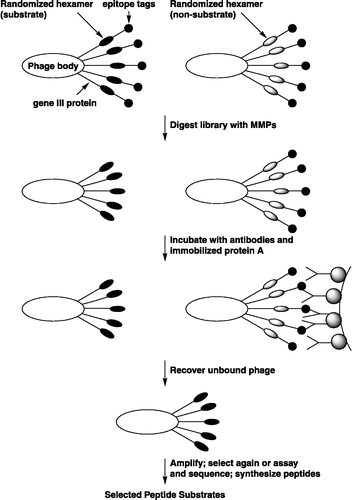
In is shown a diagram of the fTC phage (for clarity, not drawn to scale). The gene III protein extends from phage body from the C to the N terminus. At the N termini are the peptide epitopes and the random hexamer domains that may or may not act as substrates, depending on the encoded peptide sequence. The phage were treated in solution with MMPs, and those carrying substrate sequences (Left) were cleaved in the hexamer site, whereas non-substrates (Right) were not. The entire digest was treated with antibodies to the tethers and captured by using a resin that carries protein A. The protein A-antibody-phage complexes were precipitated by centrifugation, and the phage that was not bound by the antibodies remained in solution and could be recovered for amplification. After amplification, the phage could be analyzed or subjected to further rounds of selection.
The differences in the approaches of monovalent and polyvalent phage as protease substrate discovery tools invite comparison of the two systems. While the monovalent systems have been shown to be quite useful, the polyvalent system possesses certain advantage: (a) all phage can act as substrate phage in the polyvalent system. In monovalent systems, it is estimated that only 10% of the phage particles carry one copy of the recombinant pIII protein. This increases the effective substrate concentration in the polyvalent system, thus increasing the sensitivity of the system (due to higher concentration of substrate at a given level of phage). (b) Since 90% of the monovalent phage do not carry pIII fusions, the non-recombinant phage lacking the tether must be removed prior to selection. This is accomplished by immobilizing the recombinant phage in microtiter plates coated with tether binding protein and treating the phage with protease while immobilized. Polyvalent phage, being 100% recombinant, is digested in solution rather than immobilized on a solid surface. The advantages of this are: (a) there is little restriction on number of phage that can be screened in solution, but the surface system limits the number of phage that can be routinely immobilized on micro-titer plates. Scale up of the solution phage system is thus very convenient when significantly larger libraries are prepared; (b) protease resistance of tether binding protein is not an issue; (c) solution proteolysis offers more precise control of cleavage conditions. This has proven especially useful in the quantitative dot-blot assay [Citation103].
The major disadvantage of the polyvalent system is the appearance of non-reactive phage clones, which does not occur in the monovalent system because of the pre-binding step which essentially eliminates clones with defective epitopes. Nothing about the polyvalent method, however, precludes the use of a binding step in later rounds to eliminate non-reactive clones. Thus, a strategy using a combination of conditions favoring multivalent and monovalent interactions may be used to advantage [Citation104].
Future development and screening of phage-displayed peptide library should lead to the discovery of novel ligands for many purposes, including the identification of new drug candidates [Citation105].
Multiple-enzyme/multiple-reagent assay
Based on the fluorogenic triple-helical peptide (fTHP) assay, Rasmussen et al [Citation106] reported on a novel technique that can be used to simultaneously measure activity levels for MMPs. The technique, termed the multiple-enzyme/multiple-reagent assay system (MEMRAS), relies on the use of reagents such as substrates with varying selectivity profiles against a group of enzymes. When reaction rates are measured by following a change in fluorescence with time, for mixtures of enzymes, an equation with unknown concentrations for each activity is generated for each reagent used. Simultaneously solving the set of equations leads to a solution for the unknown concentrations.
They have successfully applied a two-by-two MEMRAS to measure activity levels for mixtures of MMPs such as collagenase 3 and gelatinase A.
The five steps in the experiment and analysis are described below. First, in step 1, the enzyme standard dilutions, mixed-enzyme samples, and reagents were added to a 96-well plate. More specifically, four combinations of concentrations of collagenase 3 and gelatinase A were placed in twice-duplicated wells. To sets of duplicated wells were added reagent A and reagent B. Other wells were used for standard curves of collagenase 3 and gelatinase A versus reagents A and B. In step 2, the plate reader recorded the fluorescence values every 5 min for 5 h. In step 3, the values of the slope of the net fluorescence versus time curves were plotted versus enzyme concentration for the standard enzyme dilutions, for each enzyme/reagent pair. In step 4, the net fluorescence versus time was plotted for the mixed-enzyme samples when reacted with each reagent. Finally, in steps 5 and 6, the slopes from steps 3 and 4 were combined into a set of two equations in two unknowns and solved for the “unknown” enzyme concentrations in the mixed-enzyme samples.
The two-by-two MEMRAS system can quantify how much of the total activity is due to collagenase 3 and how much is due to gelatinase A. The measurement error of the MEMRAS system is significantly less (10-fold) than the error arising from a single-reagent system.
The substrates developed, even if not very selective, also became useful when the MEMRAS technique was developed. As opposed to a single reagent that is highly selective for a given MMP, MEMRAS relies on using as many reagents as there are enzymes, and the reagents do not necessarily have to be extremely selective. This indicates that the MEMRAS system is a unique solution to the problem that substrates are never quite selective enough to directly assay activities in biological fluids.
Activity-based profiling assay
Activity-based proteomic probes (ABPP) is a chemical strategy that utilizes active site-directed probes to record variations in the activity of enzymes in whole proteomes. ABPP typically possess three general elements: (a) a binding group that promotes interactions with the active sites of specific classes of enzymes, (b) a reactive group for covalent label with the active-site of enzymes, and (c) a reporter group (e.g., fluorophore or biotin) for the visualization and affinity purification of probe-labeled enzymes. Using this technique, Saghatelian et al [Citation107] designed a rhodamine-tagged hydroxamate benzophenone probe (HxBP-Rh) for MMPs by incorporating a zinc-chelating hydroxamate and a benzophenone photocrosslinker group, which promoted selective binding and modification of MMP active sites, respectively. HxBP-Rh labeled active MMPs but not their zymogen or inhibitor-bound forms. HxBP-Rh were applied to identify MMPs up-regulated in invasive cancer cells and to evaluate the inhibitor sensitivity of MMPs in whole proteomes. With great sensitivity, HxBP-Rh was able to detect active MMP-2 at concentrations as low as 3 nM in tissue proteomes. Despite the potential of this technique, no endogenous active MMPs have been identified yet.
Another activity based profiling assay of MMPs relies on the use of group-specific inhibitor affinity sorbents for the selective enrichment prior to standard proteomic approaches. The use of MMP inhibitor affinity sorbents with immobilized affinity ligands allows selective enrichment of proteins based on common structural or functional properties, prior to separation and analysis, thereby serving as an effective sample cleanup step resulting in enhanced sensitivity. Freije et al [Citation108] applied this technique to synovial fluid from a rheumatoid arthritis patient followed by gelatin zymography. Using a broad-spectrum MMP inhibitor with nM affinity (TAPI-2), they revealed a strong enrichment of distinct MMPs from this biological sample that were not clearly visible in the original sample. This resulted in an increased extraction yield for all tested MMPs. For MMP-1, -7, -8, -10, -12, and -13 extraction yields of at least 98.8% were obtained, while for MMP-9 an extraction yield of at least 96.1% was reached. This technique is a powerful tool to study low abundance, active MMPs in complex biological samples.
Conclusions and future perspectives
A wide range of methods are available for detecting MMPs activity, however, most of the current approaches are limited by some disadvantages.
A fundamental problem with bioassays is that they lack specificity, i.e. they are unable to distinguish between two enzymes that degrade the same substrate, thus leading to the inaccurate measurements with elevated enzyme levels [Citation109]. Radio isotopic assay is time-consuming, difficult to automate, and generates waste disposal problems. Fluorogenic substrates have the advantage of allowing continuous monitoring of activity and are thus suitable for mechanistic studies, but their use in high throughput screening is often limited by their high cost, low stability and limited solubility in aqueous solvents [Citation110]. Western blotting requires knowledge of the MMPs native substrate and the availability of anti-MMPs antibodies. And what is more, it is expensive and time-consuming [Citation111]. Zymography is more suitable for screening of MMP inhibitors; however, zymography gives a measure of total potential enzymatic activity but does not allow determination of the net level of activity present in a sample. As compared with the above mentioned methods, the immuno capture assay seems better that can meet the sensitive, selective and rapid requirements.
The recently developed phage-displayed technique enables the high-throughput screening of large combinatorial chemical libraries in a short period feasible. Activity-based profiling assay offers a powerful tool to study low abundance, active MMPs directly in complex biological samples. Multiple-enzyme/multiple-reagent assay provides us the unique solution to the problem that substrates are never quite selective enough to directly assay activities in biological fluids. These techniques, still developing, need further validation in the future. In addition, a combinational use of the above mentioned assay methods, such as fluorescence quenching immunoassay [Citation112], zymography incorporating fluorescent substrates [Citation32,Citation34,Citation35,Citation113,Citation114], will combine the advantages and avoid the disadvantages of the single method. This is also a promising way to assay MMPs and their inhibitors in the future.
Acknowledgements
This work was supported by the Doctoral Foundation of Ministry of Education of the People's Republic of China (Grant No. 20060422029) and the Shandong Province Natural Foundation Research Grant (Grant no. 21310005).
References
- Vihinen P, Ala-aho R, Kähäri VM. Matrix metalloproteinases as therapeutic targets in cancer. Curr Cancer Drug Targets 2005; 5: 203–220
- Loftus IM, Naylor AR, Bell PR, Thompson MM. Matrix metalloproteinases and atherosclerotic plaque instability. Br J Surg 2002; 89: 680–694
- Leppert D, Lindberg RLP, Kappos L, Leib SL. Matrix metalloproteinases: Multifunctional effectors of inflammation in multiple sclerosis and bacterial meningitis. Brain Res Rev 2001; 36: 249–257
- Nagase H, Visse R, Murphy G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res 2006; 69: 562–573
- Mannello F, Tonti G, Papa S. Matrix metalloproteinase inhibitors as anticancer therapeutics. Curr Cancer Drug Targets 2005; 5: 285–298
- Lombard C, Saulnier J, Wallach J. Assays of matrix metalloproteinases (MMPs) activities: A review. Biochimie 2005; 87: 265–272
- Paemen L, Martens E, Norga K, Masure S, Roets E, Hoogmartens J, Opdenakker G. The gelatinase inhibitory activity of tetracyclines and chemically modified tetracycline analogues as measured by a novel microtiter assay for inhibitors. Biochem Pharmacol 1996; 52: 105–111
- Koritsas VM, Atkinson HJ. An assay for detecting nanogram levels of proteolytic enzymes. Anal Biochem 1995; 227: 22–26
- Ratnikov B, Deryugina E, Leng J, Marchenko G, Dembrow D, Strongin A. Determination of matrix metalloproteinase activity using biotinylated gelatin. Anal Biochem 2000; 286: 149–155
- Baragi VM, Shaw BJ, Renkiewicz RR, Kuipers PJ, Welgus HG, Mathrubutham M, Cohen JR, Rao SK. A versatile assay for gelatinases using succinylated gelatin. Matrix Biology 2000; 19: 267–273
- Ratnikov BI, Deryugina EI, Strongin AY. Gelatin zymography and substrate cleavage assays of matrix metalloproteinase-2 in breast carcinoma cells overexpressing membrane type-1 matrix metalloproteinase. Lab Invest 2002; 82(11)1583–1590
- Kleiner DE, Stetler-Stevenson WG. Quantitative zymography: detection of picogram quantities of gelatinases. Anal Biochem 1994; 218(2)325–329
- Leber TM, Balkwill FR. Zymography: A single-step staining method for quantitation of proteolytic activity on substrate gels. Anal Biochem 1997; 249: 24–28
- Yu W, Woessner JF. Heparin-enhanced zymographic detection of matrilysin and collagenases. Anal Biochem 2001; 293: 38–42
- Gerlach RF, Demacq C, Jung K, Tanus-Santos JE. Rapid separation of serum does not avoid artificially higher matrix metalloproteinase (MMP)-9 levels in serum versus plasma. Clin Biochem 2007; 40: 119–123
- Souza-Tarla CD, Uzuelli JA, Machado AA, Gerlach RF, Tanus-Santos JE. Methodological issues affecting the determination of plasma matrix metalloproteinase (MMP)-2 and MMP-9 activities. Clin Biochem 2005; 38: 410–414
- Gerlach RF, Uzuelli JA, Souza-Tarla CD, Tanus-Santos JE. Effect of anticoagulants on the determination of plasma matrix metalloproteinase (MMP)-2 and MMP-9 activities. Anal Biochem 2005; 344: 147–149
- Iijima T, Minami Y, Nakamura N, Onizuka M, Morishita Y, Inadome Y, Noguchi M. MMP-2 activation and stepwise progression of pulmonary adenocarcinoma: Analysis of MMP-2 and MMP-9 with gelatin zymography. Pathol Intern 2004; 54: 295–301
- Makowski GS, Ramsby ML. Calibrating gelatin zymograms with human gelatinase standards. Anal Biochem 1996; 236: 353–356
- Croall DE, Katherin M, Harold H. Casein zymography of calpains using a 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-imidazole buffer. Anal Biochem 2002; 304(1)129–132
- Resa PF, Mira E, Quesada AR. Enhanced detection of casein zymography of matrix metalloproteinases. Anal Biochem 1995; 224: 434–435
- Gogly B, Groult N, Hornebeck W, Godeau G, Pellat B. Collagen zymography as a sensitive and specific technique for the determination of subpicogram levels of interstitial collagenase. Anal Biochem 1998; 255: 211–216
- Troeberg L, Nagase H. Measurement of matrix metalloproteinase activities in the medium of cultured synoviocytes using zymography. Meth Mol Biol 2003; 225: 77–87
- Frederiks WM, Mook OR. Metabolic mapping of proteinase activity with emphasis on in situ zymography of gelatinases: Review and protocols. J Histochem Cytochem 2004; 52(6)711–722
- George SJ, Johnson JL. In situ zymography. Meth Mol Biol 2001; 151: 411–415
- Kurschat P, Wickenhauser C, Groth W, Krieg T, Mauch C. Identification of activated matrix metalloproteinase-2 (MMP-2) as the main gelatinolytic enzyme in malignant melanoma by in situ zymography. J Pathol 2002; 197(2)179–187
- Nemori R, Yamamoto M, Kataoka F, Hashimoto G, Arakatsu H, Shiomi T, Okada Y. Development of in situ zymography to localize active matrix metalloproteinase-7 (matrilysin-1). J Histochem Cytochem 2005; 53(10)1227–1234
- Galis ZS, Sukhova GK, Libby P. Microscopic localization of active proteases by in situ zymography: Detection of matrix metalloproteinase activity in vascular tissue. FASEB J 1995; 9(10)974–980
- Mungall BA, Pollitt CC. In situ zymography: topographical considerations. J Biochem Biophys Meth 2001; 47: 169–176
- Oliver GW, Leferson JD, Stetler-Stevenson WG, Kleiner DE. Quantitative reverse zymography: Analysis of picogram amounts of metalloproteinase inhibitors using gelatinase A and B reverse zymograms. Anal Biochem 1997; 244: 161–166
- Hawkes SP, Li H, Taniguchi GT. Zymography and reverse zymography for detecting MMPs, and TIMPs. Meth Mol Biol 2001; 151: 399–410
- Hattori S, Fujisaki H, Kiriyama T, Yokoyama T, Irie S. Real-time zymography and reverse zymography: A method for detecting activities of matrix metalloproteinases and their inhibitors using FITC labeled collagen and casein as substrates. Anal Biochem 2002; 301: 27–34
- Watanabe K, Hattori S. Real-time dual zymographic analysis of matrix metalloproteinases using fluorescein-isothiocyante-labeled gelatin and texas-red-labeled casein. Anal Biochem 2002; 307: 390–392
- Crawford BD, Pilgrim DB. Ontogeny and regulation of matrix metalloproteinase activity in the zebrafish embryo by in vitro and in vivo zymography. Dev Biol 2005; 286: 405–414
- Bremer C, Tung CH, Weissleder R. In vivo molecular target assessment of matrix metalloproteinase inhibition. Nat Med 2001; 7: 743–748
- Bawadi HA, Antunes TM, Shih F, Losso JN. In vitro inhibition of the activation of pro-matrix metalloproteinase 1 (pro-MMP-1) and pro-matrix metalloproteinase 9 (pro-MMP-9) by rice and soybean Bowman-Birk inhibitors. J Agric Food Chem 2004; 52: 4730–4736
- Verheijen JH, Nieuwenbroek NM, Beekman B, Hanemaaijer R, Verspaget HW, Ronday HK, Bakker AH. Modified proenzymes as artificial substrates for proteolytic enzymes: Colorimetric assay of bacterial collagenase and matrix metalloproteinase activity using modified pro-urokinase. Biochem J 1997; 323(3)603–609
- Hanemaaijer R, Visser H, Kontiiner Y, Koolwijk P, Verheijen JH. A novel and simple immunocapture assay for determination of getatinase-B (MMP-9) activities in biological fluids: Saliva from patients with Sjogren's syndrome contain increased latent and active gelatinase-B levels. Matrix Biology 1998; 17: 657–665
- Hanemaaijer R, Sier CF, Visser H, Scholte L, van Lent N, Toet K, Hoekman K, Verheijen JH. MMP-9 activity in urine from patients with various tumors, as measured by a novel MMP activity assay using modified urokinase as a substrate. Ann N Y Acad Sci 1999; 878: 141–149
- Capper SJ, Verheijen J, Smith L, Sully M, Visser H, Hanemaaijer R. Determination of gelatinase-A (MMP-2) activity using a novel immunocapture assay. Ann N Y Acad Sci 1999; 878: 487–490
- Yoshida D, Watanabe K, Noha M, Takahashi H, Teramoto A. Suppression of matrix metalloproteinase activity by SI-27: Detection by a new activity assay with S-2444, a specific chromogenic peptide. J Neuro Oncol 2002; 58: 1–11
- Hao JL, Nagano T, Nakamura M, Kumagai N, Mishima H, Nishida T. Effect of galardin on collagen degradation by pseudomonas aeruginosa. Exp Eye Res 1999; 69: 595–601
- Kurien BT, Scofield RH. Western blotting. Methods 2006; 38(4)283–293
- Enric E, Trini T, Manuel R, Senén V, Lluis PJ, Maria B. Use of western blotting filtration to detect UV-cross-linked protein: RNA complexes. Anal Biochem 2006; 353(1)138–140
- Obata K, Iwata K, Okada Y, Kohrin Y, Ohuchi E, Yoshida S, Shinmei M, Hayakawa T. A one-step sandwich enzyme immunoassay for human matrix metalloproteinase 3 (stromelysin-1) using monoclonal antibodies. Clin Chim Acta 1992; 211(1–2)59–72
- Zhang J, Fujimoto N, Iwata K, Sakai T, Okada Y, Hayakawa T. A one-step sandwich enzyme immunoassay for human matrix metalloproteinase 1 (interstitial collagenase) using monoclonal antibodies. Clin Chim Acta 1993; 219(1–2)1–14
- Fujimoto N, Mouri N, Iwata K, Ohuchi E, Okada Y, Hayakawa T. A one-step sandwich enzyme immunoassay for human matrix metalloproteinase 2 (72-kDa gelatinase/type IV collagenase) using monoclonal antibodies. Clin Chim Acta 1993; 221(1–2)91–103
- Fujimoto N, Zhang J, Iwata K, Shinya T, Okada Y, Hayakawa T. A one-step sandwich enzyme immunoassay for tissue inhibitor of metalloproteinases-2 using monoclonal antibodies. Clin Chim Acta 1993; 220(1)31–45
- Ohuchi E, Azumano I, Yoshida S, Iwata K, Okada Y. A one-step sandwich enzyme immunoassay for human matrix metalloproteinase 7 (matrilysin) using monoclonal antibodies. Clinica Chimica Acta 1996; 244: 181–198
- Matsuki H, Fujimoto N, Iwata K, Knauper V, Okada Y, Hayakawa T. A one-step sandwich enzyme immunoassay for human matrix metalloproteinase 8 (neutrophil collagenase) using monoclonal antibodies. Clinica Chimica Acta 1996; 244: 129–143
- Wang T, Aoki T, Iwata K, Takata T, Uchida T, KnaÈuper V, Llano E, Okada Y, Bartlett JD. One-step sandwich enzyme immunoassay using monoclonal antibodies for detection of human enamelysin (MMP-20). Eur J Oral Sci 2000; 108: 530–537
- Aoki T, Yonezawa K, Ohuchi E, Fujimoto N, Iwata K, Shimada T, Shiomi T, Okada Y, Seiki M. Two-step sandwich enzyme immunoassay using monoclonal antibodies for detection of soluble and membrane-associated human membrane type 1-matrix metalloproteinase. J Immunoassay Immunochem 2002; 23(1)49–68
- Bergmann U, Michaelis J, Oberhoff R, Knäuper V, Beckmann R, Tschesche H. Enzyme linked immunosorbent assays (ELISA) for the quantitative determination of human leukocyte collagenase and gelatinase. J Clin Chem Clin Biochem 1989; 27(6)351–359
- Yoshioka H, Oyamada I, Usuku G. An assay of collagenase activity using enzyme-linked immunosorbent assay for mammalian collagenase. Anal Biochem 1987; 166(1)172–177
- Clark IM, Powell LK, Wright JK, Cawston TE, Hazleman BL. Monoclonal antibodies against human fibroblast collagenase and the design of an enzyme-linked immunosorbent assay to measure total collagenase. Matrix 1992; 12(6)475–480
- Clark IM, Wright JK, Cawston TE, Hazleman BL. Polyclonal antibodies against human fibroblast collagenase and the design of an enzyme-linked immunosorbent assay to measure TIMP-collagenase complex. Matrix 1992; 12(2)108–115
- Clark IM, Powell LK, Ramsey S, Hazleman BL, Cawston TE. The measurement of collagenase, tissue inhibitor of metalloproteinases (TIMP), and collagenase-TIMP complex in synovial fluids from patients with osteoarthritis and rheumatoid arthritis. Arthritis Rheum 1993; 36(3)372–379
- Plumpton TA, Clark IM, Plumpton C, Calvin J, Cawston TE. Development of an enzyme-linked immunosorbent assay to measure total TIMP-1 (free TIMP-1 and TIMP-1 in combination with matrix-metalloproteinases) and measurement of TIMP 1 and CRP in serum. Clin Chim Acta 1995; 240: 137–154
- Walakovits LA, Moore VL, Bhardwaj N, Gallick GS, Lark MW. Detection of stromelysin and collagenase in synovial fluid from patients with rheumatoid arthritis and posttraumatic knee injury. Arthritis Rheum 1992; 35(1)35–42
- Doughty JR, Goldberg RL, Ganu V, Melton RA, Hu SI, Di Pasquale G. A stromelysin assay for the assessment of metalloprotease inhibitors on human aggregated proteoglycan. Agents Actions 1993; 39: 151–153
- Hollander AP, Heathfield TF, Webber C, Iwata Y, Bourne R, Rorabeck C, Poole AR. Increased damage to type II collagen in osteoarthritic articular cartilage detected by a new immunoassay. J Clin Invest 1994; 93(4)1722–1732
- Maliszewska M, Mader M, Scholl U, Azeh I, Hardeland R, Felgenhauer K, Beuche W, Weber F. Development of an ultrasensitive enzyme immunoassay for the determination of matrix metalloproteinase-9 (MMP-9) levels in normal human cerebrospinal fluid. J Neuroimmunol 2001; 116: 233–237
- Lein M, Nowak L, Jung K, Koenig F, Lichtinghagen R, Schnorr D, Loening SA. Analytical aspects regarding the measurement of metalloproteinases and their inhibitors in blood. Clin Biochem 1997; 30(6)491–496
- Jung K, Gerlach RF, Tanus-Santos JE. Preanalytical pitfalls of blood sampling to measure true circulating matrix metalloproteinase 9 and tissue inhibitors of matrix metalloproteinases. Clin Chim Acta 2006; 373: 180–181
- Chu Q, Lopez M, Hayashi K, Ionescu M, Billinghurst RC, Johnson KA, Poole AR, Markel MD. Elevation of a collagenase generated type II collagen neoepitope and proteoglycan epitopes in synovial fluid following induction of joint instability in the dog. Osteoarthritis Cartilage 2002; 10(8)662–669
- Fosang AJ, Stanton H, Little CB, Atley LM. Neoepitopes as biomarkers of cartilage catabolism. Inflamm Res 2003; 52: 277–282
- Ryzhakova OS, Solov'eva NI. The assay of tissue collagenase activity using fluorescein isothiocyanate labeled collagen. Biomed Khim 2005; 51(4)432–438
- Steven FS, Lowther DA. Insoluble collagen II. The use of fluorescein labelled polymeric collagen fibrils in a very sensitive assay procedure for enzymes degrading insoluble collagen. Connect Tissue Res 1975; 4(1)7–10
- Homer KA, Beighton D. Fluorometric determination of bacterial protease activity using fluorescein isothiocyanate-labeled proteins as substrates. Anal Biochem 1990; 191(1)133–137
- St-Pierre Y, Desrosiers M, Tremblay P, Estève PO, Opdenakker G. Flow cytometric analysis of gelatinase B (MMP-9) activity using immobilized fluorescent substrate on microspheres. Cytometry 1996; 25(4)374–380
- Kholodovich VV, Kara DI, Gershkovich AA, Kibirev VK, Karabut LV, Klimenko IV, Korneliuk AI. New donor-acceptor pairs for fluorogenic substrates with intramolecular fluorescence energy transfer for thrombin and trypsin. Bioorg Khim 1998; 24(3)179–185
- Knight CG. Fluorimetric assays of proteolytic enzymes. Meth Enzymol 1995; 248: 18–34
- Gershkovich AA, Kholodovych VV. Fluorogenic substrates for proteases based on intramolecular fluorescence energy transfer (IFETS). J Biochem Biophys Meth 1996; 33(3)135–162
- Fields GB. Using fluorogenic peptide substrates to assay matrix metalloproteinases. Methods Mol Biol 2001; 151: 495–518
- Peppard J, Pham Q, Clark A, Farley D, Sakane Y, Graves R, George J, Norey C. Development of an assay suitable for high-throughput screening to measure matrix metalloprotease activity. Assay Drug Dev Technol 2003; 1(3)425–433
- Bickett DM, Green MD, Berman J, Dezube M, Howe AS, Brown PJ, Roth JT, Mcgeehan GM. A high throughput fluorogenic substrate for interstitial collagenase (MMP-1) and gelatinase (MMP-9). Anal Biochem 1993; 212(1)58–64
- Knight CG, Willenbrock F, Murphy G. A novel coumarin-labelled peptide for sensitive continuous assays of the matrix metalloproteinases. FEBS Lett 1992; 296(3)263–266
- Netzel-Arnett S, Mallya SK, Nagase H, Birkedal-Hansen H, Wart HE. Continuously recording fluorescent assays optimized for five human matrix metalloproteinases. Anal Biochem 1991; 195(1)86–92
- Beekman B, Drijfhout JW, Bloemhoff W, Ronday HK, Tak PP, TeKoppele JM. Convenient fluorometric assay for matrix metalloproteinase activity and its application in biological media. FEBS Lett 1996; 390: 221–225
- Beekman B, El B, Drijfhout JW, Ronday HK, TeKoppele JM. Highly increased levels of active stromelysin in rheumatoid synovial fluid determined by a selective fluorogenic assay. FEBS Lett 1997; 418: 305–309
- Neumann U, Kubota H, Frei K, Ganu V, Leppert D. Characterization of Mca-Lys-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, a fluorogenic substrate with increased specificity constants for collagenases and tumor necrosis factor converting enzyme. Anal Biochem 2004; 328: 166–173
- George J, Teear ML, Norey CG, Burns DD. Evaluation of an imaging platform during the development of a FRET protease assay. J Biomol Screen 2003; 8(1)72–80
- Itoh M, Osaki M, Chiba T, Masuda K, Akizawa T, Yoshioka M, Seiki M. Flow injection analysis for measurement of activity of matrix metalloproteinase-7 (MMP-7). J Pharm Biomed Anal 1997; 15: 1417–1426
- Ambrose WP, Semin DJ, Robbins DL, Orden AV, Kashem MA, Hamilton SA, Nelson RM, Jett JH, Keller RA. Detection system for reaction-rate analysis in a low-volume proteinase-inhibition assay. Anal Biochem 1998; 263: 150–157
- Geoghegan KF, Emery MJ, Martin WH, McColl AS, Daumy GO. Site-directed double fluorescent tagging of human renin and collagenase (MMP-1) substrate peptides using the periodate oxidation of N-terminal serine. An apparently general strategy for provision of energy-transfer substrates for proteases. Bioconjugate Chem 1993; 4: 537–544
- Lauer-Fields JL, Kele P, Sui G, Nagase H, Leblanc RM, Fields GB. Analysis of matrix metalloproteinase triple-helical peptidase activity with substrates incorporating fluorogenic L- or D-amino acids. Anal Biochem 2003; 321: 105–115
- Lauer-Fields JL, Broder T, Sritharan T, Chung L, Nagase H, Fields GB. Kinetic analysis of matrix metalloproteinase activity using fluorogenic triple-helical substrates. Biochemistry 2001; 40: 5795–5803
- Lauer-Fields JL, Fields GB. Triple-helical peptide analysis of collagenolytic protease activity. Biol Chem 2002; 383(7–8)1095–1105
- Terato K, Nagai Y, Kawanishi K, Yamamoto S. A rapid assay method of collagenase activity using 14C-labeled soluble collagen as substrate. Biochim Biophys Acta 1976; 445(3)753–762
- Cawston TE, Barrett AJ. A rapid and reproducible assay for collagenase using [1-14C]acetylated collagen. Anal Biochem 1979; 99(2)340–345
- Sunada H, Nagai Y. A rapid micro-assay method for gelatinolytic activity using tritium-labeled heat-denatured polymeric collagen as a substrate and its application to the detection of enzymes involved in collagen metabolism. J Biochem(Tokyo) 1980; 87(6)1765–1771
- Gisslow MT, McBride BC. A rapid sensitive collagenase assay. Anal Biochem 1975; 68(1)70–78
- Brownell J, Earley W, Kunec E, Morgan BA, Olyslager B, Wahl RC, Houck DR. Comparison of native matrix metalloproteinases and their recombinant catalytic domains using a novel radiometric assay. Arch Biochem Biophys 1994; 314(1)120–125
- Manicourt DH, Lefebvre V. An assay for matrix metalloproteinases and other proteases acting on proteoglycans, casein, or gelatin. Anal Biochem 1993; 215(2)171–179
- Johnson-Wint B. A quantitative collagen film collagenase assay for large numbers of samples. Anal Biochem 1980; 104(1)175–181
- Dean DD, Woessner JF. A sensitive, specific assay for tissue collagenase using telopeptide-free [3H] acetylated collagen. Anal Biochem 1985; 148(1)174–181
- Matthews DJ, Wells JA. Substrate phage: Selection of protease substrates by monovalent phage display. Science 1993; 260(5111)1113–1117
- Ohkubo S, Miyadera K, Sugimoto Y, Matsuo K, Wierzba K, Yamada Y. Identification of substrate sequences for membrane type-1 matrix metalloproteinase using bacteriophage peptide display library. Biochem Biophys Res Commun 1999; 266: 308–313
- Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: A vast library of peptides for identifying ligands. Proc Nati Acad Sci USA 1990; 87: 6378–6382
- Ding L, Coombs GS, Strandberg L, Navre M, Corey DR, Madison EL. Origins of the specificity of tissue-type plasminogen activator. Proc Natl Acad Sci USA 1995; 92: 7627–7631
- Smith MM, Shi L, Navre M. Rapid identification of highly active and selective substrates for stromelysin and matrilysin using bacteriophage peptide display libraries. J Biol Chem 1995; 270(12)6440–6449
- Deng SJ, Bickett DM, Mitchell JL, Lambert MH, Blackburn RK, Carter HL, 3rd, Neugebauer J, Pahel G, Weiner MP, Moss ML. Substrate specificity of human collagenase 3 assessed using a phage-displayed peptide library. J Biol Chem 2000; 275(40)31422–31427
- Samoylova TI, Morrison NE, Globa LP, Cox NR. Peptide phage display: Opportunities for development of personalized anti-cancer strategies. Anticancer Agents Med Chem 2006; 6(1)9–17
- Matthias P. Phage display systems and their applications. Appl Microbiol Biotechnol 2006; 70(1)2–11
- Huber D, Beckwith J. Phage display extends its reach. Nat Biotechnol 2006; 24(7)793–794
- Rasmussen FH, Yeung N, Kiefer L, Murphy G, Lopez-Otin C, Vitek MP, Moss ML. Use of a multiple-enzyme/multiple-reagent assay system to quantify activity levels in samples containing mixtures of matrix metalloproteinases. Biochemistry 2004; 43(11)2987–2995
- Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. PNAS 2004; 101(27)10000–10005
- Freije JR, Klein T, Ooms JA, Franke JP, Bischoff R. Activity-based matrix metallo-protease enrichment using automated, inhibitor affinity extractions. J Proteome Res 2006; 5: 1186–1194
- Catterall JB, Cawston TE. Assays of matrix metalloproteinases (MMPs) and MMP inhibitors: Bioassays and immunoassays applicable to cell culture medium, serum, and synovial fluid. Methods Mol Biol 2003; 225: 353–364
- Quesada AR, Barbacid MM, Mira E, Fernández-Resa P, Márquez G, Aracil M. Evaluation of fluorometric and zymographic methods as activity assays for stromelysins and gelatinases. Clin Exp Metast 1997; 15: 26–32
- Zucker S, Mancuso P, DiMassimo B, Lysik RM, Conner C, Wu CL. Comparison of techniques for measurement of gelatinases/type IV collagenases: Enzyme-linked immunoassays versus substrate degradation assays. Clin Exp Metast 1994; 12(1)13–23
- Baker SN, Brauns EB, McCleskey TM, Burrell AK, Baker GA. Fluorescence quenching immunoassay performed in an ionic liquid. Chem Commun (Camb) 2006; 27: 2851–2853
- Yi CF, Gosiewska A, Burtis D, Geesin J. Incorporation of fluorescent enzyme substrates in agarose gel for in situ zymography. Anal Biochem 2001; 291(1)27–33
- Le QT, Ohashi A, Hirose S, Katunuma N. Reverse zymography using fluorogenic substrates for protease inhibitor detection. Electrophoresis 2005; 26(6)1038–1045