Abstract
Carbonic anhydrases (CAs) are metalloenzymes, and classified into the evolutionarily distinct α, β, γ, δ, ζ, and η classes. α-CAs are present in many living organisms. β- and γ-CAs are expressed in most prokaryotes and eukaryotes, except for vertebrates. δ- and ζ-CAs are present in phytoplanktons, and η-CAs have been found in Plasmodium spp. Since the identification of α- and β-CAs in Caenorhabditis elegans, the nematode CAs have been considered as an emerging target in research focused on antiparasitic CA inhibitors. Despite the presence of α-CAs in both helminths and vertebrates, structural studies have revealed different kinetic and inhibition results. Moreover, lack of β-CAs in vertebrates makes this enzyme as an attractive target for inhibitory studies against helminthic infection. Some CA inhibitors, such as sulfonamides, have been evaluated against nematode CAs. This review article aims to present comprehensive information about the nematode CAs and their inhibitors as potential anthelminthic drugs.
Introduction
Carbonic anhydrases (CAs, EC 4.2.1.1) are classified into six evolutionarily distinct classes: α, β, γ, δ, ζ, and ηCitation1,Citation2. The active sites of these enzymes contain most commonly a zinc ion (Zn2+), which plays a crucial role in the catalytic activity of the metalloenzymesCitation3. ζ and γ-CAs contain cadmium (ζ), iron (γ), or cobalt (γ) as cofactorsCitation3,Citation4 instead of Zn2+. CAs catalyze the reversible hydration of carbon dioxide (CO2) to bicarbonate ions (HCO3−) and protons (H+). α-CAs are present in many prokaryotic and eukaryotic organisms, such as vertebrates, invertebrates, and plants. In total, 13 enzymatically active α-CAs have been discovered in mammals, including: CA I, CA II, CA III, CA IV, CA VA, CA VB, CA VI, CA VII, CA IX, CA XII, CA XIII, CA XIV, and CA XVCitation5. β-CAs are expressed in many archaea, prokaryotes, and eukaryotes (fungi, algae, plants, protozoans, arthropods, and nematodes)Citation5–10. γ-CAs are found in archaea, plants, and some bacteria. δ- and ζ-CAs are present in many species of marine phytoplanktonCitation5. The newly discovered η-CAs are the only genus specific CAs, present in Plasmodium spp. (the causative protozoan of malaria), and contain Zn2+ in their catalytic active siteCitation2. CAs catalyze several important metabolic and biochemical functions, such as pH homeostasisCitation11, electrolyte transferCitation12, calcificationCitation13, gluconeogenesisCitation14, lipogenesisCitation15, and ureagenesisCitation16. β-CA is a critical metalloenzyme that catalyzes many biological pathways in both prokaryotes and eukaryotes including protozoans, insects, and nematodes. β-CA produces bicarbonate for carboxylases in Corynebacterium glutamicumCitation17, urease in H. pyloriCitation18, and cyanase to detoxify cyanate in Escherichia coliCitation19 and Ascaris lumbricoidesCitation20. Crystal structures of β-CAs demonstrated that two cysteines and one histidine are conserved and are ligated to a zinc ion in the catalytic active site of the enzymeCitation21. A study from 2010 introduced two metazoan β-CAs that were encoded by two different β-CAs genes (y116a8c(0).28 and bca-1) in Caenorhabditis elegansCitation6,Citation22. In another study of 2010, a β-CA gene (DmBCA) was detected from Drosophila melanogasterCitation6.
α-CAs are generally present in all metazoan species (vertebrates and invertebrates), whereas β-CAs are found only in invertebrate metazoansCitation6 (arthropods and nematodes). Nematodes are categorized into several phyla: Annelida (segmented worms), Chaetognatha (arrow worms), Gnathostomulid (jaw worms), Hemichordata (acorn/tongue worms), Nematoda (roundworms), Nematomorpha (horsehair worms), Nemertea (ribbon worms), Onychophora (velvet worms), Phoronida (horseshoe worms), Platyhelminthes (flatworms), Priapulida (phallus worms), and Sipuncula (peanut worms). Most of α- and β-CA-containing nematodes (both pathogenic and nonpathogenic) have been classified into Nematoda and Platyhelminthes phyla, respectivelyCitation23. The details of α- and β-CA expressing nematodes, platyhelminthes, annelids, and hemichordates are shown in . Among different phyla, some groups, such as Nematoda and Platyhelminthes, are very important as zoonotic helminths in both human and veterinary medicine. Some phyla, such as Hemichordata and Annelida, are generally considered as nonpathogenic nematodes. However, CAs have also been discovered in a few annelids, namely in Hirudo medicinalisCitation24, Riftia pachyptilaCitation25, and Osedax bone wormsCitation26.
Table 1. α- and β-CA expressing nematoda, platyhelminthes, annelida, and hemichordate.
In this review article, we present recent discoveries regarding nematode α- and β-CAs. Moreover, the inhibition studies of these enzyme families are described as they represent plausible targets for designing novel anti-infective drugs.
α-CA expressing nematodes
A wide range of different α-CA proteins are expressed in nematodes. Despite only one representative α-CA protein ID shown for each species in , α-CA protein sequences can differ both within and between nematodes. Previous studies have identified α-CAs from some nematodes, including C. elegans and Ostertagia ostertagiCitation27–29. The genome of C. elegans encodes for six α-CAs, namely CAH-1, CAH-2, CAH-3, CAH-4, CAH-5, and CAH-6Citation29, with CAH-4 present as two isoforms CAH-4a and CAH-4b. In 2011, Fasseas et al. concluded that only CAH-3, CAH-4, and CAH-5 possess the three conserved zinc-binding histidine residues which are also present in active human CAs. CAH-3 was shown to be an active enzyme, and CAH-4 was found to be active in vitro, while the other four CAs (CAH-1, CAH-2, CAH-5, and CAH-6) were presented as CA-related proteins (CARPs) without any CA activity (). They also discovered that the silencing of cah-3 and cah-4 genes seemed to affect the lifespan of C. elegans.
Figure 1. Multiple sequence alignment (MSA) of α-CA protein sequences from nematodes. MSA of 43 and one α-CA protein sequences from nematodes and Klebsiella pneumoniae (outgroup), respectively, aligned using Clustal Omega algorithm from EMBL-EBI database (http://www.ebi.ac.uk/Tools/msa/clustalo/)Citation45. MSA was conducted on 60 amino acids of α-CA protein sequences starting three amino acids prior to the first highly conserved histidine. The three highly conserved histidines locate in the catalytic active site of α-CAs and participate in binding with the Zn2+ ion (shown by red arrows).

O. ostertagi (Brown Stomach Worm) is a parasitic nematode (helminth) and the causative agent of ostertagiosis in cattle. The CA gene from O. ostertagi has been named as OoCACitation27. The nucleotide sequence of OoCA gene is 78% and 55% identical with the cah-6 and CA3 genes of C. elegans and human, respectively. Studies have suggested that O. ostertagi CA may play a critical role in the immediate early developmental events following exsheathment initiation. Exsheathment is the first step at the beginning of O. ostertagi infection, and involves the casting of the second larval stage cuticle, which is retained by the infective third-stage larvae. Inhibition of this O. ostertagi development process may hinder the formation of the infective forms of O. ostertagi. In addition, it has been previously shown that ethoxzolamide affected the development of H. contortus via some changes in the excretory cells and esophagus during exsheathmentCitation30. The potential of parasitic α-CAs as drug targets is limited by possible or even probable effects on host α-CAs, which predispose to various adverse effects.
β-CA expressing nematodes
β-CAs from nematodes are particularly interesting, because these enzymes are not found in vertebrates (including humans). Inhibition of nematode β-CAs presents possibilities to treat or restrict many helminthic infections with minimal side effects on the hosts. Noteworthy is the fact that even though the β-CAs and other CA groups catalyze the same reaction, their protein structure is different. First, the Zn2+ ion in the active site of β-CAs is coordinated by two cysteines and one histidine instead of three histidines ()Citation31. Second, β-CAs can be found in many oligomeric states, whereas α-CAs mainly occur as monomers and γ-CAs as trimers. Thus far, a variety of multimeric crystal structures, including dimeric, tetrameric, hexameric, and octameric β-CAs have been reportedCitation32–34. The monomeric components of a dimeric β-CA bind to each other usually by noncovalent interactions, and in some cases via a short polypeptide linker. The latter case is called a “pseudo-dimer”Citation35. Tetrameric and octameric β-CAs are formed whenever dimers form dimer-of-dimers and dimer-of-dimer-of-dimers, respectively. The most frequently available quaternary structure of β-CAs is the tetrameric state. However, a dimeric β-CA seems to be the fundamental structural unit in β-CA protein structures.
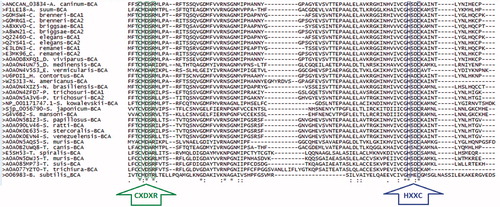
Figure 2. Multiple sequence alignment (MSA) of β-CA protein sequences from nematodes. MSA of 31 and one β-CA protein sequences from nematodes and Bacillus subtilis (outgroup), respectively, aligned using Clustal Omega algorithm from EMBL-EBI database (http://www.ebi.ac.uk/Tools/msa/clustalo/)Citation45. MSA was conducted on 115 amino acids of β-CA protein sequences starting three amino acids prior to the first highly conserved sequence (CXDXR; C: Cysteine, D: Aspartic acid, R: Arginine, D: any residue). The first (CXDXR) and second (HXXC; H: Histidine, C: Cysteine, X: any residue) highly conserved sequences locate in the catalytic active site of β-CAs and are coordinated with the Zn2+ ion (shown by green and blue arrows, respectively).
Phylogenetic analysis of nematode α- and β-CAs
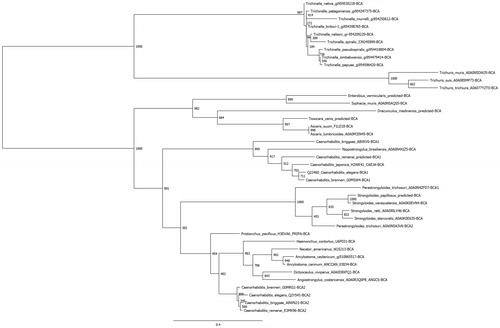
Nematode α-CAs and β-CAs were identified by BLASTP searches of the NCBI and UniProt databases using previously annotated members of both groups as query sequences, and all parameters as default. For each group, retrieved sequences were aligned using Clustal Omega and visually inspected for regions of incompleteness or poor quality. For those species containing a suspect sequence, the whole genome was downloaded and utilizing the exonerate program the remaining proposed high-quality sequences used as templates to predict gene and protein sequences. Subsequently, 14 genomes were analyzed for suspect sequences in the α-CA group and eight predictions kept, and ten genomes for the β-CA group with five predictions kept. In total, 54 α-CAs and 41 β-CAs protein sequences were aligned independently and used to perform phylogenetic analyses. The PhyML program was used to perform the phylogenetic analyses, utilizing the LG model and 1000 bootstraps; the results were visualized using FigTree and are presented as trees in and . Super-computing resources from the Center for Science and Computing of the Ministry of Finland were used to perform these analyses. Even at the amino acid level, the sequences for the nematode CAs were significantly different, which is evidenced in low bootstrap values within some subclades. Conversely, most branch points delineating subgroups of like organisms are well supported. This could imply that these species have possessed their CA sequences for a long time, and due to the fact that nontruncated versions of the sequences were used, ancillary domains accompanying the CA domain are providing a variation in function or localization of protein by species.
Figure 3. Phylogenetic analysis of nematode α-CAs. A total of 54 nematode α-CAs were aligned using Clustal Omega and used to perform a phylogenetic analysis using the LG model in the PhyML program, with 1000 bootstraps. Node values indicate bootstrap values. Any sequences which were predicted from the full genomic sequence are labeled as “predicted”.
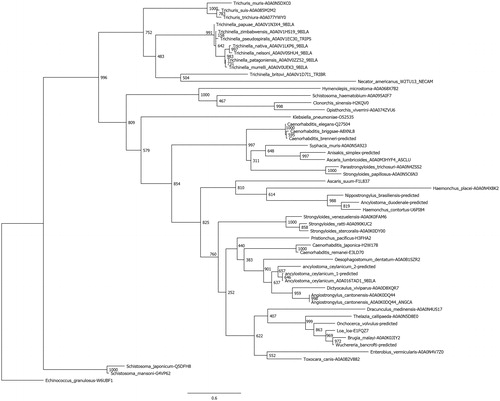
Figure 4. Phylogenetic analysis of nematode β-CAs. A total of 41 nematode β-CAs were aligned using Clustal Omega and used to perform a phylogenetic analysis using the LG model in the PhyML program, with 1000 bootstraps. Node values indicate bootstrap values. Any sequences which were predicted from the full genomic sequence are labeled as “predicted”.
Subcellular location of nematode α- and β-CAs
The plant β-CAs have been localized to the cytoplasm of cells and thylakoid space of chloroplastic stromaCitation36. In cyanobacteria, β-CA is localized in the carboxysome organelle, and is therefore involved in the CO2-concentration processCitation36,Citation37. Subcellular localization studies have indicated that DmBCA is probably a mitochondrial enzymeCitation6,Citation7,Citation38. The prediction of subcellular localization of α- and β-CAs from nematodes was performed using TargetP 1.1 Server (http://www.cbs.dtu.dk/services/TargetP/)Citation39. The results are shown in . The prediction results revealed that many nematode α-CAs contain signal peptides which target them to the secretory pathway. The prediction results further revealed that some nematode β-CAs including those in C. brenneri (G0MSW4 and G0MRG1), C. briggsae (A8WN21), C. remanei (E3MK96), E. vermicularis (A0A0N4V5S3), H. contortus (U6PDI1), Nippostrongylus brasiliensis (A0A0N4XIZ5), S. kowalevskii (NP_001171747.1), S. muris (A0A0N5AQS5), T. spiralis (E5SH53), T. muris (A0A0N5DWJ5), and T. trichiura (A0A077YZT0) contain a mitochondrial targeting sequence. In fact, both α- and β-CAs from C. brenneri seem to contain a mitochondrial targeting sequence.
Inhibitory studies on nematode α-CAs
To evaluate the effect of a CA inhibitor, acetazolamide, on a nematode species, it was first tested against live C. elegans. The test revealed that this inhibitor could not penetrate the nematode cuticleCitation28. Additionally, thiobendazole-5-sulfonamide (a sulfonamide derivative of thiobendazole) was tested, and it inhibited efficiently (Ki 9.5 nM) the CAH-4b of C. elegansCitation40. Thiobendazole is widely used as an antiparasitic agent against both human and animal parasitic infections. The mechanism of action for thiobendazole is poorly understood, but it has been suggested that it interferes with the formation of cytoplasmic microtubules and cytoskeletons. Moreover, 2-(hydrazinocarbonyl)-3-substituted-phenyl-1H-indole-5-sulfonamides have been tested against C. elegans CAH-4b. Some of the tested compounds, including acetazolamide, ethoxzolamide, and a series of sulfonamide derivatives possessing various 2-, 3-, or 4-substituted phenyl groups with methyl-, halogeno-, and methoxy-functionalities, as well as the perfluorophenyl moiety, showed very significant inhibitory effects on CAH-4b with Kis between 6.0 and 13.4 nMCitation41.
Inhibitory studies on nematode β-CAs
Inhibition data available for nematode β-CAs are still very limited. The first studies on nematode enzymes were performed on C. elegans and A. lumbricoidesCitation20,Citation22. In 2009, Fasseas et al. characterized C. elegans β-CA for the first timeCitation22. They discovered two β-CAs in C. elegans, BCA-1, and Y116A8C.28, of which the latter was found to possess catalytic activity with kcat and kcat/Km of 2.77 × 104 s−1 and 6383 × 105M−1s−1, respectively. However, the sequence for BCA-1 was obviously incorrect, suggesting that both β-CAs from C. elegans might be active enzymesCitation6. The RNAi studies of C. elegans β-CA did not reveal any visible phenotype, while silencing the β-CA gene in fruit fly Drosophila melanogaster caused complete infertility in femalesCitation38. Hence, this result suggested crucial biochemical and physiological roles for β-CAs in insects, and possibly all invertebrates. β-CA has been characterized recently from A. lumbricoides, the causative agent of zoonotic ascariasis. The enzyme was identified using bioinformatic and computational biology methods. A. lumbricoides recombinant β-CA protein (AlBCA) was produced in Sf9 insect cells, and the kinetic parameters were investigated. Based on the resultsCitation20, AlBCA possesses high catalytic activity with Km 6.0 × 105 s−1 and kcat/Km 4.3 × 107 M−1s−1. In addition, the Ki for inhibition of AlBCA by acetazolamide was 84.1 nM. Meanwhile, the kinetic and inhibition studies were also performed on the produced recombinant β-CAs from D. melanogaster (DmBCA)Citation42, Anopheles gambiae (AgaCA)Citation43, and Leishmania donovaniCitation9. These results are shown for comparison in .
Table 2. Enzyme activity and inhibition data of β-CAs from protozoa, nematode, and insects.
In vivo studies have not been conducted so far, but functional predictions of A. lumbricoides β-CA suggested that this enzyme may play important roles in bicarbonate dependent biosynthetic/metabolic pathways, such as gluconeogenesis and detoxification of metabolically produced cyanate by bicarbonate-dependent cyanase.
β-CAs from nematodes as vaccine candidates
The β-CA enzyme is highly distributed among infectious agents, including helminths. Therefore, it is a potential molecular target for controlling parasites and pests in all fields of human and veterinary medicine and agricultureCitation10. Absence of β-CA in vertebrates also makes this enzyme a potential target protein for vaccines. The detection of antigenic sites of antigens and proteins is an important step in designing an effective vaccine candidate. For this purpose, 31 β-CA protein sequences from nematodes were analyzed with the European Molecular Biology Open Software Suite (EMBOSS) program Antigenic (http://emboss.bioinformatics.nl/cgi-bin/emboss/antigenic), which is based on the Kolaskar and Tongaonkar methodCitation44. The antigenic site prediction results revealed that the second highly conserved sequence (HXXC) represents an epitope with the highest score for a probable antigenic site among the nematode β-CAs (). On the other hand, a previous homology modeling study on β-CA from A. caninum defined that this epitope is located in the active site of enzyme and is mainly buried (). In addition, most β-CAs are intracellular proteins, which make them inaccessible for immunological responses of the host. Therefore, β-CA inhibitors should be considered as a better option for developing new treatment strategies against parasitic or helminthic infections.
Figure 5. Accessibility identification of the predicted antigenic epitope of β-CA from A. caninum. The molecular surface of the homology model of A. caninum β-CA is shown as solid gray, and the second highly conserved sequence (HXXC) as the target epitope is buried from the surface of the protein. The exposed and buried residues of epitope are shown with red and green spheres and numbered. Figure adopted with author’s permission from open accessCitation10.
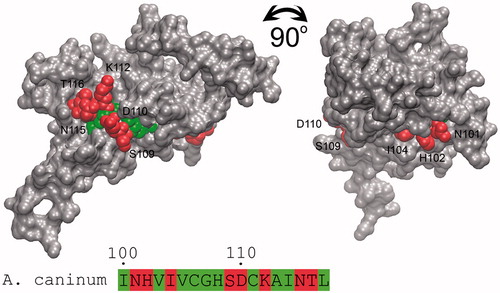
Table 3. Predicted antigenic sites of 31 β-CAs from nematodes.
Conclusion
As the overall conclusion, inhibitory studies have demonstrated that acetazolamide is able to efficiently inhibit β-CA from a nematode, A. lumbricoides. Other studies have also shown that both acetazolamide and ethoxzolamide significantly inhibit an α-CA from C. elegans (CAH-4b). Even though the literature on nematode β-CAs is still rather limited, β-CAs can be considered more attractive than α-CAs as potential targets for anti-helminthic drugs. This is based on the fact that β-CAs are absent from the proteomes of vertebrates. Therefore, β-CAs could represent a helminthic-specific drug targets with minimal side effects on the infected vertebrate host.
Declaration of interest
The authors declare that they have no competing interests.
To perform the original studies, RZE received a scholarship support from the Ministry of Science, Research and Technology, and National Institute of Genetic Engineering and Biotechnology of Islamic Republic of Iran. The studies were also supported by research grants from the Academy of Finland to SP, Finnish Funding Agency for Innovation (TEKES) to SP, Finnish Cultural Foundation (Pirkanmaa Regional Fund and Maili Autio Fund) to HRB and RZE, Sigrid Juselius Foundation to SP, Jane and Aatos Erkko Foundation to SP, Tampere Tuberculosis Foundation to SP, and Competitive Research Funding of the Tampere University Hospital.
References
- Zimmerman SA, Ferry JG. The beta and gamma classes of carbonic anhydrase. Curr Pharm Des 2008;14:716–21
- Del Prete S, Vullo D, Fisher GM, et al Discovery of a new family of carbonic anhydrases in the malaria pathogen Plasmodium falciparum – the eta-carbonic anhydrases. Bioorg Med Chem Lett 2014;24:4389–96
- Ferry JG. The gamma class of carbonic anhydrases. Biochim Biophys Acta 2010;1804:374–81
- Lane TW, Saito MA, George GN, et al Biochemistry: a cadmium enzyme from a marine diatom. Nature 2005;435:42
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81
- Syrjanen L, Tolvanen M, Hilvo M, et al Characterization of the first beta-class carbonic anhydrase from an arthropod (Drosophila melanogaster) and phylogenetic analysis of beta-class carbonic anhydrases in invertebrates. BMC Biochem 2010;11:28
- Zolfaghari Emameh R, Barker H, Tolvanen ME, et al Bioinformatic analysis of beta carbonic anhydrase sequences from protozoans and metazoans. Parasit Vectors 2014;7:38
- Zolfaghari Emameh R, Syrjanen L, Barker H, et al Drosophila melanogaster: a model organism for controlling Dipteran vectors and pests. J Enz Inhib Med Chem 2015;30:505–13
- Syrjanen L, Vermelho AB, Rodrigues Ide A, et al Cloning, characterization, and inhibition studies of a beta-carbonic anhydrase from Leishmania donovani chagasi, the protozoan parasite responsible for leishmaniasis. J Med Chem 2013;56:7372–81
- Zolfaghari Emameh R, Barker H, Hytonen VP, et al Beta carbonic anhydrases: novel targets for pesticides and anti-parasitic agents in agriculture and livestock husbandry. Parasit Vectors 2014;7:403
- Browning JA, Wilkins RJ. Mechanisms contributing to intracellular pH homeostasis in an immortalized human chondrocyte cell line. Comp Biochem Physiol A Mol Integr Physiol 2004;137:409–18
- Lee MG, Ohana E, Park HW, et al Molecular mechanism of pancreatic and salivary gland fluid and HCO3 secretion. Physiol Rev 2012;92:39–74
- Benesch R. Carbonic anhydrase and calcification. Ann NY Acad Sci 1984;429:457–8
- Dodgson SJ, Cherian K. Rat renal proximal tubular gluconeogenesis: possible involvement of nonmitochondrial carbonic anhydrase isozymes. Arch Biochem Biophys 1990;282:1–7
- Lynch CJ, Fox H, Hazen SA, et al Role of hepatic carbonic anhydrase in de novo lipogenesis. Biochem J 1995;310:197–202
- Dodgson SJ. Inhibition of mitochondrial carbonic anhydrase and ureagenesis: a discrepancy examined. J Appl Physiol 1987;63:2134–41
- Mitsuhashi S, Ohnishi J, Hayashi M, Ikeda M. A gene homologous to beta-type carbonic anhydrase is essential for the growth of Corynebacterium glutamicum under atmospheric conditions. Appl Microbiol Biotechnol 2004;63:592–601
- Nishimori I, Onishi S, Takeuchi H, Supuran CT. The alpha and beta classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr Pharm Des 2008;14:622–30
- Guilloton MB, Lamblin AF, Kozliak EI, et al A physiological role for cyanate-induced carbonic anhydrase in Escherichia coli. J Bacteriol 1993;175:1443–51
- Zolfaghari Emameh R, Kuuslahti M, Vullo D, et al Ascaris lumbricoides beta carbonic anhydrase: a potential target enzyme for treatment of ascariasis. Parasit Vect 2015;8:479
- Tripp BC, Smith K, Ferry JG. Carbonic anhydrase: new insights for an ancient enzyme. J Biol Chem 2001;276:48615–18
- Fasseas MK, Tsikou D, Flemetakis E, Katinakis P. Molecular and biochemical analysis of the beta class carbonic anhydrases in Caenorhabditis elegans. Mol Biol Rep 2010;37:2941–50
- Blair JE, Ikeo K, Gojobori T, Hedges SB. The evolutionary position of nematodes. BMC Evol Biol 2002;2:7
- Riehl B, Schlue WR. Evidence for two isoforms of carbonic anhydrase II in the leech (Hirudo medicinalis) central nervous system. Comp Biochem Physiol B 1993;106:717–18
- Sanchez S, Andersen AC, Hourdez S, Lallier FH. Identification, sequencing, and localization of a new carbonic anhydrase transcript from the hydrothermal vent tubeworm Riftia pachyptila. FEBS J 2007;274:5311–24
- Tresguerres M, Katz S, Rouse GW. How to get into bones: proton pump and carbonic anhydrase in Osedax boneworms. Proc Biol Sci 2013;280:20130625
- DeRosa AA, Chirgwin SR, Williams JC, Klei TR. Isolation and characterization of a gene encoding carbonic anhydrase from Ostertagia ostertagi and quantitative measurement of expression during in vivo exsheathment. Vet Parasitol 2008;154:58–66
- Hall RA, Vullo D, Innocenti A, et al External pH influences the transcriptional profile of the carbonic anhydrase, CAH-4b in Caenorhabditis elegans. Mol Biochem Parasitol 2008;161:140–9
- Fasseas MK, Tsikou D, Flemetakis E, Katinakis P. Molecular and biochemical analysis of the alpha class carbonic anhydrases in Caenorhabditis elegans. Mol Biol Rep 2011;38:1777–85
- Davey KG, Sommerville RI, Rogers WP. The effect of ethoxyzolamide, an analog of insect juvenile hormone, nor-adrenaline and iodine on changes in the optical path difference in the excretory cells and esophagus during exsheathment in Haemonchus contortus. Int J Parasitol 1982;12:509–13
- Cox EH, McLendon GL, Morel FM, et al The active site structure of Thalassiosira weissflogii carbonic anhydrase 1. Biochemistry 2000;39:12128–30
- Kimber MS, Pai EF. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. EMBO J 2000;19:1407–18
- Smith KS, Ferry JG. Prokaryotic carbonic anhydrases. FEMS Microbiol Rev 2000;24:335–66
- Strop P, Smith KS, Iverson TM, et al Crystal structure of the “cab”-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J Biol Chem 2001;276:10299–305
- Rowlett RS. Structure and catalytic mechanism of β-carbonic anhydrases. Sub-Cell Biochem 2014;75:53–76
- Rowlett RS. Structure and catalytic mechanism of the beta-carbonic anhydrases. Biochim Biophys Acta 2010;1804:362–73
- Fukuzawa H, Suzuki E, Komukai Y, Miyachi S. A gene homologous to chloroplast carbonic anhydrase (icfA) is essential to photosynthetic carbon dioxide fixation by Synechococcus PCC7942. Proc Natl Acad Sci USA 1992;89:4437–41
- Syrjanen L, Valanne S, Kuuslahti M, et al. β carbonic anhydrase is required for female fertility in Drosophila melanogaster. Front Zool 2015;12:19
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 2000;300:1005–16
- Crocetti L, Maresca A, Temperini C, et al A thiabendazole sulfonamide shows potent inhibitory activity against mammalian and nematode alpha-carbonic anhydrases. Bioorg Med Chem Lett 2009;19:1371–5
- Guzel O, Innocenti A, Hall RA, et al Carbonic anhydrase inhibitors. The nematode alpha-carbonic anhydrase of Caenorhabditis elegans CAH-4b is highly inhibited by 2-(hydrazinocarbonyl)-3-substituted-phenyl-1H-indole-5-sulfonamides. Bioorg Med Chem 2009;17:3212–15
- Syrjanen L, Parkkila S, Scozzafava A, Supuran CT. Sulfonamide inhibition studies of the beta carbonic anhydrase from Drosophila melanogaster. Bioorg Med Chem Lett 2014;24:2797–801
- Syrjanen L, Kuuslahti M, Tolvanen M, et al The beta-carbonic anhydrase from the malaria mosquito Anopheles gambiae is highly inhibited by sulfonamides. Bioorg Med Chem 2015;23:2303–9
- Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett 1990;276:172–4
- Sievers F Higgins DG. Clustal omega. Curr Prot Bioinformatics 2014;48:3.13.1–16
- Howe KL, Bolt BJ, Cain S, et al WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res 2016;44:D774–80
- Brown GR, Hem V, Katz KS, et al Gene: a gene-centered information resource at NCBI. Nucleic Acids Res 2015;43:D36–42
