Abstract
New catechol-containing chemical entities have been investigated as matrix metalloproteinase inhibitors as well as antioxidant molecules. The combination of the two properties could represent a useful feature due to the potential application in all the pathological processes characterized by increased proteolytic activity and radical oxygen species (ROS) production, such as inflammation and photoaging. A series of catechol-based molecules were synthesized and tested for both proteolytic and oxidative inhibitory activity, and the detailed binding mode was assessed by crystal structure determination of the complex between a catechol derivative and the matrix metalloproteinase-8. Surprisingly, X-ray structure reveals that the catechol oxygens do not coordinates the zinc atom.
Introduction
Matrix metalloproteinases (MMPs) are a family of zinc-containing endopeptidases, capable to process all the extracellular matrix components. They can be classified in collagenases (MMP-1, MMP-8, MMP-13, MMP-18), gelatinases (MMP-2, MMP-9), and metalloelastase (MMP-12), based on the substrate that they process.
Their enzymatic activity is finely regulated in physiological conditions: MMPs are responsible for tissue regeneration and remodeling, as well as bioavailability of pro-factors that need proteolytic cut activation. Endogenous molecules such as tissue inhibitors of metalloproteinases (TIMPs) are responsible for the physiological fine regulation of the numerous isoformsCitation1. However, several external factors (e.g. solar UV irradiation) or pathological conditions (cancer, metastasis, or chronic inflammation) are able to stimulate an overexpression of specific MMPs. Therefore, due to their implications in complex pathological processes, MMPs continue to be considered a pivotal target for therapeutic interventionCitation2.
The selectivity still represents the main challenge in the MMP inhibitor design: the importance of selective targetingCitation3,Citation4 has already been previously proved in MMP inhibition strategyCitation5 especially because in the last decade, a number of inhibitors failed in clinical trial phasesCitation6, mainly due to their broad spectrum activity.
Photoaging represents another process in which the proteolytic activity of MMPs has a notorious and critical role; collagenases, in particular, are responsible for the decomposition of particular types of collagen and other proteins in the extracellular matrix of the dermisCitation7–10. The breakdown of dermal collagen and elastin is purported to be one of the major contributing factors to loss of skin’s firmness and elasticity. Physiologically, the human skin expresses a number of MMPs, including MMP-1, -2, -3, -8, -9, and -13, all of which are capable of attacking native fibrillar collagen.
Chronic exposure to solar UV radiation is an accelerator factor for photoaging. UV irradiation is known to provoke oxidative stress through the generation of reactive oxygen species (ROS), which are responsible to interfere with physiological pathways and ultimately activate the overexpression of a number of proteolytic enzymes such as MMPs in skin cells, capable to destroy the dermal connective tissue. In addition, ROS can regulate pro-MMPs activationCitation11–15.
Thus, the antioxidant activity associated with the inhibition of MMPs could represent a promising strategy to obtain new chemical entities capable to reduce photoaging and prevent wrinkles and damage of the skin, and be effective for other pathological conditions where oxidative stress occurs.
MMP inhibitors (MMPIs) are usually characterized by a backbone that interacts with the specificity pocket S1′, coupled to a metal chelator portion that binds to the catalytic Zinc ion. To expand the library of potential MMPIs and to overcome some limitations of the hydroxamic acid moietyCitation16,Citation17, other zing binding groups (ZBGs) as essential portion in MMP inhibitor molecule of new synthesis have been developedCitation18–22.
Recently, a series of rosmarinic acid derivatives has been identified as micromolar MMP-1 inhibitorsCitation23. In addition, phenolic and polyphenolic compounds have already been used as antioxidants, and their activity seems to impact several different pathways. In particular, one of the most tested hypothesis is that they exert protective effects against cancer and other chronic diseases by reducing ROS levelsCitation13. Natural compounds derived from green tea (EGCG), mainly known for their antioxidant properties, have shown relevance when applied on wrinkles or fine lines caused by aging, due to their ability to enhance collagen levels and to inhibit MMPsCitation24. Many other natural compounds, such as phytosterol, fucosterol, resveratrol, soy isoflavones, alpha-tocopherol, and vitamin E and C, are also able to inhibit MMPs and ROS and reduce the degradation of skinCitation25–28. Some MMPIs have been studied in topical cosmetic compositions to counteract the effect of photo and chronological skin aging. For example, their use has been reported in combination with UV blockers (e.g. octinoxate and zinc oxide)Citation12, natural estrogen (e.g. 17-beta estradiol or an estrogen-like steroid)Citation12, or antioxidants (Alpha Lipoic Acid, ALA)Citation29.
In this study, we present a new class of MMPIs obtained by combining a catecholic portion with a backbone able to reach the S1′ specific pocket of the enzyme isoforms. All these compounds were tested toward MMP-2, -8, and -9 whereas their in vitro antioxidant activity was evaluated by DPPH assay. In order to better understand the interactions between MMPs and the chemical inhibitor, the X-ray structure of the complex between MMP-8 and one compound of the series was resolved.
Results and discussion
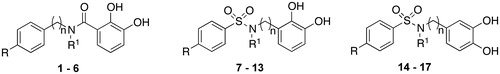
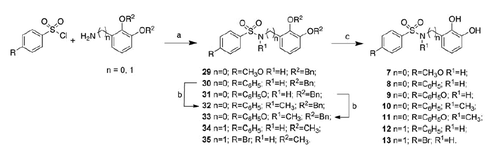
Starting from the promising inhibitory activity values obtained testing catechol on different MMPsCitation30, we decided to further investigate catecholic derivatives by focusing our attention on the backbone that, in the classical structure activity relationship (SAR) of MMPIs, blocks the substrate access to the active site. Therefore, we report herein the synthesis and biological activity of three set of analogs () in which the catechol is connected through an amide (compounds 1–6) or a sulfonamide (compounds 7–17) group to a series of alkylaryl or aryl moieties in order to reach the S1′ specificity pocket of the different enzyme isoforms.
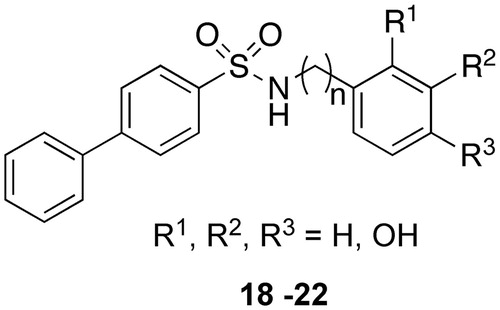
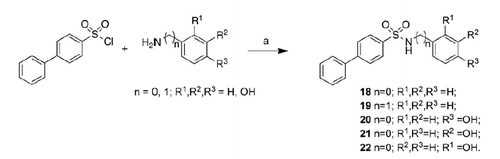
Most compounds present a diphenyl or a phenoxyphenyl residue, based on the results obtained by our previous works in which these groups were linked to typical ZBGsCitation3,4,19,22,31–34. The relative position of sulfonamide linking group as well as the importance of hydroxyl groups were evaluated. To this aim, we prepared a fourth set of compounds 18–22 () characterized by the lack of hydroxyl groups or the presence of a single hydroxyl group in different position of the aromatic ring.
Compounds 1–6 were obtained via condensation of dibenzyloxybenzoic acid or corresponding chloride with the appropriate aniline or benzylamine and following by debenzylation through catalytic hydrogenation (Scheme 1). N-(4-diphenylmethyl)-N-methyl-2,3-dibenzyloxybenzamide (26) was obtained through methylation of the sulfonamide nitrogen of the dibenzylate intermediate (25).
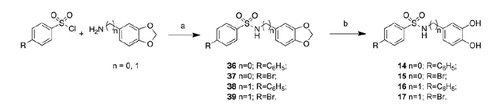
Compounds 7–13 and 18–22 were synthesized via condensation of the suitable aniline or benzyl amine with the appropriate sulfonyl chloride (Schemes 2 and 3). Subsequent deprotection under H2 atmosphere in the presence of 10% Pd/C or with BBr3 was needed to obtain the final compounds. N-(2,3-dihydroxy-phenyl)-N-methyl-4-phenyl sulfonamide (10) and N-(2,3-dihydroxy-phenyl)-N-methyl-4-phenoxy-phenyl sulfonamide (11) were obtained through methylation of the sulfonamide nitrogen of the dibenzylate intermediate.
Compounds 14–17 were synthesized via condensation of 3,4-(methylenedioxy)aniline or benzylamine with the appropriate sulfonyl chloride (Scheme 4). Subsequent deprotection with BBr3 was needed to obtain the final compounds.
All synthesized compounds 1–22 were tested against MMP-2, -8, and -9 ( and ). Compound 1 shows a very weak activity against all considered MMPs. As expected, the introduction of a diphenyl moiety (2) provides more potency and selectivity on MMP-2. A significant increase of activity toward all MMPs is obtained by inserting a methylene spacer (3), while the methylation of the amide nitrogen (4) reduces it. The introduction of an oxygen atom between the phenyl rings results in an increase of activity (5 versus 2) or selectivity for MMP-2 (6 versus 3).
Table 1. MMP activity values expressed as IC50 (μM) and antioxidant activity values expressed as EC50 in DPPH assay and ARP.
Table 2. MMP activity values expressed as IC50 (μM) and antioxidant activity values expressed as EC50 in DPPH assay and ARP.
As well as for amide derivatives, compounds 7 and 8 still maintain good activity values toward MMP-2, showing an increased activity also on MMP-8 and -9. The introduction of the more flexible phenoxy phenyl structure (9) results in a slight increase of activity toward all the enzyme isoforms, with IC50 values around 6 μM against all tested MMPs.
The methylation of the sulfonamide nitrogen still confirms its detrimental effects on MMP-8 and -9 (10 and 11), while no loss in activity results for these N-methyl derivatives against MMP-2. Compound 10 stands out for its very interesting selectivity toward MMP-2. The insertion of a methylene between the sulfonamide moiety and the catecholic portion (12) allows to obtain inhibition activity in the low micromolar range toward MMP-2, -8, and -9 and even the substitution of the phenyl ring with a bromine atom (13) maintains high activity values against all isoforms.
Moving the hydroxyl groups from the 2,3 to the 3,4 positions of the aromatic ring, it results a slight increase of activity when the sulfonamide moiety is directly linked to the phenyl residue (8 versus 14); in this case, the substitution of phenyl with bromine (15) reduces the activity on MMP-8 and MMP-9. The introduction of a methylene spacer into the structure of 14 gives 16 with lower activity toward all isoforms, whereas the same modification for 15 affords 17 showing reduced activity only against MMP-2.
In order to evaluate whether the presence of hydroxyl groups was essential for inhibitory activity, we prepared the mono- (20–22) or non-substituted (18, 19) analogs (). However, these compounds show a complete loss of activity against all isoforms.
The in vitro antioxidant activity of these catecholic derivatives was also evaluated by DPPH assay ( and ), considered as one of the standard colorimetric method for the evaluation of antioxidant properties of pure compounds and is routinely practiced for assessment of free radical scavenging potential of an antioxidant molecule. Experiments were performed also with gallic acid, a naturally occurring plant phenol as the reference substance.
Several studies suggest that structure–antioxidant activity relationship for phenolic compounds depends on the position of hydroxyl groups, the presence of other functional groups in the whole molecule, and their conjugation to hydroxyl groupsCitation35.
For compounds 1–17, antioxidant activity could be mainly ascribed to the reducing power of the o-dihydroxy structure; elimination of the hydroxy groups (18 and 19) or their substitution with methoxy ones (data not shown) results in a complete loss of activity. Derivatives with hydroxyl groups in positions 2, 3 show a better antioxidant activity with respect to the 3, 4 substitutions (8 versus 14).
The reduction of the o-dihydroxy structure to phenol (20–22) results in a decrease of the activity strongly dependent from the substituent position: only the 2-substituted analog (22) maintains an interesting antioxidant activity, while no or low activity is observed in the 3- or 4-substituted derivative, respectively (20 and 21). Therefore, it can be argued that the presence of two phenolic groups is not the only factor determining the antioxidant activity of our derivatives.
Methylation of the nitrogen results in a reduction of activity for both the amidic (4 versus 3) and sulfonamide analogs (10 versus 8 and 11 versus 9). The presence of a methylene spacer increases ARP in the amide series (3 versus 2), while a strong reduction is observed for sulfonamide derivatives (14 versus 16).
Catechol was identified as a promising ZBGCitation30 but the introduction of aryl moieties aimed to reach the S1′ specificity pocket of MMP enzymes does not lead to the expected potency improvement. In addition, at a first glance it is not obvious to derive a structure activity relationship (SAR) for the catechol analog series. To elucidate the binding mode of the catechol compounds, the inhibitor 14 was co-crystallized with MMP-8. Crystals of the complex MMP-8:14 were grown as described in the Experimental Section. Surprisingly, the analysis of the Fo-Fc electron density map in the region of binding of 14 shows the presence of the catechol and its quinone oxidized form, both competing for the same binding site (S1′ site). The refinement of the occupancy factors of the two forms reveals the predominance of the catechol with respect to quinone (77% versus 23%). The binding mode of the two forms in the S1′ site is different at the level of the aromatic ring containing the zinc binding functions. As evidenced in , only the quinone, with one of its oxygen atoms, bind the zinc ion with a distance of 1.84 Å giving rise to a distorted tetrahedral coordination, while one of the catechol oxygens makes H-bonds with the catalytic important E198 side-chain and the NH of A163, belonging to the antiparallel β-strand.
Scheme 1. Reagents and conditions: (a) anhydrous THF, r.t. or DMAP, DCC, CH2Cl2, r.t.; (b) dry DMF, NaH, CH3I, 0 °C; (c) H2, 10% Pd-C, CH3OH/THF 2:5, r.t.
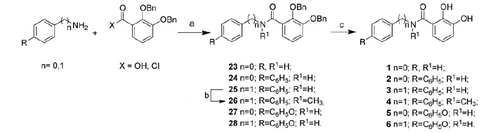
Figure 3. Hydrogen bond network of the catechol form (A) and the oxidized quinone form (B) of 14 (distances in Å).
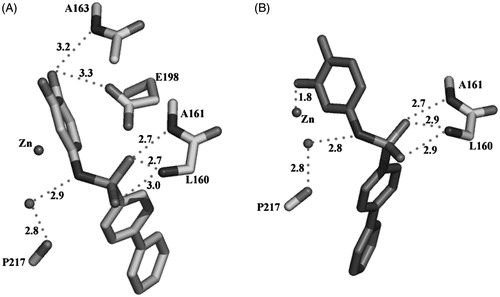
The sulfonamide NH group of both forms is engaged in a H-bond with a water molecule further bridged to the P217 CO group. The sulfonamide junction adopts a g-conformation in both forms (−86° and −102°, respectively), with one of the two oxygens turned toward the upper rim, giving rise to H-bonds with the A161 and L160 NH groups. Finally, the diphenyl group deeply protrudes into the S1′ pocket, with its terminal part facing the charged R222 side-chain (closest distance of 3.1 Å), at the end of the pocket.
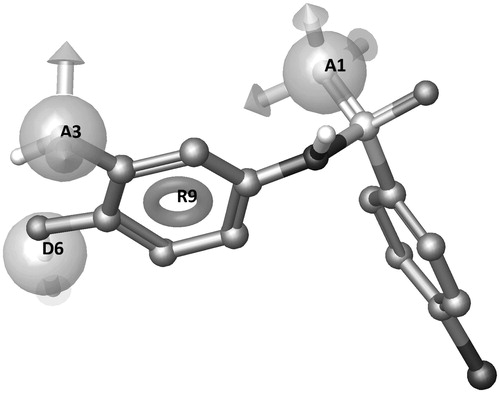
Integrating different computational approaches, we could identify the ligand features responsible for ligand–protein interaction and explain how small modifications in ligand structures can influence the inhibition potency. Through a pharmacophore-based analysis, the features that are needed for the MMP inhibition have been identified (): one aromatic ring (R9), two acceptor H-bond groups (A1 and A3, respectively), and one donor H-bond group (D6). Compound 15 was automatically selected as the reference structure, with a Fit-value of 3.00.
The pharmacophore model highlights the importance of both hydroxyl groups for the MMP-8 inhibition and of their positions in relation to the aromatic ring. The resulting QSAR model works well in discriminating actives versus inactives (), is robust and predictive (correlation values for the MMP-8 inhibition model, R2: 0.8, Q2: 0.8, F: 51.3, P: 3.3e-006, RMSE: 0.4, Pearson’s R: 0.9).
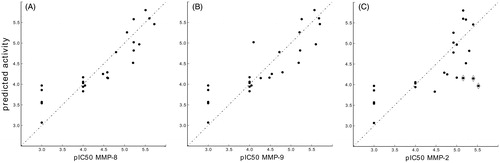
The activity prediction is well correlated even with MMP-9 and MMP-2 () experimental values; however, interestingly, the pharmacophore QSAR model underestimates the potency of compounds 3, 5, and 6 toward MMP-2. To investigate this difference and to characterize the interactions corresponding to the identified features, we integrate the pharmacophore-based analysis with a structure-based study.
Surprisingly, the X-ray structure of 14 in complex with MMP-8 reveals that the catechol oxygens contribute to a dense water network around the ligand (see ). Noteworthy, the quinone oxidized form of the ligand was found in the crystal complex, but not in the solution used for inhibition assay (see supporting information); therefore, we have considered activity values not related to the oxidized forms, which have been neglected in the computational studies. Apparently, catechol oxygen atoms prefer to enter in the water network rather than coordinate the zinc ion, suggesting a relevant role of water molecules in the binding and activity of catechol analogs toward MMP enzymes. It is well-known the role of water molecules in ligand–protein interaction, and this role becomes fundamental in the case of weak bindersCitation36. To clarify the role of the water molecules, the X-ray complex MMP-8:14 has been prepared and minimized with explicit waters with MacroModel Embrace MinimizationCitation37. The minimized complex reveals the interactions corresponding to the pharmacophore features: the donor feature (D6) is located on the OH group establishing an H-bond interaction with E198 side chain; the acceptor group A1 directly H-bonds with L160 NH and A161 NH; the acceptor group A3 is connected through two water molecules to H162 side chain; the aromatic feature (R9) is located above the zinc ion ensuring a good orientation for D6 and A3 interactions.
Figure 6. (A) MMP-8:14 X-ray complex (IC50: 3.1 μM, predicted total energy −395 kJ/mol), (B) MMP-2:14 modeled complex (IC50: 7.0 μM, predicted total energy −379 kJ/mol), and (C) MMP-9:14 modeled complex (IC50: 2.7 μM, predicted total energy −405 kJ/mol). The ligand is represented as sticks and the water molecules as spheres. The network of water molecules is depicted with H-bonds reported as dashed lines and water clusters are encircled.
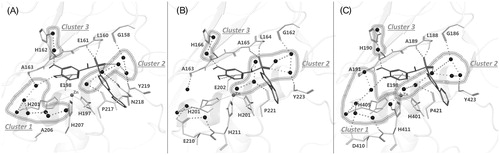
In addition, three relevant clusters of water molecules mediate the interaction between the ligand and MMP-8 (): (i) the oxygen bound to D6 is connected through seven water molecules to A163 CO on the top, H201 CO on the side and H207 side chain on the bottom; (ii) five water molecules connect the sulfonamide oxygen with P217 CO, N218 side chain and Y219 NH on the bottom, and G158 CO on the top; (iii) two water molecules mediate the interaction between the A3 feature oxygen and H162 side chain.
The structure determination of an enzyme bound to a potentially therapeutic inhibitor is usually aimed to design optimized leads: starting from the identification of interactions involved in binding, moving to the analysis of possible additional or alternative interactions that can improve the potency. However, the complex described here highlights difficulties of theoretical prediction of binding energies and optimization process due to the fact that the binding involves hydrogen bonding through water molecules whose positions can change.
This observation prompted us to investigate the influence of the solvent in the binding of compound 14 with MMP-2 and MMP-9 (). We can observe similar interactions in all three complexes, but a different hydration by the solvent (). In the MMP-2:14 complex, the oxygen bound to D6 is not connected with the water molecule cluster 1 (). This difference is due to a sequence difference: A206 in MMP-8 is aligned to E210 in MMP-2. E210 side chain takes the place of the water molecule present in MMP-8:14 complex and shifts the water cluster 1 far away from the ligand (). In the case of MMP-9, we found in this position D410 (); since it is shorter than the glutamate present in this position in MMP-2, the cluster 1 water network is restored.
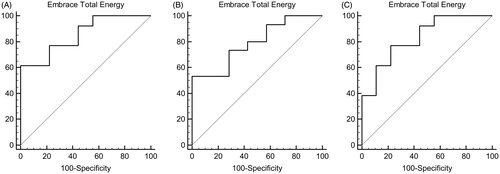
The structure-based analyses, taking into account all possible ligand–protein interactions and water contribution, ensure a more accurate prediction of the binding. Differently from what observed with the ligand-based analysis, the predicted binding energies of MMP-2:3, 5, and 6 (−303 kJ/mol, −362 kJ/mol and −366 kJ/mol, respectively) are in a good correlation with the experimental pIC50. The ROC curves of all tested compounds toward MMP-8, MMP-2, and MMP-9 are shown in .
Conclusions
A new series of catechol-based compounds has been synthesized and tested for MMP inhibitory activity as well as antioxidant properties. Structure–activity relationship studies were implemented by X-ray analysis of the co-crystallization complex between MMP-8 and one of these new chemical entities (compound 14). Computational studies have been performed to identify a pharmacophore model and rationalize the influence of chemical structure on inhibition potency.
The activity of these molecules on both MMPs and ROS encloses a potential therapeutic strategy able to reduce the impact of these two important targets in degradation processes such as photoaging or inflammation, as well as to provide an additional treatment for degenerative phenomenon involving pathological enzymatic overexpression.
Experimental section
Chemistry
Melting points were determined in open capillaries on a Gallenkamp electrothermal apparatus. Mass spectra were recorded on a HP MS 6890-5973 MSD spectrometer, electron impact 70 eV, equipped with a HP ChemStation or with an Agilent LC–MS 1100 Series LC–MSD Trap System VL spectrometer, electrospray ionization (ESI). 1H-NMR spectra were recorded using the suitable deuterated solvent on a Varian Mercury 300 NMR Spectrometer. Chemical shifts (δ) are expressed as parts per million (ppm) and the coupling constants (J) in Hertz (Hz). Microanalyses of solid compounds were carried out with a Eurovector Euro EA 3000 model analyzer; the analytical results are within ± 0.4% of theoretical values. Flash column chromatography was performed using Geduran silica gel 60 Å (45–63 μm). Chemicals were purchased from Aldrich Chemicals (Milan, Italy) and were used without any further purification.
Preparation of 2,3-dibenzyloxy-N-4-bromophenylbenzamide, 23 and 25
To a solution of 2,3-dibenzyloxybenzoicacid (30) (2.27 mmol) in anhydrous toluene (35 mL), thionyl chloride (23.7 mmol) was added dropwise at 0 °C under nitrogen atmosphere. The mixture was refluxed for 2 h and concentrated under reduced pressure. The resulting oil was dissolved in dry THF (30 mL) and a solution of aniline or 4-phenylbenzylamine (7.12 mmol) in dry THF (1 mL) was added dropwise at 0 °C. After 12 h at room temperature, the mixture was concentrated under reduced pressure and diluted with CH2Cl2. The organic layer was washed with HCl 1N, brine, dried over Na2SO4, and concentrated under reduced pressure. The residue was purified by column chromatography (n-hexane/ethyl acetate = 9:1) to give the title products.
2,3-Dibenzyloxy-N-phenylbenzamide (23)
53% yield; 1H NMR (CDCl3): δ = 5.16 and 5.20 (2 s, 4H, 2 CH2Ph), 7.03–7.09, 7.20–7.53, and 7.83–7.87 (m, 18H, aromatics), 10.01 (s, 1H, NH); MS (ESI) m/z (%): 432 [M + Na]+ (100)
2,3-Dibenzyloxy-N-(4-diphenyl)methylbenzamide (25)
71% yield; 1H NMR (CDCl3): δ 4.54 (s, 2H, CH2N), 4.99, and 5.15 (2 s, 4H, 2 CH2Ph), 7.07–7.59 and 7.79–7.83 (m, 22H, aromatics), 8.38 (t, 1H, NH); MS (ESI): m/z (%): 522 [M + Na]+ (100), 340 (7).
2,3-Dibenzyloxy-N-4-bromophenylbenzamide
99% yield; mp: 122–124 °C 1H NMR (CDCl3): 5.10 and 5.20 (2 s, 4H, 2 CH2Ph), 7.10–7.50, and 7.80–7.86 (d, 17H, aromatics), 10.0 (s, NH); MS (ESI) m/z: 488 [M + 2 + Na]+ (100), 486 [M + Na]+ (100).
Preparation of 2,3-dibenzyloxy-N-(4-diphenyl)benzamide (24)
A suspension of 2,3-dibenzyloxy-N-4-bromophenylbenzamide (0.56 mmol), benzeneboronic acid (1.6 mmol), Cs2CO3 (1.2 mmol), and [Pd(PPh3)4] (0.024 mmol) in anhydrous toluene (15 mL) was stirred at 95 °C overnight under nitrogen atmosphere. Then, the mixture was diluted with 1N HCl (1 mL) and ethyl acetate (1.3 mL) at room temperature and filtered through a Celite pad. The resulting solution was washed with a saturated NaHCO3 solution (3 × 15 mL), brine, dried over Na2SO4, and concentrated under reduced pressure. The residue was purified by column chromatography (9.4:0.5:0.1 petroleum ether/ethyl acetate/IPA) to give the title product.
74% yield; mp: 121–124 °C; 1H NMR (CDCl3): δ = 5.18 and 5.21 (2 s, 4H, 2 CH2-Ph), 7.20–7.50, 7.70–7.73, 7.83–7.86, and 8.23–8.26 (d, 22H, aromatics), 10.0 (s, NH). MS(ESI) m/z (%): 508 [M + Na]+(100).
Preparation of compounds 27 and 28
DCC (2.04 mmol) was added to a solution of 2,3-dibenzyloxybenzoic acid (30) (1.5 mmol) in dichloromethane (30 mL) at 0 °C under nitrogen atmosphere. After 15 min, a solution of 4-DMAP (1.35 mmol) and 4-phenoxyaniline or 4-phenoxybenzylamine (1.38 mmol) in dichloromethane (4 mL) was added dropwise. The resulting mixture was allowed to warm to room temperature and stirred for further 22 h. The resulting precipitate was filtered off, and the organic layer was washed with a saturated NH4Cl solution (3 × 20 mL) and twice with brine, then dried over Na2SO4, filtered and the solvent removed in vacuo to yield a brown solid. Purification by flash chromatography (8:1.9:0.1 hexane/chloroform/IPA) and crystallization (THF/hexane) yielded the title compounds as a white powder.
2,3-Dibenzyloxy-N-(4-phenoxyphenyl)benzamide (27)
73% yield; 1H NMR (CDCl3): δ = 5.18 and 5.21 (2 s, 4H, 2 CH2Ph), 6.89–6.99, 7.05–7.10, 7.21–7.53, and 7.83–7.88 (m, 22H, aromatics), 9.99 (s, 1H, NH).
2,3-Dibenzyloxy-N-(4-phenoxyphenyl)methylbenzamide (28)
69% yield; 1H NMR (CDCl3): δ = 4.47 (d, 2H, CH2N), 4.99 and 5.15 (2 s, 4H, 2 CH2Ph), 6.88–7.48, and 7.76–7.82 (m, 22H, aromatics), 8.31 (t, 1H, -OH); MS (ESI) m/z (%): m/z 514 [M-H]− (100), 408 (9), 315 (9), 223 (9).
General procedure for the preparation of 29–31, 34–39, and 18–22
A solution of the suitable sulfonyl chloride (1.4 mmol) in CH2Cl2 (5 mL) was added to a solution of the appropriate aniline (1.5 mmol) and Et3N (2.8 mmol) in CH2Cl2 (5 mL). The mixture was refluxed for 4 h, cooled, diluted with CH2Cl2 (10 mL), washed with 1N HCl and brine, dried over Na2SO4, and then evaporated in vacuo. The crude product was purified by flash column chromatography on silica gel using different mixtures as eluent (indicated in brackets) or was used for the next step without any purification.
N-(2,3-dibenzyloxy)phenyl-4-methoxyphenylsulfonamide (29)
(Petroleum ether/ethyl acetate 9:1, 51% yield); 1H NMR (CDCl3): δ = 3.79 (s, 3H, OCH3), 4.78 and 5.07 (2 s, 4H, 2 OCH2Ph), 6.70–6.97 (m, 4H, aromatics), 7.04 (bb, 1H, NH), 7.15–7.42 (m, 11H, aromatics), 7.63–7.68 (m, 2H aromatics); MS (ESI) m/z (%): 498 [M + Na]+ (100), 407 (6), 304 (15).
N-(2,3-dibenzyloxy)phenyl-4-diphenylsulfonamide (30)
(Petroleum ether/ethyl acetate/methylene chloride 9:0.5:0.5, 70% yield); 1H NMR (CDCl3): δ = 4.79 and 5.07 (2 s, 4H, 2 OCH2Ph), 6.72–6.75, 6.94–7.00 (m, 2H, aromatics), 7.11 (bb, 1H, NH), 7,20–7.62, and 7.76–7.80 (m, 20H, aromatics); MS (ESI) m/z (%): 544 [M + Na]+ (100), 304 (75).
N-(2,3-dibenzyloxy)phenyl-4-phenoxyphenylsulfonamide (31)
(Petroleum ether/chloroform/IPA 9:0.9:0.1, 81% yield); 1H NMR (CDCl3): δ = 4.81 and 5.09 (2 s, 4H, 2 OCH2), 6.72–6.75, 6.90–7.04, 7.16–7.47, 7.63–7.77 (m, 22H, aromatics and 1H, NH); MS (ESI) m/z (%): 560 [M + Na]+ (100), 469 (5).
N-(2,3-dimethoxyphenyl)methyl-4-diphenylsulfonamide (34)
98% yield; mp: 120–121 °C; 1H NMR (CDCl3): δ = 3.73 (s, 3H, OCH3), 3.76 (s, 3H, OCH3), 4.19 (d, J = 6.0 Hz, 2H, CH2NH), 5.30 (t, J = 6.0 Hz, 1H, NH), 6.72–6.79, 6.85–6.92, 7.38–7.50, 7.55–7.65, 7.83–7.88 (m, 12H, aromatics); 13C NMR (CDCl3): δ 43.6, 56.0, 60.9, 112.6, 121.7, 124.3, 127.5, 127.7, 127.9, 128.7, 129.3, 130.0, 138.8, 139.6, 145.4, 147.1, 152.6.
N-(2,3-dimethoxyphenyl)methyl-4-bromophenylsulfonamide (35)
88% yield; mp: 125 °C; 1H NMR (CDCl3): δ 3.72 (s, 3H, OCH3), 3.80 (s, 3H, OCH3), 4.15 (d, J = 6.0 Hz, 2H, CH2NH), 5.28 (t, J = 6.0 Hz, 1H, NH), 6.64–6.68, 6.76–6.80, 6.85–6.91, 7.47–7.51, 7.57–7.61 (m, 7H, aromatics); 13C NMR (CDCl3): δ 43.8, 56.0, 60.8, 112.7, 121.7, 124.3, 127.4, 128.8, 129.6, 132.2, 139.4, 147.0, 152.6.
N-(1,3-benzodioxol-5-yl)-4-diphenylsulfonamide (36)
(CH2Cl2, 56% yield); mp: 145–146 °C; 1H NMR (CDCl3): δ = 5.93 (s, 2H, -CH2), 6.46–6.50, 6.62–6.66, 6.72–6.74 (m, 3H, aromatic), 6.85 (bb, 1H, NH), 7.37–7.49, 7.55–7.67, 7.79–7.84 (m, 9H, aromatics); 13C NMR (CDCl3): δ 101.8, 105.8, 108.5, 117.2, 127.5, 127.8, 128.1, 128.8, 129.3, 130.2, 137.6, 139.4, 146.0, 146.3, 148.3.
N-(1,3-benzodioxol-5-yl)-4-bromophenylsulfonamide (37)
62% yield; mp: 140–141 °C; 1H NMR (CDCl3): δ = 5.94 (s, 2H, CH2), 6.41–6.45, 6.61–6.64, 6.67–6.81 (m, 3H, aromatics), 6.97 (bb, 1H, NH), 7.54–7.62 (m, 4H, aromatics); 13C NMR (CDCl3): δ = 101.9, 105.8, 108.6, 116.6, 117.3, 128.4, 129.1, 132.6, 137.9, 146.6, 148.4.
N-(1,3-benzodioxol-5-ylmethyl)-4-diphenylsulfonamide (38)
(CH2Cl2, 98% yield); mp: 159–160 °C; 1H NMR (CDCl3): δ = 4.08 (d, J = 6.0 Hz, 2H, CH2NH), 4.86 (t, J = 6.0 Hz, 1H, NH), 5.87 (s, 2H, CH2), 6.63–6.70, 7.39–7.52, 7.58–7.63, 7.67–7.62, 7.88–7.93 (m, 12H, aromatics); 13C NMR (CDCl3): δ = 45.7, 101.4, 108.4, 108.7, 121.6, 127.6, 127.9, 127.9, 128.7, 129.3, 130.1, 138.7, 139.5, 145.8, 147.6, 148.1.
N-(1,3-benzodioxol-5-ylmethyl)-4-bromophenylsulfonamide (39)
(CH2Cl2, 97% yield); mp: 148–149 °C; 1H NMR (CDCl3): δ = 4.04 (d, J = 6.0 Hz, 2H, CH2NH), 4.84 (t, J = 6.0 Hz, 1H, NH), 5.93 (s, 2H, –CH2–), 6.60–6.70, 7.59–7.71 (m, 7H, aromatics); 13C NMR (CDCl3): δ = 45.4, 101.5, 108.5, 108.6, 121.7, 127.9, 128.9, 129.8, 132.6, 139.3, 147.6, 148.2.
N-phenyl-4-diphenylsulfonamide (18)
55% yield; mp: 126–127 °C; 1H NMR (CDCl3): δ = 6.97 (bb, 1H, NH), 7.10–7.15, 7.22–7.29, 7.40–7.48, 7.53–7.58, 7.62–7.66, 7.83–7.86 (m, 14H, aromatics); 13C NMR (CDCl3): δ = 121.8, 125.6, 127.7, 127.09 128.1, 128.0, 128.8, 129.3, 129.6, 136.7, 137.7, 139.3, 146.1. MS (ESI) m/z (%): 308 [M-H]− (18), 244 (100). HR-MS [(C19H17NO2S-H)]−, m/z 308.0744 (calc. 308.0751).
N-benzyl-4-diphenylsulfonamide (19)
75% yield; mp: 134–135 °C; 1H NMR (CDCl3): δ = 4.19 (d, J = 6.0 Hz, 2H, CH2NH), 4.74 (t, J = 6.0 Hz, 1H, NH), 7.20–7.31, 7.41–7.53, 7.59–7.63, 7.69–7.73, 7.91–7.95 (m, 14H, aromatics); 13C NMR (CDCl3): δ = 47.6, 127.6, 127.9, 128.0, 128.1, 128.2, 128.4, 128.8, 129.0, 129.3, 136.4, 138.6, 139.5, 145.9. MS (ESI) m/z (%): 322 [M-H]− (5), 153 (100). HR-MS [(C19H17NO2S-H)]−, m/z 322.0903 (calc. 322.0907).
N-(4-hydroxyphenyl)-4-diphenylsulfonamide (20)
63% yield; mp: 212–214 °C; 1H NMR ([D6]DMSO): δ = 6.58–6.61, 6.84–6.88, 7.40–7.82 (m, 13H, aromatics), 9.30 and 9.78 (bb, 2H, –OH and –NH); 13C NMR ([D6]DMSO): δ = 116.0, 124.4, 127.5, 127.6, 127.8, 128.9, 129.0, 129.5, 138.8, 138.9, 144.4, 155.3. MS (ESI) m/z (%): 324 [M-H]− (100). HR-MS [(C18H15NO3S + Na)]+, m/z 348.0665 (calc. 348.0665).
N-(3-hydroxyphenyl)-4-diphenylsulfonamide (21)
71% yield; mp: 258–259 °C; 1H NMR ([D6]DMSO): δ = 5.85–6.30, 6.58–6.63, 7.31–7.73 (m, 13H, aromatics), 8.51 (bb, 2H, –OH and –NH); 13C NMR ([D6]DMSO): δ = 105.6, 107.8, 112.5, 126.6, 126.8, 127.2, 127.3, 127.5, 128.2, 128.8, 129.4, 139.8, 141.7, 157.8. MS (ESI) m/z (%): 324 [M-H]− (100). HR-MS [(C18H15NO3S-H)]−, m/z 324.0694 (calc. 324.0700).
N-(2-hydroxyphenyl)-4-diphenylsulfonamide (22)
55% yield; mp: 155–157 °C; 1H NMR ([D6]DMSO): δ = 6.66–6.73, 6.89–6.94, 7.14–7.17, 7.38–7.50, 7.68–7.80 (m, 13H, aromatics), 9.43 (bb, 2H, OH and NH); 13C NMR ([D6]DMSO): δ = 116.0, 119.4, 124.5, 125.0, 126.6, 127.4, 127.5, 127.8, 128.9, 129.5, 138.8, 140.0, 144.3, 150.7. MS (ESI) m/z (%) 324 [M-H]− (95), 217 (100); m/z (%): 348 [M + Na]+ (100). HR-MS [(C18H15NO3S-H)]−, m/z 324.0695 (calc. 324.0700).
General procedure for the preparation of 26, 32 and 33
3a or 30 or 31 (1 mmol) was dissolved in anhydrous DMF (20 mL) under argon atmosphere at 0 °C. Then, NaH (3 mmol, 95% powder) and, after 30 min, CH3I (50 mL) were carefully added and the mixture was stirred overnight at room temperature. After evaporation of volatiles, the residue was dissolved in ethyl acetate and washed with 1N HCl (3 × 50 mL) and brine (3 × 40 mL), dried over Na2SO4, filtered, and concentrated in vacuo giving a yellow oil, that was used for the next step without any purification.
N-(4-diphenylmethyl)-N-methyl-2,3-dibenzyloxybenzamide (26)
97% yield; 1H NMR (CDCl3): δ = 2,76 (s, 3H, CH3N), 3.00 (s, 2H, CH2N), 5.15 and 5.17 (2 s, 4H, 2 CH2Ph), 6.92–7.55 (m, 22H, aromatics); MS (ESI) m/z (%) 536 [M + Na]+ (100).
N-(2,3-dibenzyloxyphenyl)-N-methyl-4-diphenylsulfonamide (32)
90% yield; 1H NMR ([D6]DMSO): δ = 3.10 (s, 3H, CH3N), 4.99 and 5.11 (2 s, 4H, 2 OCH2Ph), 6.76–6.82, 6.95–7.02, and 7.20–7.88 (m, 22H, aromatics); MS (ESI) m/z (%): 558[M + Na]+ (100), 304 (7).
N-(2,3-dibenzyloxyphenyl)-N-methyl-4-phenoxyphenylsulfonamide (33)
84% yield; 1H NMR ([D6]DMSO): δ = 3.11 (s, 3H, CH3N), 4.99, and 5.10 (2 s, 4H, 2 OCH2Ph), 6.76–6.79, 6.94–7.05, 7.18–7.43, and 7.71–7.76 (m, 22H, aromatics); MS (ESI) m/z (%): 574 [M-Na]+ (100), 319 (16), 228 (6).
General procedure for the preparation of compounds 1–6
A mixture of the appropriate dibenzyloxybenzamide 23–28 (0.38 mmol) and 10% Pd-C (24 mg) in CH3OH/THF 2:5 (14 mL) was stirred at room temperature for 20 h under H2 atmosphere at 6 atm. The mixture was filtered through a Celite pad and concentrated in vacuo to give the title product as a white solid.
2,3-Dihydroxy-N-4-phenylbenzamide (1)
72% yield; mp: 114–117 °C; 1H NMR (CDCl3): δ = 5.82 (s, 1H, NH), 6.82–6.87, 7.05–7.12, 7.19–7.25 and 7.38–7.60 (m, 8H, aromatics), 7.95 (s, 1H, OH), 12.34 (s, 1H, OH); 13C NMR (CDCl3): δ = 114.3, 116.1, 118.6, 119.0, 121.3, 125.5, 129.2, 136.4, 146.1, 149.3, 168.4. MS (ESI) m/z (%): 228 [M-H]− (100). HR-MS [(C13H11NO3-H)]−, m/z 228.0663 (calc. 228.0663).
N-(4-diphenyl)-2,3-dihydroxybenzamide (2)
38% yield; mp: 226–228; 1H NMR (CDCl3): δ = 5.92 (s, 1H, NH), 6.81–6.91, 7.09–7.12, 7.25–7.69, 7.76–7.88 7.98—8.01 and 8.07–8.26 (m, 12H, aromatics), 9,53 (s, 1H, OH), 12,32 (s, 1H, OH); MS (ESI): m/z (%): 304 [M-H]− (100). HR-MS [(C19H15NO3-H)]-, m/z 304.0972 (calc. 304.0972).
N-(4-diphenyl)methyl-2,3-dihydroxybenzamide (3)
79% yield; mp: 111–113 °C; 1H NMR (CDCl3): δ = 4.68 (d, 2H, CH2N, J = 5.49), 5.80 (s, 1H, OH), 6.62 (bb, 1H, NH), 6.73–6.78, 6.89–6.91, 7.05–7.09, 7.34–7.47 and 7.57–7.71 (m, 12H, aromatics), 12.68 (s, 1H, OH); 13C NMR (CDCl3): δ = 43.4, 113.8, 115.9, 118.2, 118.7, 127.1, 127.5, 127.7, 128.3, 128.8, 136.2, 140.5, 141.0, 146.0, 149.2, 169.9. MS (ESI): m/z (%): 318 [M-H]− (100). HR-MS [(C20H17NO3-H)]−, m/z 318.1127 (calc. 318.1136).
N-(4-diphenyl)methyl-N-methyl-2,3-dihydroxybenzamide (4)
33% yield; mp: 160–163 °C; 1H NMR ([D6]DMSO): δ = 3.14 (s, 3H, CH3), 4.81 (s, 2H, CH2), 5.80 (bb, 1H, OH), 6.70–6.75, 6.89–7.01, 7.33–7.66 (m, 12H, aromatics), 10.2 (bb, 1H, OH); MS (ESI) m/z (%): 356 [M-H + Na+]− (100). HR-MS [(C21H19NO3 + H)]+, m/z 334.1435 (calc. 334.1438).
2,3-dihydroxy-N-(4-phenoxyphenyl)benzamide (5)Citation38
84% yield; mp: 164–165 °C; 1H NMR (CDCl3): 5,82 (s, 1H, NH), 6.81–6.87, 7.00–7.15, 7.14–7.39, and 7.51–7.55 (m, 12H, aromatics), 7,93 (s, 2H, OH); 13C NMR (CD3OD): δ = 116.2, 118.1, 118.2, 118.5, 118.5, 118.8, 122.9, 123.1, 129.5, 133.1, 146.0, 148.4, 154.1, 157.5, 168.2. MS (ESI) m/z (%) 320 [M-H]− (100). HR-MS [(C19H15NO4-H)]−, m/z 320.0923 (calc. 320.0928).
N-(4-phenoxyphenyl)methyl-2,3-dihydroxy-benzamide (6)Citation38
79% yield; mp: 113–114 °C; 1H NMR (CDCl3): δ = 4.60 (d, J = 5.66, CH2N, 2H), 5.82 (s, 1H, OH), 6.59 (bb, 1H, NH), 6.72–6.91, 6.97–7.14, and 7.29–7.37 (m, 12H, aromatics), 12.67 (s, 1H, OH); 13C NMR (CD3OD): δ = 42.0, 115.3, 117.3, 118.2, 118.3, 118.3, 118.5, 122.9, 128.8, 129.4, 133.6, 145.9, 148.9, 156.4, 157.4, 170.0. MS (ESI) m/z (%): 334 [M-H]− (100). HR-MS [(C20H17NO4-H)]−, m/z 334.1074 (calc. 334.1085).
General procedure for the preparation of compounds 7–11
To a stirred suspension of the appropriate intermediate (29–33) (0.41 mmol) in THF/MeOH (16 mL, 3:1), 10% Pd/C (26 mg) was added. After stirring overnight under hydrogen atmosphere (6 atm), the reaction mixture was filtered through a pad of Celite and concentrated under reduced pressure affording the desired product.
N-(2,3-dihydroxyphenyl)-4-methoxyphenylsulfonamide (7)
82% yield; mp: 147–149 °C; 1H NMR ([D6]DMSO): δ = 3.78 (s, 3H, CH3), 6.53–6.59, 6.98–7.01, 7.64–7.67 (m, 7H, aromatics), 8.87 (bb, 3H, 2 OH, NH); MS (ESI) m/z (%): 294 [M-H]− (100), 171 (59). 13C NMR ([D6]DMSO): δ = 56.0, 112.8, 114.5, 114.7, 118.8, 125.6, 129.4, 132.6, 138.7, 146.0, 162.7. HR-MS [(C13H13NO5S-H)]−, m/z 294.0436 (calc. 294.0442).
N-(2,3-dihydroxyphenyl)-4-diphenylsulfonamide (8)Citation38
79% yield; mp: 182–183 °C; 1H NMR (DMSO-d6): δ 6.45–6.63, 7.38–7.50, 7.69–7.82 (m, 12H, aromatics), 9.00 (bb, 3H, 2- OH, -NH); 13C NMR ([D6]DMSO): δ = 113.0, 115.0, 118.8, 125.4, 127.5, 127.9, 128.9, 129.5, 138.9, 138.9, 139.9, 144.3, 146.1. MS (ESI): m/z 340 [M-H]− (93), 217 (100). HR-MS [(C18H15NO4S-H)]−, m/z 340.0643 (calc. 340.0649).
N-(2,3-dihydroxyphenyl)-4-phenoxyphenylsulfonamide (9)Citation38
80% yield; mp: 176–178 °C; 1H NMR (CDCl3): δ = 2.20–2.60 (bb, 3H, 2-OH and -NH), 6.54–6.58, 6.74–6.85, 6.93–6.96, 7.09–7.14, 7.28–7.33, 7.62–7.65 (m, 12H, aromatics); MS (ESI) m/z (%): 380 [M + Na]+ (100). 13C NMR (CD3OD): δ = 112.2, 114.8, 116.8, 118.6, 119.8, 124.5, 124.7, 129.3, 129.8, 133.3, 138.2, 145.3, 155.3, 161.5. HR-MS [(C18H15NO5S-H)]−, m/z 356.0594 (calc. 356.0598).
N-(2,3-dihydroxyphenyl)-N-methyl-4-diphenylsulfonamide (10)
57% yield; mp: 170–172 °C; 1H NMR (CDCl3): δ = 3.22 (s, 3H, NCH3), 5.99–6.02, 6.62–6.67, 6.87–6.89, and 7.43–7.71 (m, 14H, aromatics and -OH); MS (ESI) m/z (%): 354 [M-H]− (100), 217 (61). 13C NMR (CD3OD): δ = 37.6, 114.8, 118.4, 119.4, 126.9, 128.0, 128.1, 128.3, 128.7, 138.8, 139.2, 143.3, 145.4, 146.1. HR-MS [(C19H17NO4S-H)]−, m/z 354.0799 (calc. 320.0806).
N-(2,3-dihydroxyphenyl)-N-methyl-4-phenoxyphenylsulfonamide (11)
68% yield; mp: 150–152 °C; 1H NMR (CDCl3): δ = 3.16 (m, 3H, NCH3), 4.99 (bb, 2H, OH), 5.99–6.02, 6.65–7.54 (m, 12H, aromatics); MS (ESI) m/z (%): 370 [M-H]− (100); m/z 340 [M + Na]+ (100), 233 (81). 13C NMR (CD3OD): δ = 37.5, 114.7, 116.9, 118.3, 119.4, 119.9, 124.6, 128.0, 129.9, 130.0, 131.8, 143.3, 146.1, 155.3, 161.7. HR-MS [(C19H17NO5S-H)]−, m/z 370.0749 (calc. 370.0755).
General procedure for the preparation of compounds 12–17
To a solution of the appropriate sulfonamide 34–39 (1.36 mmol) in dry CH2Cl2 (20 mL) was added a 1M solution of boron tribromide (3.26 mmol) in CH2Cl2, at 0 °C and under nitrogen atmosphere. After 1 h at room temperature, methanol (5 mL) was added dropwise carefully at 0 °C, and the mixture was stirred for additional 10 min. The crude was diluted with a 1:1 mixture of AcOEt/H2O, the organic phase was washed with brine, dried, and purified by flash chromatography on silica gel affording the desired product.
N-(2,3-dihydroxyphenyl)methyl-4-diphenylsulfonamide (12)
55% yield; mp: 154–155 °C; 1H NMR (CD3OD): δ = 4.12 (s, 2H, CH2), 6.47–6.52, 6.58–6.63, 7.32–7.46, 7.57–7.66, 7.81–7.87 (m, 12H, aromatics); 13C NMR (CD3OD): δ 42.2, 114.2, 119.1, 120.1, 123.6, 127.1, 127.2, 127.4, 128.2, 128.9, 139.3, 139.6, 143.3, 144.8, 145.2. MS (ESI) m/z (%): 354 [M-H]− (7), 234 (100), HR-MS [(C19H17NO4S-H)]−, m/z 354.0811 (calc. 354.0806).
N-(2,3-dihydroxyphenyl)methyl-4-bromophenylsulfonamide (13)
75% yield; mp: 179–180 °C; 1H NMR (CD3OD): δ = 4.10 (s, 2H, CH2), 6.47–6.57, 6.61–6.64, 7.54–7.65 (m, 7H, aromatics); 13C NMR (CD3OD): δ = 42.2, 114.3, 119.1, 120.2, 123.3, 126.7, 128.6, 131.9, 141.2, 143.3, 144.7. MS (ESI) m/z (%): 356 [M-H]− (1), 236 (100), HR-MS [(C13H12BrNO4S-H)]−, m/z 355.9591 (calc. 355.9598).
N-(3,4-dihydroxyphenyl)-4-diphenylsulfonamide (14)
88% yield; mp: 216 °C (dec.); 1H NMR (CD3OD): δ = 6.37–6.41, 6.58–6.66, 7.31–7.44, 7.54–7.58, 7.62–7.66, 7.72–7.76 (m, 12H, aromatics); 13C NMR (CD3OD): δ = 111.2, 114.8, 115.1, 127.0, 127.1, 127.7, 128.3, 128.9, 129.3, 138.4, 139.3, 143.4, 145.4. MS (ESI) m/z (%): 340 [M-H]− (11), 153 (100), HR-MS [(C18H15NO4S-H)]−, m/z 340.0632 (calc. 340.0649).
N-(3,4-dihydroxyphenyl)-4-bromophenylsulfonamide (15)
62% yield; mp: 209 °C (dec.); 1H NMR (CD3OD): δ = 6.30–6.34, 6.57–6.61, 7.53–7.62 (m, 7H, aromatics); 13C NMR (CD3OD): δ = 111.4, 115.0, 115.1, 127.1, 128.9, 128.9, 132.0, 138.9, 143.6, 145.4. MS (ESI) m/z (%): 342 [M-H]− (12), 157 (100). HR-MS [(C12H10BrNO4S + Na)]+, m/z 365.9400 (calc. 365.9406).
N-(3,4-dihydroxyphenyl)methyl-4-diphenylsulfonamide (16)
58% yield; mp: 189 °C (dec.); 1H NMR (CD3OD): δ 3.92 (s, 2H, CH2), 6.47–6.51, 6.59–6.66, 7.37–7.50, 7.64–7.88, 7.72–7.76, 7.83–7.87 (m, 12H, aromatics); 13C NMR (CD3OD): δ = 114.9, 115.2, 119.4, 127.2, 127.3, 127.4, 128.2, 128.7, 128.9, 139.6, 139.6, 144.7, 145.1, 145.3. MS (ESI) m/z (%): 354 [M-H]− (7), 234 (100), HR-MS [(C19H17NO4S-H)]−, m/z 354.0811 (calc. 320.0806).
N-(3,4-dihydroxyphenyl)methyl-4-bromophenylsulfonamide (17)
47% yield; mp: 124–125 °C; 1H NMR (CD3OD): δ = 3.90 (s, 2H, CH2), 6.46–6.49, 6.61–65, 7.62–7.69 (m, 7H, aromatics); 13C NMR (CD3OD): δ = 46.6, 115.0, 115.2, 126.8, 128.5, 128.6, 132.1, 140.3, 144.7, 145.1. MS (ESI) m/z (%): 356 [M-H]− (1), 236 (100), HR-MS [(C13H12BrNO4S-H)]−, m/z 355.9538 (calc. 355.9598).
DPPH assay
The DPPH radical scavenging assay was performed in 96-well microplates according to the method reported by Blois with some modificationsCitation39,Citation40. Briefly, a freshly prepared solution of DPPH in methanol (100 μM final concentration) was added to test compounds methanolic solution. The mixtures were shaken vigorously and left to stand in the dark for 30 min at room temperature, and then absorbance was read at 520 nm using a spectrophotometric plate reader (Victor 3 Perkin–Elmer). The antioxidant activity was determined as the RSA% (radical scavenging activity), calculated using following equation:
where, Ao and Ai are the DPPH absorbance in the absence or in presence of antioxidant, respectively. Different sample concentrations were used in order to obtain antiradical curves for calculating the EC50 values. The value of EC50 was expressed in terms of molar ratio of antioxidant to DPPH. ARP is inverse of EC50 value, the larger the ARP the more efficient the antioxidant. The EC50 values and statistical analyses were processed using GraphPad PrismCitation41 and are expressed as mean ± SEM of at least three independent measurements in triplicate.
MMP inhibition assays
The catalytic domains of MMP-2, -8, and -9 were purchased from Enzo Life Sciences. The assays were performed in triplicate in 96-well white microtiter plates (Corning, NBS). For assay measurements, inhibitor stock solutions (DMSO, 25 mM) were diluted to six different concentrations (1 nM–250 μM) in fluorometric assay buffer (50 mM Tris·HCl pH 7.5, 200 mM NaCl, 1 mM CaCl2, 1 μM ZnCl2, 0.05% Brij-35, and 1% DMSO). Enzyme and inhibitor solutions were incubated in the assay buffer for 15 min at room temperature before the addition of the fluorogenic substrate solution (OmniMMP = Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2, Calbiochem, 2.5 μM final concentration). After further incubation for 2–4 h at 37 °C, fluorescence was measured (λex = 340 nm, λem = 405 nm) using a Perkin–Elmer Victor V3 plate reader.
Control wells lacked inhibitor. The MMP inhibition activity was expressed in relative fluorescence units (RFU). Percent inhibition was calculated from control reactions without inhibitor, IC50 values were determined using GraphPad PrismCitation41 and are expressed as mean ± SEM of at least three independent measurements in triplicate.
Expression and purification of the protein
The truncated form M80-G242 of the catalytic domain of MMP-8 was expressed in E. coli strain BL21 (DE3). At an OD600 0.5–0.6 expression of the collagenase was induced in a 1 l culture by adding IPTG to a final concentration of 0.5 mM. Inclusion bodies isolated and purified from harvested E. coli cells were resuspended in 20 mL of 6M urea, 100 mM β-mercaptoethanol and 20 mMTris, pH 8.5, and incubated at room temperature o/n under shaking to extract the solubilized collagenase. This extract was centrifuged for 30′ at 40 000 rpm, and the supernatant was loaded onto a Mono Q-Sepharose column (GE Healthcare) previously equilibrated with the denaturating buffer.
Elution of the collagenase was carried out by applying a linear gradient of 0–1 M NaCl in the same buffer at a flow rate of 1 mL/min. The truncated form of MMP-8 was eluted at a salt concentration of 100 mM NaCl and could be purified to apparent homogeneity. A further step of purification was carried out by gel filtration using a HiLoadSuperdex 75 column (GE Healthcare) equilibrated with 6M urea, 10 mM DTT, and 20 mM Tris, pH 8.5 at a flow rate of 0.5 mL/min. The collected protein was then refolded onto a HiLoadSuperdex 75 column in buffer MES-NaOH 3 mM, pH 6.0, 100 mM NaCl, 5 mM CaCl2, 0.5 mM ZnCl2, NaN3 0.02% at a flow rate of 0.5 mL/min. The fraction containing the desalted and refolded protein was eluted after ca. 13 mL.
Protein crystallization
The inhibitor (stock solution 50 mM in DMSO) was immediately added to the fraction containing the refolded protein in the ratio 3:1 (final concentration of DMSO 1%) in order to prevent autoproteolysis during concentration. The MMP-8 protein with the inhibitor was then concentrated with Amicon-Ultra 15, to a final concentration of 6 mg/mL. Crystallization was performed by hanging-drop vapor diffusion method at 20 °C. Hanging droplets were made by mixing 2 μL of protein/inhibitor solution with 5 μL of PEG solution (10% (m/v) PEG6000, 0.2 M MES-NaOH, 0.02% NaN3, pH 6.0). Droplets were concentrated against a reservoir buffer containing 1.0–2.0 M sodium phosphate, 0.02% NaN3, pH 6.0. Crystals appeared after few days.
Data collection and processing
X-ray data were collected under cryogenic conditions (100 K) at the ID29 beamline of ESRF, Grenoble, using a wavelength of 0.976 Å and a Pilatus 6M_F detector. The crystals were flash-frozen in the nitrogen stream after transferring them for few seconds into the mother solution containing 35% PEG400. Data were integrated and scaled using the programs MOSFLM and ScalaCitation42. The statistics of collection is given in of the supporting Information.
Structure solution and refinement
Structure solution was performed with AMoReCitation43 using the coordinates of the complex between MMP-8 and a non-zinc chelating inhibitor (PDB entry 3DPE)Citation44 as the starting model. The coordinates were then refined with CNSCitation45. The statistics of refinement is summarized in of the supporting Information.
Computational studies
Ligand-based studies: ligands reported in and were manually built using the Built facility in MaestroCitation37. 3D structures, stereoisomers, tautomers, and protomers at pH 7.0 ± 0.5 were generated with LigPrepCitation37. All synthesized analogs were aligned to the crystallographic coordinates of 14 with Phase Shape ScreeningCitation37 and, using the resulting aligned conformation, a pharmacophore-based QSAR model was built with Phase program. The minimum intersite distance between the features was set to be 1 Å. Molecules with pIC50 < 4 toward MMP-8 were grouped as the inactive molecule set, while those with pIC50 > 5 were grouped as the active molecules; all actives were set to be matched during the screening. The QSAR model was constructed using molecules 1, 12, 16, 17, 18, and 21 as test set and all the others as training set. The best model has 15 as reference molecule, and is made of four features: 1 aromatic ring (R9), 2 acceptor groups (A1 and A3), and 1 donor group (D6). Excluded volume features were added around the shape of reference molecule.
Structure-based studies: the hydrogen atoms of the X-ray complex–MMP-8:14 has been added and optimized with the Protein Preparation Wizard tool from SchrödingerCitation37. Then the complex with explicit waters was minimized to a derivative convergence of 0.05 kJ/mol Å using the Polak–Ribiere Conjugate Gradient (PRCG) minimization algorithm, the OPLS2005 force field, with MacroModel Embrace MinimizationCitation37. A shell of 15 Å around the catalytic zinc was set to be free to move, another shell of 5 Å minimized applying a force constant of 200 kJ/mol Å2.
Scheme 2. Reagents and conditions: (a) Et3N, CH2Cl2, reflux; (b) dry DMF, NaH, CH3I, 0 °C; (c) THF/MeOH, 10% Pd/C, H2, r.t. or BBr3, CH2Cl2, 0 °C 1 h.
The interaction energy between the receptor and each ligand was calculated with the Interaction energy mode implemented in Embrace. The same procedure was applied to calculate the interaction energies of MMP-2:14 and MMP-9:14. Ligand coordinates were taken from the MMP-8 bound conformation of 14. Protein and water coordinates were downloaded from the Protein Data Bank (PDB): 1GKC.pdbCitation46 and 1QIB.pdbCitation47 for MMP-9 and MMP-2, respectively. Then, interaction energies were calculated for all compounds reported in and previously aligned to 14. Calculated energies were used to derive ROC curves (see ). For the ROC curve calculations molecules with pIC50 >4 were considered active and all the others inactive.
Declaration of interest
The authors report no conflict of interest.
Supplementary material available online
IENZ_1217853_Supplementary_Material.pdf
Download PDF (253.8 KB)References
- Khokha R, Murthy A, Weiss A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat Rev Immunol 2013;13:649–65.
- Vandenbroucke RE, Libert C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat Rev Drug Discov 2014;13:904–27.
- Tauro M, Laghezza A, Loiodice F, et al. Arylamino methylene bisphosphonate derivatives as bone seeking matrix metalloproteinase inhibitors. Bioorg Med Chem 2013;21:6456–65.
- Rubino MT, Agamennone M, Campestre C, et al. Biphenyl sulfonylamino methyl bisphosphonic acids as inhibitors of matrix metalloproteinases and bone resorption. ChemMedChem 2011;6:1258–68.
- Dufour A, Overall CM. Missing the target: matrix metalloproteinase antitargets in inflammation and cancer. Trends Pharmacol Sci 2013;34:233–42.
- Tauro M, McGuire J, Lynch CC. New approaches to selectively target cancer-associated matrix metalloproteinase activity. Cancer Metastasis Rev 2014;33:1043–57.
- Gupta A, Kaur CD, Jangdey M, Saraf S. Matrix metalloproteinase enzymes and their naturally derived inhibitors: novel targets in photocarcinoma therapy. Ageing Res Rev 2014;13:65–74.
- Hwang KA, Yi BR, Choi KC. Molecular mechanisms and in vivo mouse models of skin aging associated with dermal matrix alterations. Lab Anim Res 2011;27:1–8.
- Quan T, Qin Z, Xia W, et al. Matrix-degrading metalloproteinases in photoaging. J Investig Dermatol Symp Proc 2009;14:20–4.
- Yaar M, Gilchrest BA. Photoageing: mechanism, prevention and therapy. Br J Dermatol 2007;157:874–87.
- Hsieh H-Y, Lee W-C, Senadi GC, et al. Discovery, synthetic methodology, and biological evaluation for antiphotoaging activity of bicyclic [1,2,3] triazoles: in vitro and in vivo studies. J Med Chem 2013;56:5422–35.
- Ceccoli JD, Costello B, Hayward JA, Lahanas KM. Bath & Body Works Brand Management, Inc., USA, assignee. Topical compositions for inhibiting matrix metalloproteases and providing antioxidative activities patent US20120058140A1; 2012.
- Khan SB, Kong C-S, Kim J-A, Kim S-K. Protective effect of Amphiroa dilatata on ROS induced oxidative damage and MMP expressions in HT1080 cells. Biotechnol Bioprocess Eng 2010;15:191–8.
- Gupta SK. Bioderm Research, Scottsdale, AZ, USA, assignee. Matrix metalloprotease (MMP) inhibitors and their application in cosmetic and pharmaceutical composition patent US20060074108A1; 2006.
- Yoon SO, Park SJ, Yoon SY, et al. Sustained production of H(2)O(2) activates pro-matrix metalloproteinase-2 through receptor tyrosine kinases/phosphatidylinositol 3-kinase/NF-kappa B pathway. J Biol Chem 2002;277:30271–82.
- Rao BG. Recent developments in the design of specific Matrix Metalloproteinase inhibitors aided by structural and computational studies. Curr Pharm Des 2005;11:295–322.
- Skiles JW, Gonnella NC, Jeng AY. The design, structure, and therapeutic application of matrix metalloproteinase inhibitors. Curr Med Chem 2001;8:425–74.
- Campestre C, Agamennone M, Tauro M, Tortorella P. Phosphonate emerging zinc binding group in matrix metalloproteinase inhibitors. Curr Drug Targets 2015;16:1634–44.
- Giustiniano M, Tortorella P, Agamennone M, et al. Amino acid derivatives as new zinc binding groups for the design of selective matrix metalloproteinase inhibitors. J Amino Acids 2013;178381, 12 p.
- Agrawal A, Romero-Perez D, Jacobsen JA, et al. Zinc-binding groups modulate selective inhibition of MMPs. ChemMedChem 2008;3:812–20.
- Jacobsen FE, Lewis JA, Cohen SM. The design of inhibitors for medicinally relevant metalloproteins. ChemMedChem 2007;2:152–71.
- Campestre C, Agamennone M, Tortorella P, et al. N-Hydroxyurea as zinc binding group in matrix metalloproteinase inhibition: mode of binding in a complex with MMP-8. Bioorg Med Chem Lett 2006;16:20–4.
- Yuan H, Lu W, Wang L, et al. Synthesis of derivatives of methyl rosmarinate and their inhibitory activities against matrix metalloproteinase-1 (MMP-1). Eur J Med Chem 2013;62:148–57.
- Mahmood T, Akhtar N. Combined topical application of lotus and green tea improves facial skin surface parameters. Rejuvenation Res 2013;16:91–7.
- Kim MS, Oh GH, Kim MJ, Hwang JK. Fucosterol inhibits matrix metalloproteinase expression and promotes type-1 procollagen production in UVB-induced HaCaT cells. Photochem Photobiol 2013;89:911–18.
- Gweon EJ, Kim SJ. Resveratrol attenuates matrix metalloproteinase-9 and -2-regulated differentiation of HTB94 chondrosarcoma cells through the p38 kinase and JNK pathways. Oncol Rep 2014;32:71–8.
- Kousidou OC, Mitropoulou TN, Roussidis AE, et al. Genistein suppresses the invasive potential of human breast cancer cells through transcriptional regulation of metalloproteinases and their tissue inhibitors. Int J Oncol 2005;26:1101–9.
- Ricciarelli R, Maroni P, Ozer N, et al. Age-dependent increase of collagenase expression can be reduced by alpha-tocopherol via protein kinase C inhibition. Free Radic Biol Med 1999;27:729–37.
- Alleva R, Tomasetti M, Sartini D, et al. alpha-Lipoic acid modulates extracellular matrix and angiogenesis gene expression in non-healing wounds treated with hyperbaric oxygen therapy. Mol Med 2008;14:175–83.
- Rubino MT, Maggi D, Laghezza A, et al. Identification of novel matrix metalloproteinase inhibitors by screening of phenol fragments library. Arch Pharm (Weinheim, Ger) 2011;344:557–63.
- Nicolotti O, Catto M, Giangreco I, et al. Design, synthesis and biological evaluation of 5-hydroxy, 5-substituted-pyrimidine-2,4,6-triones as potent inhibitors of gelatinases MMP-2 and MMP-9. Eur J Med Chem 2012;58:368–76.
- Rubino MT, Agamennone M, Campestre C, et al. Synthesis, SAR, and biological evaluation of alpha-sulfonylphosphonic acids as selective matrix metalloproteinase inhibitors. ChemMedChem 2009;4:352–62.
- Biasone A, Tortorella P, Campestre C, et al. alpha-Biphenylsulfonylamino 2-methylpropyl phosphonates: enantioselective synthesis and selective inhibition of MMPs. Bioorg Med Chem 2007;15:791–9.
- Pochetti G, Gavuzzo E, Campestre C, et al. Structural insight into the stereoselective inhibition of MMP-8 by enantiomeric sulfonamide phosphonates. J Med Chem 2006;49:923–31.
- Mishra KK, Pal RS, Arunkumar R, et al. Antioxidant properties of different edible mushroom species and increased bioconversion efficiency of Pleurotus eryngii using locally available casing materials. Food Chem 2013;138:1557–63.
- Hummer G. Molecular binding: under water’s influence. Nat Chem 2010;2:906–7.
- LigPrep version 3.4, Maestro version 10.2, MacroModel, version 10.8, Phase version 4.3, Schrödinger, LLC, New York, NY; 2015.
- Lauro G, Tortorella P, Bertamino A, et al. Structure-based design of microsomal prostaglandin E2 synthase-1 (mPGES-1) inhibitors using a virtual fragment growing optimization scheme. ChemMedChem 2016;11:612–19.
- Mishra K, Ojha H, Chaudhury NK. Estimation of antiradical properties of antioxidants using DPPH[rad] assay: a critical review and results. Food Chem 2012;130:1036–43.
- Sharma OP, Bhat TK. DPPH antioxidant assay revisited. Food Chem 2009;113:1202–5.
- GraphPad Prism-5.0c: GraphPad Software Inc., San Diego, CA; 2009.
- Battye TG, Kontogiannis L, Johnson O, et al. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 2011;67:271–81.
- Navaza J. Amore – an automated package for molecular replacement. Acta Cryst A 1994;50:157–63.
- Pochetti G, Montanari R, Gege C, et al. Extra binding region induced by non-zinc chelating inhibitors into the S1′ subsite of matrix metalloproteinase 8 (MMP-8). J Med Chem 2009;52:1040–9.
- Brunger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 1998;54:905–21.
- Rowsell S, Hawtin P, Minshull CA, et al. Crystal structure of human MMP9 in complex with a reverse hydroxamate inhibitor. J Mol Biol 2002;319:173–81.
- Dhanaraj V, Williams MG, Ye QZ, et al. X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor. Croat Chem Acta 1999;72:575–91.