Abstract
Subtype F wild type HIV protease has been kinetically characterized using six commercial inhibitors (amprenavir, indinavir, lopinavir, nelfinavir, ritonavir and saquinavir) commonly used for HIV/AIDS treatment, as well as inhibitor TL-3 and acetyl-pepstatin. We also obtained kinetic parameters for two multi-resistant proteases (one of subtype B and one of subtype F) harboring primary and secondary mutations selected by intensive treatment with ritonavir/nelfinavir. This newly obtained biochemical data shows that all six studied commercially available protease inhibitors are significantly less effective against subtype F HIV proteases than against HIV proteases of subtype B, as judged by increased Ki and biochemical fitness (vitality) values. Comparison with previously reported kinetic values for subtype A and C HIV proteases show that subtype F wild type proteases are significantly less susceptible to inhibition. These results demonstrate that the accumulation of natural polymorphisms in subtype F proteases yields catalytically more active enzymes with a large degree of cross-resistance, which thus results in strong virus viability.
Introduction
Almost all studies on HIV-1 drug susceptibility have been performed in developed countries, where subtype B [Citation1] dominates the epidemic, even though it is not a predominant HIV subtype worldwide. Given that, it is demanding that the effectiveness of existing anti-HIV drugs against non-B HIV-1 subtypes to be examined. For example, Latin American epidemic is characterized by multiple HIV-1 subtypes, primarily subtype B and subtype F [Citation2]. Specifically, there are at least half-a-million people infected by subtype F HIV worldwide, out of which over a 100 thousands men and women in Latin America alone (http://www.hiv.lanl.gov/content/hiv-db/mainpage.html). HIV-1 protease (HIV-PR) is a key enzyme in viral propagation [Citation3] and its X-ray structure [Citation4,Citation5,Citation6] was crucial for anti-AIDS drug design.
The present work investigates the enzymatic behavior of one subtype F wild-type HIV-PR (Fwt) and its comparison with a wild-type B HIV-PR (Bwt) using six commercial inhibitors (amprenavir, indinavir, lopinavir, nelfinavir, ritonavir and saquinavir) commonly used for HIV/AIDS treatment. Differences between protease sequences of non-B and B subtypes of untreated persons are defined as subtype-specific polymorphism [Citation7]. It is known that HIV-1 protease occurs in various mutant forms and that protease-targeted drug therapy leads to the selection of specific mutations in HIV-1 protease isolated from patients [Citation8]. Some of the Fwt polymorphic mutations () occur at sites that are accessory (non-active site) mutations selected by drug therapy in subtype B isolates.
Figure 1. Studied HIV-PRs and their mutations. (a) Amino acid sequence alignment of the four HIV-PR. The mutations, either polymorphic or arisen from treatment, are marked [Citation9]: primary in black and secondary in grey. (b) Crystallographic structures of the proteases Fwt (PDB code 3P3C), Bmut (PDB code 2P3A) and Fmut (PDB code 2P3D) [Citation29]. Primary mutations are represented by black spheres, secondary by grey spheres, and other differences in white spheres.
![Figure 1. Studied HIV-PRs and their mutations. (a) Amino acid sequence alignment of the four HIV-PR. The mutations, either polymorphic or arisen from treatment, are marked [Citation9]: primary in black and secondary in grey. (b) Crystallographic structures of the proteases Fwt (PDB code 3P3C), Bmut (PDB code 2P3A) and Fmut (PDB code 2P3D) [Citation29]. Primary mutations are represented by black spheres, secondary by grey spheres, and other differences in white spheres.](/cms/asset/b08bee60-110e-4bca-9b14-7c01a6c56b7e/ienz_a_332341_f0001_b.gif)
We have also investigated the enzymatic features of two multi-resistant mutant HIV proteases obtained from patients receiving ritonavir/nelfinavir as PIs: one subtype B HIV-PR (Bmut) and one subtype F HIV-PR (Fmut). Both enzymes present primary and secondary mutation [Citation9] associated with PI administration (a). The mutations can directly change the shape or character of the binding cavity, but also might indirectly influence inhibitor binding via long-range structural perturbations of the active site, or by changing the efficiency of catalysis and the stability of the enzyme Citation10,Citation11,Citation12. Some non-active site mutations alone can increase catalytic efficiency and partially compensate for the reduction of catalytic efficiency caused by active site mutations [Citation13]. Another study revealed that some non-active site mutations might also contribute significantly to destabilization of inhibitor binding [Citation14].
Even though drug resistance mutations in the protease have been well studied, few data are currently available on the influence of polymorphic mutations on drug susceptibility. Preliminary data have indicated that the existence of polymorphic mutations can lead to early development of drug resistance in patients infected with non-B HIV subtypes [Citation15]. However, only a limited number of biochemical data is available for non-B HIV proteases (see, for example, [Citation16]), and none of these include subtype F HIV PR.
The aim of the present work is to access the biochemical differences between subtype F and B, and to gain insight into the role of the polymorphic mutations in the development of drug resistance. For that we have determined catalytic parameters and inhibition constants for six inhibitors in clinical use (amprenavir, indinavir, lopinavir, nelfinavir, ritonavir and saquinavir) as well as two more universal HIV PR inhibitors, acetyl-pepstatin and TL-3 [Citation17], (). Our results reveal that the Fwt protease is naturally more resistant to all the inhibitors assayed and has much higher vitality than the Bwt protease. In this sense, Fwt can be compared to Bmut, a multi-drug resistant protease. Furthermore, Fmut presents the highest Ki values of all four proteases, with all eight inhibitors analyzed, even though it carries the same primary mutations seen in Bmut. Throughout the analysis, an effort was made to establish a correlation between the biochemical results and the available structural data.
Materials and methods
Recombinant production of HIV proteases
All of the proteases used in these studies carry the mutation Q7K, which eliminates the most significant autocatalytic site and enhances the stability of the preparations for biochemical and biophysical studies without any effect on its enzymatic activity [Citation18]. The protease coding sequence from subtypes B mutant and both subtype F were obtained from HIV-1 vertically infected seropositive Brazilian children [Citation19]. The protocol for the recombinant production of Brazilian HIV proteases had been previously reported [Citation20].
HIV proteases activity assay
Enzymatic activity of HIV PR was studied using the quenched fluorogenic peptide substrate Arg-Glu(EDANS)-Ser-Gln-Asn-Tyr-Pro-Ile-Val-Gln-Lys(DABCYL)-Arg. Calculation of the reaction rates (nM × min− 1) from the progress curves was based on calibration with EDANS in the presence of substrate (10 μM). The assays were performed at 37°C in 100 mM sodium acetate, pH 4.7, 1 mM EDTA, 1 mM DTT, 1.0 M NaCl, 10% v/v DMSO (similar to the buffer used in [Citation21]) and 10 μM of substrate. The fluorescence was monitored in semi-micro cuvettes containing a volume of 500 μL by using a PC1 Photon Counting Spectrofluorometer (ISS) with excitation and emission wavelengths of 340 nm and 490 nm, respectively.
Measurement of proteases inhibition by commercial PIs
The inhibition experiments were performed with the following six therapeutically used protease inhibitors (PIs): amprenavir (APV), indinavir (IDV), lopinavir (LPV), nelfinavir (NFV), ritonavir (RTV) and saquinavir (SQV), and with the universal inhibitors acetyl-pepstatin (PPT) and TL-3 (). The initial reaction rate of HIV PR was adjusted to 10–20 nM × min− 1 by addition of the proper amount of enzyme solution. Different concentrations of PI were added and the reaction was followed for 15–30 min afterward. Ten initial concentrations of each of the six commercially available PIs (typically 0.0625, 0.125, 0.25, 0.5, 1, 2, 4, 8, 16 and 32 nM) were measured in duplicate. For precise Ki determination the PI concentration was adjusted for the sample been measured.
Evaluation of inhibition data
The ratio between inhibited (Vi) and uninhibited (V0) initial rates was determined using the general equation for competitive tight-binding inhibitors [Citation22,Citation23]:
where [Et] is the total concentration of the enzyme (free and bound); [It] is the total concentration of the inhibitor (free and bound); Ki,app is the apparent dissociation constant of the enzyme-inhibitor complex at a given substrate concentration. Typical inhibition curves are given in . Since under our experimental conditions [S] < KM, thus Ki,app and Ki are similar and the competition of the inhibitor with the substrate is negligible. From kcat the active PR concentrations in the corresponding equilibrium inhibition experiments were calculated and were found to be in the same range (0.1-0.2 nM) as the inhibition constants for wild-type PR, indicating the prevalence of tight-binding conditions. The Michaelis-Menten constant (KM) of WT PR was determined by measuring the substrate turnover at different substrate concentrations and fitting the data to the Michaelis-Menten equation. The poor solubility of substrate prevented the use of concentrations higher than 60 μM.
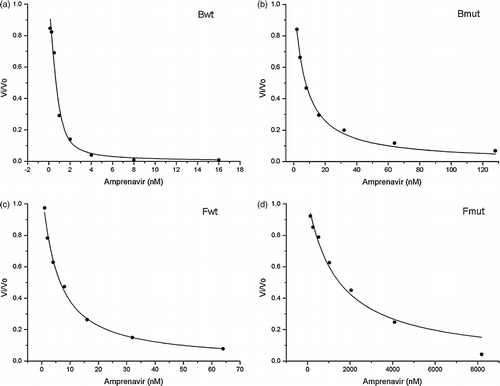
Figure 3. Inhibition curves of (a) Bwt, (b) Bmut, (c) Fwt and (d) Fmut HIV PRs by amprenavir. V/V0 values have been obtained as described in Materials and Methods. Solid curve represents the fit of Equation (1) to the experimental data. The differences in the concentration ranges of amprenavir used for inhibition of subtype B and subtype F HIV PRs and their drug-resistant mutants should be noted.
Results and discussion
Wild type F subtype protease (Fwt)
The measurements of catalytic efficiencies with subtype Bwt were performed as a reference and are in good agreement with the published data [Citation21,Citation24,Citation25]. The Fwt catalytic efficiency (kcat/KM), when compared to Bwt, demonstrated an increase of about 3 times, mainly caused by a 5-fold decrease in KM, besides a slightly decrease of 1.7-fold in kcat (). Moreover, there was an increase of at least an order of magnitude in Ki () for all the tested PIs (), with a highest increase of 301-fold for nelfinavir.
Table I. Catalytic efficiency for all four studied HIV-1 proteases.
Table II. Inhibition constants (Ki) for studied HIV-1 proteases.
These results reveal that the subtype-specific amino acid polymorphisms for Fwt are able to significantly affect both the KM and the Ki values, even though none of those mutations are located in the binding site or in the flaps (b). In all cases, an increase in the Ki values, accompanied by decrease in the KM values, is observed ( & ). This opposite effect on Ki and KM has been previously described for inhibitor resistant mutants of the B subtype [Citation11,Citation26,Citation27]. Notably, this effect occurs even for mutations that do not change the chemical nature or polarity of the binding site but are known to change its geometry [Citation11].
The Fwt carries twelve naturally occurring polymorphic mutations (). The increased values observed for the Fwt Ki are characteristic of subtype B proteases harboring primary mutations. It is known that the non-B subtypes HIV PR do not usually develop the primary mutation L90M, while the L89M substitution is very frequent among the non-B subtypes isolates [Citation28,Citation18]. In addition, the double mutation L89M/L90M is very rare in all subtypes of HIV PRs. Our previous crystallographic studies of Fwt, Bwt and Bmut proteases [Citation29] revealed that the L89M imposes the same structural effects observed for the L90M mutation. The later is associated with resistance against all therapeutically used PIs, explaining its pronounced effect in Ki.
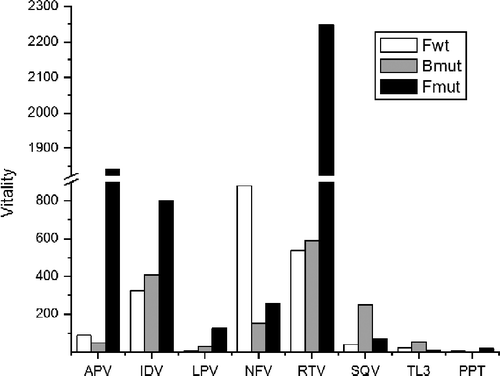
We also calculated the biochemical fitness, or vitality [Citation30], for Fwt, Bmut and Fmut using Bwt as comparison (). Fwt vitality is particularly high for nelfinavir (), an inhibitor for which L90M mutation is considered primary. The mutation to a bulkier side chain displaces this residue (at position 89 or 90) toward the active site loop, which slightly compresses the substrate/inhibitor binding cavity, leading to a reduction in both volume and flexibility of the active site. At the thermodynamic level, the origin of the differential effect appears to be related to the difficulties of conformationally constrained inhibitors to adapt to distortions in the geometry of the binding site [Citation31,Citation32]. Another polymorphic mutation, M36I, is commonly seen in response to nelfinavir and ritonavir therapy and is considered a secondary mutation for subtype B proteases [Citation33]. Structurally, this mutation causes a hardening of the flap hinges [Citation29], which could be the cause of the slightly lower value of kcat (). Most of the other polymorphic mutations do not cause detectable structural discrepancies and are away from the active site. Their effect on the Ki values, if any, must reside at the dynamic level. The polymorphisms directly reflect in the vitality values () obtained for Fwt protease. The highest vitalities were observed as a response to nelfinavir, ritonavir and indinavir, respectively. Moreover, Fwt enzyme is about 3 times more catalytically efficient than Bwt. These results demonstrate that the accumulation of PIs secondary resistant mutations in Fwt, due to natural polymorphism, yields a catalytically more active protease with a large degree of cross-resistance, which thus results in strong virus viability.
Table III. Vitality values for subtype B and F HIV proteases.
Mutant B subtype protease (Bmut)
The B subtype mutant protease, Bmut, carries eight mutations with respect to Bwt (b). Its catalytic efficiency is very close to that of Bwt due to a 3-fold simultaneous decrease in both kcat and KM, indicative of a highly viable virus. The primary mutations V82A and L90M, most probably selected by the long term administration of ritonavir and nelfinavir to the patient, are commonly observed in response to all therapeutically used PIs [Citation8]. The mutation V82A reshapes the S1/S1′ binding pocket, causing a collapse of the crevice formed between the flap and the loop containing the residue Pro81. This reshaping forces the inhibitor to change its conformation, especially the ones with phenyl groups in both P1 and P1′ [Citation11,Citation29], leading to a loss of interactions with the binding pocket. This effect is particularly pronounced with TL-3 (), with an increase of two orders of magnitude in Ki when compared to Bwt, and one order of magnitude to Fwt (). It has been proposed that a bulkier group in P1 could lead to an inhibitor less susceptible to V82A mutation [Citation11]. That is verified for nelfinavir () which contains a bicyclical group in that position and shows an increase in Ki which is about 3 times lower than the increase observed for Fwt, which does not contain V82A. The contrary effect is seen with saquinavir, which contains a small amide group in P1 (), which reflects in pronounced differences between the vitalities of Bmut and Fwt (). The secondary mutation I54V was probably selected due to ritonavir administration [Citation33]. Since no significant structural differences caused by this mutation could be detected [Citation29], its presumably affects inhibitor binding dynamically by altering the flexibility of the flaps [Citation34]. Additional secondary mutations are L63P and A71V, associated with resistance to therapy with indinavir, nelfinavir, lopinavir and ritonavir.
Mutant F subtype protease (Fmut)
Out of the clinically identified enzymes studied here, the Fmut is the one that carries the largest number of mutations, both polymorphic and selected by treatment (b). It showed a decrease of about 5 times in catalytic efficiency because of a 12-fold decrease in kcat and a 2.5-fold decrease in KM (). Among the mutations encountered in Fmut, V82A is the primary mutation generated by treatment, while L90M does not arise due to the presence of the polymorphic L89M. The structural effects of V82A/L89M mutations in the structure of Fmut are the same observed for Bmut [Citation29]. It also contains the important secondary mutation M46I, arising in response to almost all approved inhibitors, except for saquinavir [Citation33]. Additionally, I54V is commonly detected in response to amprenavir, indinavir, lopinavir, nelfinavir and saquinavir therapy, L10I to indinavir, lopinavir and nelfinavir and K20R to lopinavir and nelfinavir [Citation8]. The accumulation of these mutations yields a highly cross-resistant enzyme, which has the highest decrease in susceptibility to all the tested inhibitors (), as indicated by the high values of its inhibition constants and vitality ( and ). On the other hand, Fmut displays the lowest catalytic efficiency against the substrate used in the experimental procedures. This loss of efficiency, which still renders a viable virus, could be explained by a concomitant mutation in the natural substrate of the protease, reestablishing the activity of the mutant protease.
Implications To Hiv/aids Treatment
Our experimental results reveal that all the six commercially available PIs evaluated are significantly less effective against subtype F HIV-PR than against HIV-PR of subtype B. Furthermore, Fwt is also much less susceptible to PI inhibition as compared to available data for subtypes A and C HIV-PRs [Citation24]. A long term study may be necessary to evaluate the susceptibility of each variant to the inhibitors studied. Nevertheless, our biochemical data can serve as an indication for the treatment of individuals infected with subtype F protease. To this end, nelfinavir presents the worst vitality for Fwt and should be avoided in the treatment. In the other hand, lopinavir, followed by saquinavir, sustain only minor effects from the polymorphisms present in Fwt, and the mutations of Fmut, and should probably be elected as the first choice therapies for subtype F infected individuals.
Acknowledgements
We thank to FAPESP for grants 99/03387-4, 04/11890-8, 04/12201-1 and 06/00182-8 and to CNPq. Authors thank Dr. Jordan J. Tang (Oklahoma Medical Research Foundation, USA) for a kind gift of a plasmid of artificial HIV PR subtype B gene and Dr. Amilcar Tanuri (UFRJ, Brasil) for cDNAs of HIV Bmut, Fwt and Fmut.
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
References
- C Kuiken, B Foley, E Freed, B Hahn, P Marx, F McCutchan, L Mellors, B Korber. HIV sequence compendium 2002, Theoretical Biology and Biophysics Group Los Alamos National Laboratory, LA-UR n 03-3564; 2002.
- S Osmanov, C Pattou, N Walker, B Schwardlander, J Esparza. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J Acquir Immune Defic Syndr 2002; 29:184–190.
- AD Frankel, JA Young. HIV-1: Fifteen proteins and a RNA. Annu Rev Biochem 1998; 67:1–25.
- A Wlodawer, M Miller, MK Jaskolski, B Sathyanarayana, ET Baldwin, IM Weber, L Selk, L Clawson, J Schneider, SB Kent. Conserved folding in retroviral proteases: Crystal structure of a synthetic HIV-1 protease. Science 1989; 245:616–621.
- M Miller, J Schneider, BK Sathyanarayana, MV Toth, GR Marshall, L Clawson, L Selk, SB Kent, A Wlodawer. Structure of complex of synthetic HIV-1 protease with a substrate based inhibitor at 2.3-Å resolution. Science 1989; 246:1149–1152.
- R Lapatto, T Blundell, A Hemmings, J Overington, A Wilderspin, S Wood, JR Merson, PJ Whittle, DE Danley, KF Geoghegan, SJ Hawrylik, SE Lee, KG Scheld, PM Hobart. X-ray analysis of HIV-1 proteinase at 2.7 Å resolution confirms structural homology among retroviral enzymes. Nature 1989; 342:299–302.
- R Kantor, DA Katzenstein, B Efron, AP Carvalho, B Wynhoven, P Cane, J Clarke, S Sirivichayakul, MA Soares, J Snoeck, C Pillay, H Rudich, R Rodrigues, A Holguin, K Ariyoshi, MB Bouzas, P Cahn, W Sugiura, V Soriano, LF Brigido, Z Grossman, L Morris, A-M Vandamme, A Tanuri, P Phanuphak, JN Weber, D Pillay, PR Harrigan, R Camacho, JM Schapiro, RW Shafer. Impact of HIV-1 subtype and antiretroviral therapy on protease and reverse transcriptase genotype: Results of a global collaboration. PLoS Med 2005; 2:325–337.
- SY Rhee, WJ Fessel, AR Zolopa, L Hurley, T Liu, J Taylor, DP Nguyen, S Slome, D Klein, M Horberg, J Flamm, S Follansbee, JM Schapiro, RWJ Shafer. HIV drug resistance database. Infect Dis 2005; 192:456–465.
- VA Johnson, F Brun-Vézinet, B Clotet, HF Günthard, DR Kuritzkes, D Pillay, JM Schapiro, A Telenti, DD Richman. Update of the drug resistance mutations in HIV-1: 2007. Top HIV Med 2007; 15:119–125.
- JW Erickson, SK Burt. Structural mechanisms of HIV drug resistance. Annu Rev Pharmacol Toxicol 1996; 36:545–571.
- ET Baldwin, TN Bhat, B Liu, N Pattabiraman, JW Erickson. Structural basis of drug resistance for the V82A mutant of HIV-1 proteinase. Nat Struct Biol 1995; 2:244–249.
- S Muzammil, P Ross, E Freire. A major role for a set of non-active site mutations in the development of HIV-1 protease drug resistance. Biochemistry 2003; 42:631–638.
- HB Schock, VM Garsky, LC Kuo. Mutational anatomy of an HIV-1 protease variant conferring cross-resistance to protease inhibitors in clinical trials. Compensatory modulations of binding and activity. J Biol Chem 1996; 271:31957–31963.
- DB Olsen, MW Stahlhut, CA Rutkowski, HB Schock, AL van Olden, LC Kuo. Non-active site changes elicit broad-based cross-resistance of the HIV-1 protease to inhibitors. J Biol Chem 1999; 274:23699–23701.
- CF Perno, A Cozzi-Lepri, C Balotta, F Forbici, M Violin, A Bertoli, G Facchi, P Pezzetti, G Cadeo, G Tositti, S Pasquinucci, S Pauluzzi, A Scalzini, B Salassa, A Vincenti, AN Phillips, F Dianzani, A Apice, G Angarano, L Monno, G Ippolito, M Moroni, AA Monfortethe. Italian Cohort Naive Antiretroviral (I.CO.N.A.) study group. secondary mutations in the protease region of human immunodeficiency virus and virologic failure in drug-naïve patients treated with protease inhibitor-based therapy. J Infect Dis 2001; 184:983–991.
- A Velázquez-Campoy, S Vega, E Fleming, U Bacha, Y Sayed, HW Dirr, E Freire. Protease inhibition in African subtypes of HIV-1. AIDS Rev 2003; 5:165–171.
- T Lee, VD Le, D Lim, YC Lin, GM Morris, AL Wong, AJ Olson, JH Elder, C-H Wong. Development of a new type of protease inhibitors, efficacious against FIV and HIV variants. J Am Chem Soc 1999; 121:1145–1155.
- AM Mildner, DJ Rothrock, JW Leone, CA Bannow, JM Lull, IM Reardon, JL Sarcich, WJ Howe, CS Tomich, CW Smith, RL Heinrikson, AG Tomasselli. The HIV-1 protease as enzyme and substrate: Mutagenesis of autolysis sites and generation of a stable mutant with retained kinetic properties. Biochemistry 1994; 33:9405–9413.
- PA Brindeiro, RM Brindeiro, C Mortensen, K Hertogs, VD Vroey, NPM Rubini, FS Sion, CAM Sá, DM Machado, RCM Succi, A Tarnuri. Testing genotypic and phenotypic resistance in Human Immunodeficiency Virus type 1 isolates of clade B and other clades from children failing antiretroviral therapy. J Clin Microbiol 2002; 40:4512–4519.
- M Sanches, NH Martins, A Calazans, RM Brindeiro, A Tanuri, OAC Antunes, I Polikarpov. Crystallization of a non-B and a B mutant HIV protease. Acta Crystallogr 2004; D60:1625–1627.
- ED Matayoshi, GT Wang, GA Kraft, J Erickson. Novel fluorogenic substrates for assaying retroviral proteases by resonance energy transfer. Science 1990; 24:954–958.
- JF Morrison. Kinetics of the reversible inhibition of enzyme catalysed reactions of tight-binding inhibitors. Biochim Biophys Acta 1969; 185:269–285.
- JG Bieth. Theoretical and practical aspects of proteinase inhibition kinetics. Methods Enzymol 1995; 248:59–84.
- A Velazquez-Campoy, MJ Todd, S Vega, E Freire. Catalytic efficiency and vitality of HIV-1 proteases from African viral subtypes. Proc Natl Acad Sci USA 2001; 98:6062–6067.
- D Hoffmann, H Assfalg-Machleidt, H Nitschko, K von der Helm, U Koszinowski, W Machleidt. Rapid enzymatic test for phenotypic HIV protease drug resistance. Biol Chem 2003; 384:1109–1117.
- Y Lin, X Lin, L Hong, S Foundling, RL Heinrikson, S Thaisrivongs, W Leelamanit, D Raterman, M Shah, BD Dunn, J Tang. Effect of point mutations on the kinetics and the inhibition of human immunodeficiency virus type 1 protease: Relationship to drug resistance. Biochemistry 1995; 34:1143–1152.
- L Hong, A Treharne, JA Hartsuck, S Foundling, J Tang. Crystal structures of complexes of a peptidic inhibitor with wild-type and two mutant HIV-1 proteases. Biochemistry 1996; 35:10627–10633.
- A Calazans, R Brindeiro, P Brindeiro, H Verli, MB Arruda, LMF Gonzalez, JA Guimarães, RS Diaz, OAC Antunes, A Tanuri. Low accumulation of L90M in protease from subtype F HIV-1 with resistance to protease inhibitors is caused by the L89M polymorphism. J Infect Dis 2005; 191:1961–1970.
- M Sanches, S Krauchenco, NH Martins, A Gustchina, A Wlodawer, I Polikarpov. Structural characterization of B and non B subtypes of HIV-protease: Insights into natural susceptibility to drug resistance development. J Mol Biol 2007; 369:1029–1040.
- S Gulnik, LI Suvorov, B Liu, B Yu, B Anderson, H Mitsuya, JW Erickson. Kinetic characterization and cross-resistance patterns of HIV protease mutants selected under drug pressure. Biochemistry 1995; 34:9282–9287.
- I Luque, MJ Todd, J Gomez, N Semo, E Freire. Molecular basis of resistance to HIV-1 protease inhibition: A plausible hypothesis. Biochemistry 1998; 37:5791–5797.
- MJ Todd, I Luque, A Velazquez-Campoy, E Freire. Thermodynamic basis of resistance to HIV-1 protease inhibition: Calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry 2000; 39:11876–11883.
- VA Johnson, F Brun-Vézinet, B Clotet, HF Günthard, DR Kuritzkes, D Pillay, JM Schapiro, DD Richman. Update of the drug resistance mutations in HIV-1: 2007. Top HIV Med 2007; 15:119–125.
- JC Clemente, RE Moose, R Hemrajani, LR Whitford, L Govindasamy, R Reutzel, R McKenna, M Agbandje-McKenna, MM Goodenow, BM Dunn. Comparing the accumulation of active- and nonactive-site mutations in the HIV-1 protease. Biochemistry 2004; 43:12141–12151.