Abstract
The effects on mitochondrial respiration and complex I NADH oxidase activity of cubebin and derivatives were evaluated. The compounds inhibited the state 3 glutamate/malate-supported respiration of hamster liver mitochondria with IC50 values ranging from 12.16 to 83.96 μM. NADH oxidase reaction was evaluated in submitochondrial particles. The compounds also inhibited this activity, showing the same order of potency observed for effects on state 3 respiration, as well as a tendency towards a non-competitive type of inhibition (KI values ranging from 0.62 to 16.1 μM). A potential binding mode of these compounds with complex I subunit B8, assessed by docking calculations, is proposed.
Introduction
Cubebin was initially isolated from the crude hexane extract of the leaves of Zanthoxylum Naranjillo Griseb [Citation1]. It is a dibenzylbutyrolactone lignan, belonging to a group of compounds with considerable interest as antiviral [Citation2,Citation3], trypanocidal [Citation4], anti-inflammatory [Citation5,Citation6], analgesic [Citation7] and anti-cancer agents [Citation8].
Mitochondrial complex I (NADH:ubiquinone oxidoreductase) oxidizes the mitochondrial matrix NADH, reducing ubiquinone, and generating part of the proton electrochemical gradient required for ATP synthesis. Its structure, composed of more than 40 different subunits and including multiple distinct redox components like flavine mononucleotide and iron-sulfur clusters, is extremely intricate. The recently elucidated structure of complex I subunit B8 (CI-B8) has been proposed as being a subunit interaction site involved in regulating activity of the complex or its assembly, via assistance in redox processes [Citation9]. The NADH oxidase reaction, which is detectable in submitochondrial particles, is the first activity of the concerted operation of complex I in the respiratory chain [Citation10].
Inhibitors of mitochondrial complex I can alter either the kinetic parameters or the equilibrium between catalytically active and inactive forms of complex [Citation11]. The majority of complex I inhibitors appears to act at the enzyme-ubiquinone-junction sites and the only characteristic feature shared by them is the hydrophobic or amphiphilic character [Citation11–13].
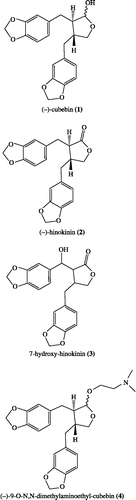
Mitochondrial complex I is involved in several human diseases, in particular neurodegenerative ones [Citation14], indicating the relevance of new inhibitors targeted at its activity. Several chemicals [Citation15], including lignans like nor-isoguaiacin and nor-dihydroguaiaretic acid [Citation16,Citation17], are established inhibitors of complex I. In the present work we have assessed the effects of the lignan cubebin and of three derivatives (see for structures) on respiration of isolated hamster liver mitochondria and NADH oxidase activity of submitochondrial particles complex I. Flexible docking calculations were performed in order to propose a potential binding mode of these compounds with the complex I subunit B8 (CI-B8).
Materials and methods
Chemicals
Ethylene glycol bis (-aminoethyl ether)-N,N,N′,N′-tetraacetic acid (EGTA), N-(2-hydroxyethyl) piperazine-N′-(2-ethanesulfonic acid) (HEPES), ADP, glutamate, malate, succinate, rotenone and NADH were obtained from Sigma Chemical (St. Louis, MO, USA). All the other reagents were of the highest commercially available grade. The stock solutions were prepared using glass-distilled deionized water.
Isolation of cubebin and preparation of its derivatives
( − )-Cubebin was isolated from Piper cubeba L. according to Bastos et al. (1996). Its chemical structure was confirmed by 1H NMR and IR data, as previously reported [Citation6]; its purity, 99%, was estimated by both HPLC and spectral data analysis. ( − )-Hinokinin, 7-hydroxy-hinokinin and ( − )-9-O-N,N-dimethylamino-ethyl-cubebin were obtained from cubebin by partial synthesis as previously described [Citation6,Citation7,Citation18].
Isolation of hamster liver mitochondria
Mitochondria were isolated by standard differential centrifugation [Citation19]. Male Hamster weighing approximately 150 g were sacrificed by cervical dislocation; livers were immediately removed, sliced in 50 mL of medium containing 250 mM sucrose, 1 mM EGTA and 10 mM HEPES-KOH, pH 7.2, and homogenized three times for 15 s at 1 min intervals with a Potter-Elvehjem homogenizer. Homogenates were centrifuged at 580 g for 5 min and the resulting supernatant was further centrifuged at 10,300 g for 10 min. Pellets were suspended in 10 mL of medium containing 250 mM sucrose, 0.3 mM EGTA and 10 mM HEPES-KOH, pH 7.2, and centrifuged at 3,400 g for 15 min. The final mitochondrial pellet was suspended in 1 mL of medium containing 250 mM sucrose and 10 mM HEPES-KOH, pH 7.2 and used within 3 h. Mitochondrial protein content was determined by the biuret reaction. The undertaken experiments comply with the current laws of the country in which they were performed.
Preparation of submitochondrial particles
For preparation of submitochondrial particles the mitochondrial pellet was frozen and stored at − 20°C. After 24 h it was thawed and suspended with the homogenization medium to 20 mg protein/mL. The mitochondrial suspension was subjected 5 times to 10 s of sonic oscillation with 30 s intervals using 80 W. The suspension was centrifuged at 9,750 g for 10 min, and the submitochondrial particles in the supernatant were isolated by centrifugation at 100,000 g during 1 h, and stored at − 70°C [Citation20].
Respiratory assay
Mitochondrial respiration was monitored polarographically with an oxygraph equipped with a Clark-type oxygen electrode (Gilson Medical Electronics, Middleton, WI, USA). Isolated hamster liver mitochondria were energized by 2.5 mM glutamate +2.5 mM malate, or by 5 mM succinate (+2.5 μM rotenone), in a standard incubation medium containing 125 mM sucrose, 65 mM KCl, and 10 mM HEPES-KOH, pH 7.4, plus 0.5 mM EGTA and 10 mM K2HPO4, at 30°C. State 3 respiration was initiated with 720 nmol ADP.
NADH oxidase activity assay
NADH oxidase activity was assayed at 30°C in submitochondrial particles, as decrease of absorbance at 340 nm employing NADH as substrate. The standard assay mixture contained 125 mM sucrose, 65 mM KCl and 10 mM HEPES-KOH, pH 7.4, plus 0.2 mM EGTA. The enzymatic reaction was started with submitochondrial particles (10 μg protein/mL). The kinetic parameters Vmax (maximum velocity), Km (Michaelis constant) and KI (inhibition constant) were calculated using the Sigraf software [Citation21].
Docking procedures
Cubebin, its derivatives and rotenone structures were built and a conformational search was performed using the MONTE CARLO method with the MMFF molecular mechanics model, as implemented in the Spartan 06 software (Spartan '06 Tutorial and User's Guide, Wavefunction, Inc., Irvine, California, 2006) [Citation22]. Structures were then fully optimized in the gas phase at the B3LYP/6-31G* level. Docking simulations were performed with GOLD 3.3 software [Citation23], which performs flexible docking using a genetic algorithm. The parameters used were originally optimized from a set of 305 complex structures with coordinates deposited in the Protein Data Bank (PDB). A population equivalent to 100, 100000 operations, 95 mutations and 95 crossovers, available in this program, was used. Docking calculations were made inside a sphere of 15 Å radius centered at the carbon gamma of the Ile53 side chain of the complex I structure, subunit B8 (PDB code 1S3A) [Citation9]. Structures were solved by NMR, and hence we have used the hydrogen atoms with their original orientations. The five orientations of highest score for each compound were selected using the GoldScore function, which allows the program to classify the orientation of molecules in a decreasing order of affinity (score, fitness) to the ligant site of the receptor.
Analysis of data
The IC50 values were estimated using the GraphPad Prism software, version 4.0 for Windows, GraphPad Software, San Diego, CA, USA.
Results
Effects of cubebin and derivatives on mitochondrial respiration
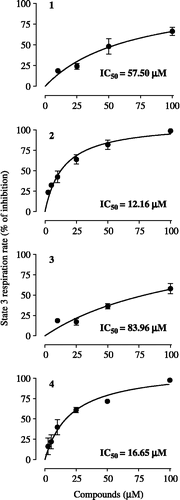
Isolated hamster liver mitochondria were energized by the respiratory chain site I substrates—glutamate/malate or by the site II substrate—succinate (basal respiration), prior to addition of ADP (state 3 respiration). The effects of cubebin and its derivatives ( − )-hinokinin, 7-hydroxy-hinokinin and ( − )-9-O-N,N-dimethylamino-ethyl-cubebin on basal and state 3 respirations were evaluated. The compounds did not stimulate basal respiration supported either by site I or site II substrates (results not shown), an indication that they do not uncouple mitochondria. However, as demonstrated by the concentration-response curves described in , cubebin and its derivatives inhibited glutamate/malate-supported state 3 respiration according to the following order of potency: ( − )-hinokinin>( − )-9-O-N,N-dimethylamino-ethyl-cubebin≫( − )-cubebin > 7-hydroxy-hinokinin (IC50 values ranging from 12.16 ± 2.19 to 83.96 ± 16.67 μM). Rotenone (1 μM) fully inhibited respiration (result not shown). Tested at 50 μM, only ( − )-9-O-N,N-dimethylamino-ethyl-cubebin also inhibited (by 58,46%) the succinate-supported state 3 respiration.
Figure 2. Concentration-response curves for the effects of cubebin and derivatives on state 3 respiration rate of isolated Hamster liver mitochondria (1.5 mg protein), incubated at 30°C with 2.5 mM glutamate + 2.5 mM malate, in a standard medium containing 125 mM sucrose, 65 mM KCl and 10 mM HEPES-KOH, pH 7.4, in presence of 0.5 mM EGTA and 10 mM K2HPO4, in a final volume of 1.8 mL. State 3 respiration was initiated with 720 nmol ADP. Data are mean ± SD of three independent experiments. Percent of inhibition in relation to control: 57.84 ± 14.69 ng atoms O/min/mg protein. Respiratory control ratio: 4.40 ± 1.13; ADP/O: 1.41 ± 0.08.
Effects of cubebin and derivatives on complex I NADH oxidase activity
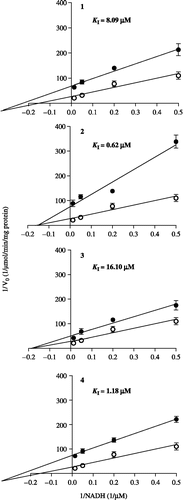
The effects of cubebin and its derivatives on complex I NADH oxidase activity were evaluated in submitochondrial particles employing 100 μM NADH as substrate, in absence of exogenously added electron acceptor. The inhibition of this activity by rotenone (1 μM, result not shown), indicates that it indeed is associated with complex I. Initial velocities (V0) were calculated from the slopes of the lines representing NADH consumption by the complex at the beginning of the activity curves (not shown). The progress of the concentration-response curves as well as their respective IC50 values are shown in . All compounds inhibited NADH oxidase activity, showing the same potency sequence observed for their inhibition of mitochondrial respiration, namely ( − )-hinokinin>( − )-9-O-N,N-dimethylamino-ethyl-cubebin≫( − )-cubebin > 7-hydroxy-hinokinin (IC50 values ranging from 1.19 ± 0.82 to 15.35 ± 4.89 μM). The type of inhibition of cubebin and derivatives were determined at concentrations corresponding to their IC50 values, and KI values were determined from double-reciprocal plots (). They presented a tendency towards a non-competitive type of inhibition in relation to the interaction of NADH with the complex. Their KI values (ranging from 0.62 to 16.1 μM) showed the same order of inhibitory potency on both mitochondrial state 3 respiration and NADH oxidase activity (see and ).
Figure 3. Concentration-response curves for the effects of cubebin and derivatives on submitochondrial particles (10 μg protein) NADH oxidase activity employing 100 μM NADH as substrate, at 30°C, in the standard medium described in legend of Figure 2, in presence of 0.2 mM EGTA (1 mL final volume). Data are mean ± SD of three independent experiments. Percent of inhibition in relation to control: V0 = 0.00637 ± 0.002 μmol/min/mg protein.
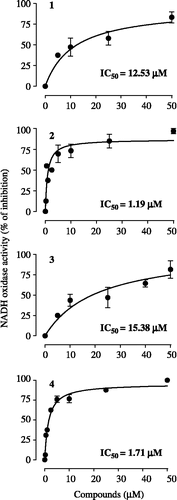
Figure 4. Double-reciprocal plots for the inhibition of submitochondrial particles NADH oxidase activity by cubebin and derivatives, assayed as described in legend of . Data are mean ± SD of three independent experiments. (○) absence of cubebin/derivatives; (•) presence of cubebin/derivatives in concentrations corresponding to the respective IC50 values, given on . Km and Vmax for control: 27.65 μM and 0.00733 μmol/min/mg protein, respectively. KI: inhibition constant.
Docking simulation of cubebin, derivatives and rotenone with CI-B8
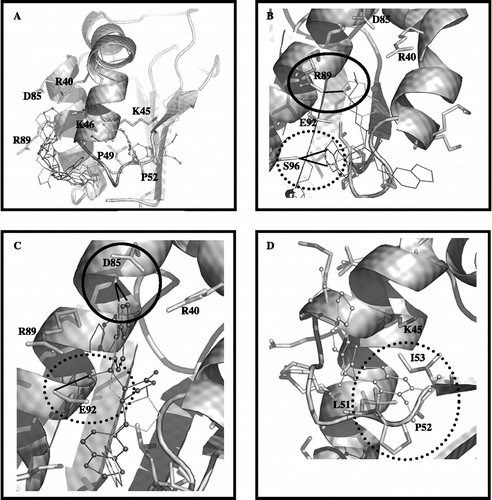
Docking simulations of cubebin, its derivatives and rotenone with complex I subunit B8 (CI-B8) are shown in . The compounds showed different orientations with regard to CI-B8 binding mode. GOLD has been parameterized to yield the 5 best docking calculations for each molecule, i.e. 5 top-ranked solutions. In line with the observed tendency for a non-competitive type of inhibition, the compounds interact with CI-B8 at the side that does not possess the highly conserved residues (–D). The software GOLD was able to discriminate among the most and less active inhibitors: the most active, 2 and 4, showed potential interaction with region of residues R89 and S96 () whereas the less active, 1 and 3, showed potential interaction with residue D85 (). The classical complex I inhibitor rotenone was docked for comparison purpose (). Although showing contact with the two conserved residues, i.e., I53 and P52, it potentially interacts with CI-B8 at the same side as the cubebin/derivatives.
Figure 5. In (A), the top-ranked solution for flexible docking between the compounds/rotenone with CI-B8. The residues having the strongest interaction with the inhibitors, are shown. In (B), the superposition between the top-ranked solutions for the most active compounds 2. (in light grey) and 4 (in dark grey). The circle demonstrates the possible hydrogen bond between residue R89 and oxygen of benzodioxol ring of compound 2; the dotted line circle demonstrates hydrogen bond donate by the S96 side chain to carboxyl oxygen of compound 2 or 4. In (C), the superposition between the top-ranked solutions for the less active compounds 1 (in white) and 3 (in black ball line). The circle demonstrates the possible contact between residue D85 and benzodioxol ring of compounds 1 and 3; the dotted line circle demonstrates possible hydrogen bond between residue E92 and hydroxyl group at C7′ of compound 3. In (D), the rotenone orientation; the dotted line circle demonstrates the possible interaction of rotenone with two conserved residues (P52 and I53).
Discussion
Previous studies demonstrated that cubebin and its three presently evaluated derivatives have prominent activity against Trypanosoma cruzi [Citation4,Citation24] and some activity against LLC-MK2 cell line (unpublished results). The IC50 values against Trypanosoma cruzi found for ( − )-hinokinin(2), ( − )-9-O-N,N-dimethylamino-ethyl-cubebin(4), ( − )-cubebin(1) and 7-hydroxy-hinokinin(3) were respectively of 0.67, 4.7, 100 and 72 μM, and of respectively 199, 393, 1510 and 1169 μM against LLC-MK2 cells. Because mitochondria have often been implicated in cell death, we have here addressed their potential involvement in these activities of cubebin and derivatives on organelles isolated from hamster livers, a classical model for in vitro studies aimed at the evaluation of the effects of chemicals on mitochondria. Cubebin and derivatives were typical inhibitors of the mitochondrial complex I, as demonstrated by the inhibitory effect on glutamate/malate-suported state 3 respiration of mitochondria and NADH oxidase activity of submitochondrial particles. The similarity of the relationship between the degree of potency for this mitochondrial effect and for those previously observed on Trypanosoma cruzi, leads us to believe that the later can be ascribed to the inhibition of the mitochondrial complex I. It is worth noting in this connection that other natural and synthetic products of important biological activity have been characterized as inhibitors of mitochondrial complex I [Citation12,Citation25].
Cubebin alone was a relatively poor inhibitor of mitochondrial state 3 respiration and of NADH oxidase activity. However, introduction of a carbonyl group in place of its hydroxyl group at C-9 giving ( − )-hinokinin, strongly increased its inhibitory effect on those mitochondrial activities. The additional introduction of a hydroxyl group at C7 of ( − )-hinokinin, resulting in 7-hydroxy-hinokinin, restored this compound's relative low potency to the level of cubebin. Introduction of a aminic group at C-9 in place of cubebin hydroxyl group (( − )-9-O-N,N-dimethylamino-ethyl-cubebin), enhanced inhibitory effect on state 3 respiration and NADH oxidase activity. Collectively, these results indicate that hydroxyl groups are unfavorable for the inhibition of complex I, suggesting at first instance, in agreement with previous report [Citation11], that the hydrophobic or amphyphilic character of inhibitors is important feature for their effects on complex I.
CI-B8 is among the most conserved complex I proteins in mammals (94%) [Citation26], and therefore is likely to play an important role in the activity of complex. Brockmann et al. (2004) proposed that the surface patch formed by the PILIRE motif, together with the strongly conserved charged residues, is one of the protein-protein interaction sites of CI-B8. The conservation degree of residues on the surface of CI-B8 of human complex I indicates that all highly conserved ones cluster in a specific domain. One face of enzyme contains almost all conserved residues, while the degree of conservation on the other side of molecule is too lower. Our theoretical results show that cubebin and derivatives may interact with residues others than those conserved ones on CI-B8. Rotenone, a classical non-competitive complex I inhibitor [Citation27], although making contact with two conserved residues in the inferior region of CI-B8, it also may interact on the other side of the subunit.
A reasonable relationship can be observed between orientation of cubebin and derivatives on CI-B8 and their inhibition of state 3 respiration and NADH oxidase activity. The most active inhibitors, compounds 2 and 4, show preferential potential interaction with the region of residues R89 and S96; in particular the most active, compound 2, can make hydrogen bond (at distance of 3.1 Å) with the residue R89 via benzodioxol oxygen atoms (). In contrast, the less active inhibitors, compounds 1 and 3, show preferential potential interaction with the residue D85 where the inter-oxygen carbon of benzodioxol ring makes contact with the carboxyl group D85 (). Therefore, the theoretical results indicate that the ring interaction with D85 could decrease the inhibitory activity of enzyme, while its interaction with the region R89 and S96 could improve the inhibitory activity. The orientation of compound 3 to region D85 may be related to the presence of a hydroxyl group at position C7′, that can favour it to donate a hydrogen bond to E92, inducing 3 to orient the benzodioxol ring in order to establish contact with D85.
These results present lignans in general as inhibitors of NADH oxidase activity of mitochondrial complex I, a finding of relevance due to the potential of these type of compounds in a pharmacological context [Citation2–8]. They indicate, in addition, potential binding groups in CI-B8, specifically R89 and S96, whose interaction via hydrogen bond may oppose NADH binding to its interacting site and/or avoid its oxidation.
Acknowledgements
We would like to thank FAPESP for the financial support.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- JK Bastos, OR Gottlieb, JS Sarti, and DS Filho. (1996). Isolation of lignans and sesquiterpenoids from leaves of Zanthoxylum naranjillo. Nat Prod Lett 9:65–70.
- JL Charlton. (1998). Antiviral activity of lignans. J Nat Prod 61:1447–1451.
- AL Piccinelli, N Mahmood, G Mora, L Poveda, F De Simone, and L Rastrelli. (2005). Anti-HIV activity of dibenzylbutyrolactone-type lignans from Phenax species endemic in Costa Rica. J Pharm Pharmacol 57:1109–1115.
- VA de Souza, R da Silva, AC Pereira, VA Royo, J Saraiva, M Montanheiro, GH de Souza, AA da Silva Filho, MD Grando, PM Donate, JK Bastos, S Albuquerque, and ML Silva. (2005). Trypanocidal activity of ( − )-cubebin derivatives against free amastigote forms of Trypanosoma cruzi. Bioorg Med Chem Lett 15:303–307.
- JK Bastos, S Albuquerque, and ML Silva. (1999). Evaluation of the trypanocidal activity of lignans isolated from the leaves of Zanthoxylum naranjillo. Planta Medica 65:541–544.
- GH Souza, AA da Silva Filho, VA de Souza, AC Pereira, VA Royo, ML Silva, R da Silva, PM Donate, JC Carvalho, and JK Bastos. (2004). Analgesic and anti-inflammatory activities evaluation of ( − )-O-acetyl, ( − )-O-methyl, ( − )-O-dimethylethylamine cubebin and their preparation from ( − )-cubebin. Fármaco 59:55–61.
- R da Silva, GH de Souza, AA da Silva, VA de Souza, AC Pereira, VA Royo, ML Silva, PM Donate, AL de Matos Araujo, JC Carvalho, and JK Bastos. (2005). Synthesis and biological activity evaluation of lignan lactones derived from ( − )-cubebin. Bioorg Med Chem Lett 15:1033–1037.
- L Li, Q Lu, Y Shen, and X Hu. (2006). Schisandrin B enhances doxorubicin-induced apoptosis of cancer cells but not normal cells. Biochem Pharmacol 71:584–595.
- C Brockmann, A Diehl, K Rehbein, H Strauss, P Schmieder, B Korn, R Kuhne, and H Oschkinat. (2004). The oxidized subunit B8 from human complex I adopts a thioredoxin fold. Structure 12:1645–1654.
- AD Vinogradov. (1998). Catalytic properties of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) and the pseudo-reversible active/inactive enzyme transition. Biochim Biophys Acta 1364:169–185.
- MV Loskovich, VG Grivennikova, G Cecchini, and AD Vinogradov. (2005). Inhibitory effect of palmitate on the mitochondrial NADH:ubiquinone oxidoreductase (complex I) as related to the active-de-active enzyme transition. Biochem J 387:677–683.
- M Degli Esposti. (1998). Inhibitors of NADH-ubiquinone reductase: An overview. Biochim Biophys Acta 1364:222–235.
- JG Okun, P Lummen, and U Brandt. (1999). Three classes of inhibitors share a common binding domain in mitochondrial complex I (NADH:ubiquinone oxidoreductase). J Biol Chem 274:2625–2630.
- CW Shults. (2004). Mitochondrial dysfunction and possible treatments in Parkinson's disease. Mitochondrion 4:641–648.
- H Ueno, H Miyoshi, M Inoue, Y Niidome, and H Iwamura. (1996). Structural factors of rotenone required for inhibition of various NADH-ubiquinone oxidoreductases. Biochim Biophys Acta 1276:195–202.
- RS Pardini, CH Kim, R Biagini, RJ Morris, and DC Fletcher. (1973). Inhibition of mitochondrial electron-transport systems by nor-isoguaiacin. Biochem Pharmacol 22:1921–1925.
- M Pavani, E Fones, D Oksenberg, M Garcia, C Hernandez, G Cordano, S Munoz, J Mancilla, A Guerrero, and J Ferreira. (1994). Inhibition of tumoral cell respiration and growth by nordihydroguaiaretic acid. Biochem Pharmacol 48:1935–1942.
- VA Royo, FF Santos, VA Souza, AC Pereira, R da Silva, AHC Vinhólis, PM Donate, ML Silva, S Albuquerque, and JK Bastos. (2003). Biological activity evaluation of dibenzilbutirolactones lignans derivatives against Leishmania braziliensis. Rev Bras Farmacogn 14:18–21.
- PL Pedersen, JW Greenawalt, B Reynafarje, J Hullihen, GL Decker, JW Soper, and E Bustamente. (1978). Preparation and characterization of mitochondria and submitochondrial particles of rat liver and liver-derived tissues. Methods Cell Biol 20:411–481.
- SA Uyemura, and C Curti. (1992). Steady-state kinetic properties of FoF1-ATPase: The pH effect. Int J Biochem 24:1743–1748.
- FA Leone, JA Baranauskas, RPM Furriel, and IA Borin. (2005). SigrafW: An easy-to-use program for fitting enzyme kinetic data. Biochem Mol Biol Educ 33:399–403.
- Spartan '06 full. 2006., Tutorial and User's Guide, Wavefunction, Inc., USA.
- ML Verdonk, JC Cole, MJ Hartshorn, CW Murray, and RD Taylor. (2003). Improved protein-ligand docking using GOLD. Proteins 52:609–623.
- J Saraiva, C Vega, M Rolon, R da Silva, ML Silva, PM Donate, JK Bastos, A Gomez-Barrio, and S de Albuquerque. (2007). In vitro and in vivo activity of lignan lactones derivatives against Trypanosoma cruzi. Parasitol Res 100:791–795.
- A Cantin, MP Lopez-Gresa, MC Gonzalez, P Moya, MA Miranda, J Primo, V Romero, E Peris, and E Estornell. (2005). Novel inhibitors of the mitochondrial respiratory chain: Oximes and pyrrolines isolated from Penicillium brevicompactum and synthetic analogues. J Agric Food Chem 53:8296–8301.
- J Hirst, J Carroll, IM Fearnley, RJ Shannon, and JE Walker. (2003). The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim Biophys Acta 1604:135–150.
- E Estornell. (2000). Mitochondrial complex I: New insights from inhibitor assays. Protoplasma 213:11–17.