Abstract
We cloned, expressed, purified, and determined the kinetic constants of the recombinant α-carbonic anhydrase (rec-MgaCA) identified in the mantle tissue of the bivalve Mediterranean mussel, Mytilus galloprovincialis. In metazoans, the α-CA family is largely represented and plays a pivotal role in the deposition of calcium carbonate biominerals. Our results demonstrated that rec-MgaCA was a monomer with an apparent molecular weight of about 32 kDa. Moreover, the determined kinetic parameters for the CO2 hydration reaction were kcat = 4.2 × 105 s−1 and kcat/Km of 3.5 × 107 M−1 ×s−1. Curiously, the rec-MgaCA showed a very similar kinetic and acetazolamide inhibition features when compared to those of the native enzyme (MgaCA), which has a molecular weight of 50 kDa. Analysing the SDS-PAGE, the protonography, and the kinetic analysis performed on the native and recombinant enzyme, we hypothesised that probably the native MgaCA is a multidomain protein with a single CA domain at the N-terminus of the protein. This hypothesis is corroborated by the existence in mollusks of multidomain proteins with a hydratase activity. Among these proteins, nacrein is an example of α-CA multidomain proteins characterised by a single CA domain at the N-terminus part of the entire protein.
Introduction
Carbonic anhydrases superfamily (CAs, EC 4.2.1.1) are metalloenzymes, which have been found in all the three domains of life (Eubacteria, Archaea, and Eukarya) and represent a very interesting example of convergent/divergent evolution phenomenon with seven known families: α-, β-, γ-, δ-, ζ-, η-, and θ-CAsCitation1–3. In fact, despite the low sequence similarity existing between the seven known CA families, they evolved analogous structures characterised by the following features: (i) catalyse a simple but physiologically relevant reaction consisting in the hydration of carbon dioxide to bicarbonate and protonsCitation4–8; (ii) the catalytically active form of the enzyme is the metal hydroxide derivativeCitation1–3; (iii) the rate determining step of the entire catalytic turnover is the formation of the metal hydroxide species of the enzyme by the transfer of a proton from the metal-coordinated water molecule to the surrounding solventCitation2–4,Citation6–17. The CA macromolecules are grouped in the seven different classes mainly on the basis of their structural fold and arrangement of the active site residues. The α-, β-, δ-, η-, and perhaps θ-CAs are characterised by a Zn(II) ion in the active site. γ-CAs are probably Fe(II) enzymes, although this family is also active with bound Zn(II) or Co(II) ionsCitation18–25. ζ-CAs are cambialistic enzymes, active both with Cd(II) or Zn(II) bound within the active siteCitation26–28. The metal ion from the CA active site is coordinated by three His residues in the α-, γ-, δ- and, probably, θ-classes; by one His, and two Cys residues in β- and ζ-CAs or by two His and one Gln residues in the η-class, with the fourth ligand being a water molecule/hydroxide ion acting as nucleophile in the catalytic cycle of the enzymeCitation1,Citation5–7,Citation29,Citation30. Some of the catalytically active α-CAs also catalyse the hydrolysis of esters/thioesters, e.g. 4-nitrophenyl acetate (4-NpA) hydrolysis, as well as other hydrolytic reactions. However, no esterase activity was detected so far for enzymes belonging to the other five CA genetic families. The tri-dimensional fold of the five CA classes is very different: α-CAs are normally monomers and rarely dimers; β-CAs are dimers, tetramers, or octamers; γ-CAs are trimersCitation19,Citation20,Citation23,Citation31. The only ζ-CA crystallised so far has three slightly different active sites on the same polypeptide chain, whereas no X-ray crystal structures of δ-, η-, and θ-CAs are available so far. All CAs identified in animal systems belong to α-classCitation32,Citation33. CAs identified in plants and algae belong to the α-, β-, γ-, δ-, and θ-classes; fungi encode for α- and β-CAs; protozoa encode for α-, β-, or η-CAs; bacteria encode for enzymes belonging to the α-, β-, and γ-CA classesCitation4–7,Citation12,Citation34,Citation35. In metazoans, the α-CA family is largely represented. As described in the literature, CAs play a pivotal role in the deposition of calcium carbonate biominerals in at least 30 metazoan calcifying speciesCitation36–41. In fact, during calcium carbonate formation, the metazoan CAs are involved in the process of acid–base regulation, calcification and mineralisationCitation39,Citation41 providing inorganic carbon at the site of calcificationCitation41 and/or determining the precipitation of calcium carbonateCitation42–44. Recently, we characterised and determined the kinetic constants of the CA purified from the mantle tissue of the bivalve Mediterranean mussel, Mytilus galloprovincialis. The protein was indicated with the acronym MgaCA and has been assigned to the α-class of the CA superfamily with the following kinetic parameters for the CO2 hydration reaction: kcat = 4.1 × 105 s−1 and kcat/Km of 3.6 × 107 M−1 × s−147. The enzyme activity was poorly inhibited by the sulfonamide acetazolamide, with a KI of 380 nM. Intriguingly, MgaCA had a molecular weight of 50 kDa, which is roughly two times higher than that of a typical monomeric α-class enzyme (25 kDa)Citation45. Here, using the recombinant DNA technology, we prepared and heterologously expressed the recombinant CA (in the text indicated as rec-MgaCA) starting from N-amino terminal sequence of the native MgaCA. The catalytic properties of the rec-MgaCA were compared with those obtained for the native enzyme. Our results demonstrated that the rec-MgaCA was a monomer with an apparent molecular weight of 32 kDa and the following kinetic parameters for the CO2 hydration reaction: kcat = 4.2 × 105 s−1 and kcat/Km of 3.5 × 107 M−1 × s−1. From the comparison of the SDS-PAGE, the protonography, and the kinetic analysis performed on the native and recombinant enzyme, we hypothesised that probably the native MgaCA is a multidomain protein containing a single CA domain, which allows the carbon dioxide hydration reaction.
Materials and methods
Gene identification
The rec-MgaCA gene of M. galloprovincialis (accession number: ALF62133.1) was identified running the protein “BLAST” program and using the amino acid sequence “SWGYGNDNGP” as query sequence, which is the N-amino terminal sequence of the native MgaCA previously determined by the Edman degradation performed on the blotted enzymeCitation45.
Construct preparation, protein expression and purification
The GeneArt Company, specialized in gene synthesis, designed the synthetic M. galloprovincialis gene encoding for the α-CA, and containing four base pair sequences (CACC) necessary for directional cloning at the 5′ end of the rec-MgaCA gene. The fragment was subsequently cloned into the expression vector pET100/D-TOPO (Invitrogen, Waltham, MA), creating the plasmid pET100D-Topo/rec-MgaCA. In order to confirm the integrity of the M. galloprovincialis gene and the fact that no errors occurred at the ligation sites, the vector containing the fragment was sequenced. Escherichia coli ArcticExpress (DE3)RIL competent cells were transformed with pET100/D-Topo/rec-MgaCA, grown at 37 °C, induced with 1 mM IPTG. Zn(SO4) was added after 30 min and after additional growth for 16 h, cells were harvested and disrupted by sonication at 4 °C in 20 mM buffer phosphate, pH 8.0. Following sonication, the sample was centrifuged at 1200g at 4 °C for 30 min. The supernatant was dialysed against 0.02 M phosphate buffer (pH 8.0) containing 0.01 M imidazole at 4 °C and loaded onto a His-select HF Nickel affinity column (1.0 by 1.0 cm, GE Healthcare). The column was equilibrated with 0.02 M phosphate buffer (pH 8.0) containing 0.01 M imidazole and 0.5 M KCl at a flow rate of 1.0 ml/min. The rec-MgaCA elution was performed with 0.02 M phosphate buffer (pH 8.0) containing 0.5 M KCl and 0.3 M imidazole at a flow rate of 1.0 ml/min. Active fractions (1 ml) were collected and combined for a total volume of 5 ml. Subsequently, they were dialysed, concentrated, and analysed by SDS-PAGE. At this stage of purification, the enzyme was at least 95% pure and the obtained recovery was of 1.0 mg of the rec-MgaCA.
Sequence analysis
Multialignment of amino acid sequences was performed using the program MUSCLE (MUltiple Sequence Comparison by Log-Expectation), a new computer program for creating multiple alignments of protein sequenceCitation46.
SDS-PAGE
Sodium dodecyl sulfate SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed as described by Laemmli using 12% gelsCitation47.
Protonography
Wells of 12% SDS-PAGE gel were loaded with bCA and rec-MgaCA mixed with loading buffer without 2-mercaptoethanol and without boiling the samples, in order to avoid protein denaturation. The gel was run at 150 V until the dye front ran off the gel. Following the electrophoresis, the 12% SDS-PAGE gel was subject to protonography to detect the bCA and rec-MgaCA hydratase activity on the gel as described by Capasso and coworkersCitation46,Citation48,Citation49.
Enzyme kinetics
An Applied Photophysics (United Kingdom) stopped-flow instrument has been used for assaying the CA-catalysed CO2 hydration activityCitation50. Bromothymol blue (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 – 20 mM TRIS (pH 8.3) as buffer, and 20 mM Na2SO4 for maintaining constant the ionic strength (this anion is not inhibitory and has a KI > 200 mM against this enzyme), following the initial rates of the CA-catalysed CO2 hydration reaction for a period of 10–100 s. The CO2 concentrations ranged from 1.7 to 17 mM for the determination of the kinetic parameters and inhibition constants. For each measurement at least six traces of the initial 5–10% of the reaction have been used for determining the initial velocity. The uncatalysssed rates were determined in the same manner and subtracted from the total observed rates. Stock solutions of inhibitor (1–10 mM) were prepared in distilled-deionized water and dilutions up to 0.01 μM were done thereafter with distilled-deionised water. Inhibitor and enzyme solutions were pre-incubated together for 15 min at room temperature prior to assay, in order to allow for the formation of the E-I complex. The inhibition constants were obtained by non-linear least-squares methods using the Cheng–Prusoff equation whereas the kinetic parameters for the uninhibited enzymes from the Lineweaver–Burk plots, as reported earlier, and represent the mean from at least three different determinations.
Results and discussion
Identification of the full amino acid sequence of the M. galloprovincialis α-CA
Using the Basic Local Alignment Search Tool (BLAST) and as query sequence the amino acid sequence “SWGYGNDNGP” deduced by the Edman degradation carried out on the blotted native MgaCACitation45, the full amino acid sequence of the M. galloprovincialis α-CA deposited in the NCBI (National Center for Biotechnology Information, USA) library of the protein/enzyme sequences has been identified. The result of BLAST analysis showed that the first amino acid sequence of the top library sequences was the carbonic anhydrases II from M. galloprovincialis with a “Query score” and “Identity” of 100% (). As shown in , the full nucleotide sequence encoding for the native MgaCA showed an open reading frame of 255 amino acid residues containing the conserved three histidines, His94, His96, and His119 (hCA I numbering system), which coordinate the Zn(II) ion crucial for catalysis; and the two gate-keeper residues, the Glu106 and Thr199. The mussel enzyme had a residue of lysine as substituent of the proton shuttle residue His64 (), characteristic of the human isozymes, explaining the relatively low catalytic activity of the native MgaCA, with the following kinetic parameters for the CO2 hydration reaction: kcat = 4.1 × 105 s−1 and kcat/Km of 3.6 × 107 M−1 × s−1 Citation45, with respect to hCA II, which is considered as one of the most active among the α-CAs and the other CA-classesCitation51.
Figure 1. Blast output reporting the CA library sequences. By going down the list, it is possible to see less than perfect matches, slowly degrading as the corresponding score decreases and the E-value increases. The E-value is an assessment of the statistical significance of the score. E-value close to 1 are a warning that the alignment is not reliable.
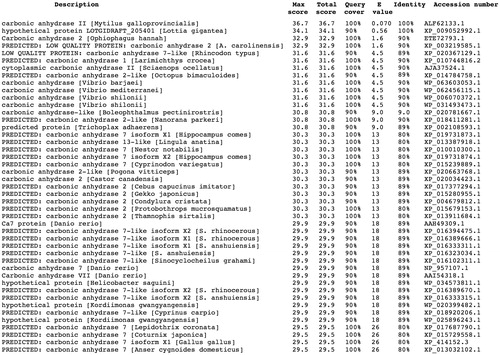
Figure 2. Alignment of α-CA sequences from Mytilus galloprovincialis (MgaCA, Accession number: ALF62133.1) Corallium rubrum (CruCA4, Accession number: KU557746), Homo sapiens I (hCAI, Accession number: NP001729) and II (hCAII, Accession number: P00918), hCA I numbering system was used. The zinc ligands (His94, 96 and 119) and the gate-keeper residues (Glu106 and Thr199) are conserved in aligned sequences; while the proton shuttle residue (His64) is preserved only in the human enzymes. The asterisk (*) indicates identity at all aligned positions; the symbol (:) relates to conserved substitutions, while (.) means that semi-conserved substitutions are observed. Multialignment was performed with the program Muscle, version 3.1.

Production of the recombinant enzyme (rec-MgaCA)
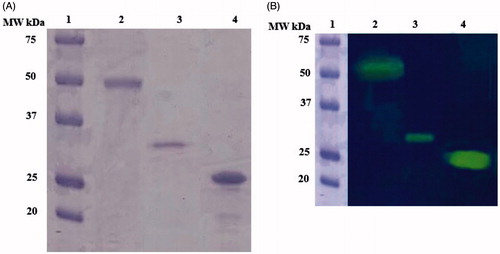
The recombinant rec-MgaCA was prepared designing a synthetic gene as described in the section ‘‘Materials and methods’’ and heterologously expressed as a His-Tag fusion protein using the method reported earlier for several CAsCitation52. The recombinant enzyme was recovered in the soluble fraction of the E. coli ArcticExpress (DE3)RIL cells extract obtained after sonication and centrifugation. Using an affinity column (His-select HF Nickel Affinity Gel), rec-MgaCA was purified to apparent homogeneity, as indicated by SDS-PAGE and protonography (, lane 3). The total amount of metalloenzyme recovered was 1 mg. The rec-MgaCA showed a band of about 32 kDa (monomeric form) under reducing condition (, lane 3). Intriguingly, the native MgaCA showed a molecular weight of about 50 kDa (, lane 2), while the commercial bovine CA (α-CA) had a molecular weight of about 26 kDa (, lane 4). Considering the fact that the molecular weight of the rec-MgaCA without the His-Tag is about 30 kDa, its dimer should have a molecular weight of about 60 kDa. As shown in , lane 2, it is readily apparent that the native enzyme, MgaCA, showed a molecular weight of approximately 50 kDa, which is 10 kDa lower than that proposed for the dimer (60 kDa). From this analysis, we propose that probably the native enzyme is a multidomain protein characterised by a CA domain present at its N-amino terminal sequence and another domain of about 20 kDa at the C-terminus. Our hypothesis is corroborated by the existence of particular α-CAs in mollusks, called nacreins. Nacrein has been identified for the first time in the Japanese pearl oyster Pinctada fucataCitation53. This protein showed a M.W. of 50 kDa and is involved in the nacreous layer formation of shell and pearl. It possesses a hydratase activity because it has a CA domain at the N-terminus part of the entire molecule.
Kinetic analysis comparison
The kinetic parameters for the CO2 hydration catalysed by the rec-MgaCA were measured and compared to the human isoforms hCA I, hCA IICitation54 as well as to the previous CAs cloned in Stylophora pistillata, STPCA (SpiCA1)Citation43,Citation55 and STPCA2 (SpiCA2); CruCA4 from Corallium rubrumCitation40,Citation43 and the native CA purified from the mantle of M. galloprovincialis (MgaCA) ()Citation45. The kinetic constants of the MgaCA and rec-MgaCA are two times higher than the hCA I enzyme. This is very intriguing since mussel α-CA respect to the hCA I lacked a His64, which is involved in the transfer of a proton from the water coordinated to the Zn(II) ion to the environment with the function to accelerate the rate of the catalytic cycle. Curiously, the recombinant enzyme (rec-MgaCA) has a very similar kinetic and acetazolamide inhibition features, which are comparable with those of the enzyme isolated from the living mussels (within the limits of the experimental error). Thus, it is possible that in the conditions of the assay the recombinant enzyme dimerizes or, as we described in the previously paragraph, the MgaCA isolated from the mussels could have a CA domain and another domain, which is not connected to the catalytic function, and as thus, should not be a dimer but a multidomain protein. Of course, work is in progress in our laboratories to verify the multidomain nature of the native MgaCA.
Table 1. Kinetic parameters for the CO2 hydration reaction catalysed by the rec-MgaCA, the purified native mussel CA (MgaCA), the Homo sapiens CA isoforms (hCA I and hCA II) and coral CA isoforms (SpiCA1 and SpiCA2 from Stylophora pistillata; CruCA4 from Corallium rubrum). Acetazolamide (AAZ) inhibition data are also shown.
Conclusions
As described in the literature, mollusks contain multidomain proteins with hydratase activityCitation53,Citation56. For example, the nacrein is physiologically involved in the nacreous layer formation of shell and pearlCitation53. It showed a MW of 50 kDa and has been identified for the first time in the Japanese pearl oyster Pinctada fucata. Nacrein is able to convert the carbon dioxide to bicarbonate and protons because it has a single CA domain at the N-terminus part of the entire protein. Successively, two novel nacrein-like proteins with CA catalytic function and playing a key role in shell biomineralisation were identified from the shell-forming mantle of the Pacific oyster, Crassostrea gigasCitation57. Again, the CA encoded by the genome of Tridacna gigas represents an example of a α-CA multidomain protein with two CA domains. In fact, this α-CA is a glycoprotein, which has MW of 70 kDa and contains two complete carbonic anhydrase domains within the protein, one at the N-terminus, the other at the C-carboxy-terminal parts of the proteinCitation56. The dual domain structure could have arisen from the fusion of two separate CA genes or by a duplication of a single gene followed by a fusion eventCitation58. Interesting to note that the dual domain CAs have also been previously reported for two algal species, Dunaliella salina and Porphyridium purpureumCitation59,Citation60. These observations and the results obtained from the SDS-PAGE, protonography, and kinetic analysis give strength to our hypothesis that probably the native MgaCA is a multidomain protein with a single CA domain at the N-terminus of the protein. Moreover, the heterologous expression in E. coli of the recombinant protein resulted in a valid method for producing a discrete amount of the active rec-MgaCA. This will make possible the use of the biocatalyst either free or immobilised in the CO2 biomimetic capture process.
Acknowledgements
This research was financed by the grant “SMART GENERATION – Sistemi e tecnologie sostenibili per la generazione di energia – PON03PE_00157_1, OR3 – Bio-sistemi di cattura ed utilizzazione della CO2”. We also thank the Distinguished Scientist Fellowship Program (DSFP) of King Saud University, Riyadh, Saudi Arabia.
Disclosure statement
The authors report no conflicts of interest.
Additional information
Funding
References
- Supuran CT. Structure and function of carbonic anhydrases. Biochem J 2016;473:2023–32.
- Del Prete S, Vullo D, Fisher GM, et al. Discovery of a new family of carbonic anhydrases in the malaria pathogen Plasmodium falciparum – the eta-carbonic anhydrases. Bioorg Med Chem Lett 2014;24:4389–96.
- Kikutani S, Nakajima K, Nagasato C, et al. Thylakoid luminal theta-carbonic anhydrase critical for growth and photosynthesis in the marine diatom Phaeodactylum tricornutum. Proc Natl Acad Sci U S A 2016;113:9828–33. sss
- Capasso C, Supuran CT. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin Ther Targets 2015;19:1689–704.
- Capasso C, Supuran CT. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr Med Chem 2015;22:2130–9.
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from Bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2015;30:325–32.
- Capasso C, Supuran CT. An overview of the carbonic anhydrases from two pathogens of the oral cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr Top Med Chem 2016;16:2359–68.
- Ozensoy Guler O, Capasso C, Supuran CT. A magnificent enzyme superfamily: carbonic anhydrases, their purification and characterization. J Enzyme Inhib Med Chem 2016;31:689–94.
- Supuran CT, Capasso C. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J Enzyme Inhib Med Chem 2016;31:1254–60.
- Del Prete S, Vullo D, De Luca V, et al. Sulfonamide inhibition studies of the beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:1115–20.
- Del Prete SD, Luca VD, Simone G, et al. Cloning, expression and purification of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. J Enzyme Inhib Med Chem 2016;1–6.
- Supuran CT, Capasso C. The η-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63.
- Del Prete S, De Luca V, Supuran CT, Capasso C. Protonography, a technique applicable for the analysis of eta-carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:920–4.
- De Simone G, Di Fiore A, Capasso C, Supuran CT. The zinc coordination pattern in the eta-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett 2015;25:1385–9.
- De Luca V, Del Prete S, Supuran CT, Capasso C. Protonography, a new technique for the analysis of carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:277–82.
- Alafeefy AM, Abdel-Aziz HA, Vullo D, et al. Inhibition of human carbonic anhydrase isozymes I, II, IX and XII with a new series of sulfonamides incorporating aroylhydrazone-, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazinyl- or 2-(cyanophenylmethylene)-1,3,4-thiadiazol-3(2H)-yl moieties. J Enzyme Inhib Med Chem 2015;30:52–6.
- Del Prete S, Vullo D, De Luca V, et al. Cloning, expression, purification and sulfonamide inhibition profile of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. Bioorg Med Chem Lett 2016;26:4184–90.
- Pinard MA, Lotlikar SR, Boone CD, et al. Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg Med Chem 2015;23:4831–8.
- Ferraroni M, Del Prete S, Vullo D, et al. Crystal structure and kinetic studies of a tetrameric type II beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr D Biol Crystallogr 2015;71:2449–56.
- De Simone G, Monti SM, Alterio V, et al. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2015;25:2002–6.
- Zolnowska B, Slawinski J, Pogorzelska A, et al. Carbonic anhydrase inhibitors. Synthesis, and molecular structure of novel series N-substituted N'-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)guanidines and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Eur J Med Chem 2014;71:135–47.
- De Luca L, Ferro S, Damiano FM, et al. Structure-based screening for the discovery of new carbonic anhydrase VII inhibitors. Eur J Med Chem 2014;71:105–11.
- Di Fiore A, Capasso C, De Luca V, et al. ray structure of the first ‘extremo-alpha-carbonic anhydrase’, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D Biol Crystallogr 2013;69:1150–9.
- Supuran CT. Structure-based drug discovery of carbonic anhydrase inhibitors. J Enzyme Inhib Med Chem 2012;27:759–72.
- Supuran CT. Carbonic anhydrases-an overview. Curr Pharm Des 2008;14:603–14.
- Supuran CT. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin Drug Discov 2017;12:61–88.
- Bhatt A, Mahon BP, Cruzeiro VW, et al. Structure-activity relationships of benzenesulfonamide-based inhibitors towards carbonic anhydrase isoform specificity. Chembiochem 2017;18:213–22.
- Alterio V, Langella E, Viparelli F, et al. Structural and inhibition insights into carbonic anhydrase CDCA1 from the marine diatom Thalassiosira weissflogii. Biochimie 2012;94:1232–41.
- Buzas GM, Supuran CT. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam Ioan Puşcaş (1932-2015)). J Enzyme Inhib Med Chem 2016;31:527–33.
- Carta F, Supuran CT, Scozzafava A. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med Chem 2014;6:1149–65.
- Lomelino CL, Mahon BP, McKenna R, et al. Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms I, II, IX and XII of a thioureido analog of SLC-0111. Bioorg Med Chem 2016;24:976–81.
- Aspatwar A, Tolvanen ME, Ortutay C, Parkkila S. Carbonic anhydrase related proteins: molecular biology and evolution. Subcell Biochem 2014;75:135–56.
- Supuran CT. Carbonic anhydrases as drug targets-an overview. Curr Top Med Chem 2007;7:825–33.
- Capasso C, Supuran CT. Sulfa and trimethoprim-like drugs – antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2014;29:379–87.
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat 2013;23:693–704.
- Le Roy N, Jackson DJ, Marie B, et al. The evolution of metazoan α-carbonic anhydrases and their roles in calcium carbonate biomineralization. Front Zoology 2014;11:75.
- Bertucci A, Innocenti A, Zoccola D, et al. Carbonic anhydrase inhibitors: inhibition studies of a coral secretory isoform with inorganic anions. Bioorg Med Chem Lett 2009;19:650–3.
- Bertucci A, Innocenti A, Zoccola D, et al. Carbonic anhydrase inhibitors: inhibition studies of a coral secretory isoform by sulfonamides. Bioorg Med Chem 2009;17:5054–8.
- Bertucci A, Moya A, Tambutte S, et al. Carbonic anhydrases in anthozoan corals – a review. Bioorg Med Chem 2013;21:1437–50.
- Bertucci A, Tambutté S, Supuran CT, et al. A new coral carbonic anhydrase in Stylophora pistillata. Mar Biotechnol 2011;13:992–1002.
- Bertucci A, Zoccola D, Tambutte S, et al. Carbonic anhydrase activators. The first activation study of a coral secretory isoform with amino acids and amines. Bioorg Med Chem 2010;18:2300–3.
- Drake JL, Mass T, Haramaty L, et al. Proteomic analysis of skeletal organic matrix from the stony coral Stylophora pistillata. Proc Natl Acad Sci U S A 2013;110:3788–93.
- Le Goff C, Ganot P, Zoccola D, et al. Carbonic anhydrases in cnidarians: novel perspectives from the Octocorallian Corallium rubrum. PLos One 2016;11:e0160368.
- Mass T, Drake JL, Peters EC, et al. Immunolocalization of skeletal matrix proteins in tissue and mineral of the coral Stylophora pistillata. Proc Natl Acad Sci USA 2014;111:12728–33.
- Perfetto R, Del Prete S, Vullo D, et al. Biochemical characterization of the native alpha-carbonic anhydrase purified from the mantle of the Mediterranean mussel, Mytilus galloprovincialis. J Enzyme Inhib Med Chem 2017;32:632–9.
- Del Prete SD, Luca V, Supuran CT, Capasso C. Protonography, a technique applicable for the analysis of η-carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:920–4.
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792–7.
- Del Prete S, De Luca V, Iandolo E, et al. Protonography, a powerful tool for analyzing the activity and the oligomeric state of the gamma-carbonic anhydrase identified in the genome of Porphyromonas gingivalis. Bioorg Med Chem 2015;23:3747–50.
- Vullo D, De Luca V, Del Prete S, et al. Sulfonamide inhibition studies of the gamma-carbonic anhydrase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Bioorg Med Chem Lett 2015;25:3550–5.
- Khalifah RG. The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 1971;246:2561–73.
- Vullo D, Nishimori I, Scozzafava A, Supuran CT. Carbonic anhydrase activators: activation of the human cytosolic isozyme III and membrane-associated isoform IV with amino acids and amines. Bioorg Med Chem Lett 2008;18:4303–7.
- Del Prete S, De Luca V, De Simone G, et al. Cloning, expression and purification of the complete domain of the η-carbonic anhydrase from Plasmodium falciparum. J Enzyme Inhib Med Chem 2016;31:54–9.
- Miyamoto H, Miyoshi F, Kohno J. The carbonic anhydrase domain protein nacrein is expressed in the epithelial cells of the mantle and acts as a negative regulator in calcification in the mollusc Pinctada fucata. Zoolog Sci 2005;22:311–15.
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81.
- Moya A, Tambutte S, Bertucci A, et al. Carbonic anhydrase in the scleractinian coral Stylophora pistillata – characterization, localization, and role in biomineralization. J Biolog Chem 2008;283:25475–84.
- Leggat W, Dixon R, Saleh S, Yellowlees D. A novel carbonic anhydrase from the giant clam Tridacna gigas contains two carbonic anhydrase domains. FEBS J 2005;272:3297–305.
- Song X, Wang X, Li L, Zhang G. Identification two novel nacrein-like proteins involved in the shell formation of the Pacific oyster Crassostrea gigas. Mol Biol Rep 2014;41:4273–8.
- Fisher M, Gokhman I, Pick U, Zamir AA. Salt-resistant plasma membrane carbonic anhydrase is induced by salt in Dunaliella salina. J Biol Chem 1996;271:17718–23.
- Mitsuhashi S, Miyachi S. Amino acid sequence homology between N- and C-terminal halves of a carbonic anhydrase in Porphyridium purpureum, as deduced from the cloned cDNA. J Biol Chem 1996;271:28703–9.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5.