Abstract
Dengue virus (DENV) is the leading mosquito-transmitted viral infection in the world. With more than 390 million new infections annually, and up to 1 million clinical cases with severe disease manifestations, there continues to be a need to develop new antiviral agents against dengue infection. In addition, there is no approved anti-DENV agents for treating DENV-infected patients. In the present study, we identified new compounds with anti-DENV replication activity by targeting viral replication enzymes – NS5, RNA-dependent RNA polymerase (RdRp) and NS3 protease, using cell-based reporter assay. Subsequently, we performed an enzyme-based assay to clarify the action of these compounds against DENV RdRp or NS3 protease activity. Moreover, these compounds exhibited anti-DENV activity in vivo in the ICR-suckling DENV-infected mouse model. Combination drug treatment exhibited a synergistic inhibition of DENV replication. These results describe novel prototypical small anti-DENV molecules for further development through compound modification and provide potential antivirals for treating DENV infection and DENV-related diseases.
Introduction
Dengue virus (DENV) is responsible of worldwide arthropodborne viral infection, which globally represents a serious human health concern. DENV is the etiological agent of disease in more than hundred countries with up to 3 billion people exposed to the risk of infection in the tropical regionsCitation1–3. DENV has expanded its global range with sustained outbreaks in South America and Asia, with these epidemics accompanied by increased disease severity.
DENV are single-stranded, positive sense RNA viruses belonging to the Flaviviridae family. The DENV family can be viewed as falling in four related, but antigenically distinct, DENV 1–4 serotypes. The carriers of DENV to humans are the mosquitoes Aedes aegypti and Aedes albopictus. The DENV infection causes a variety of illness, including asymptomatic or subclinical disease, dengue fever (DF) symptoms and the most severe dengue haemorrhagic fever (DHF) and dengue shock syndrome (DSS) which cause millions of infections around the worldCitation4–8. DENV has a 10.7 kb, positive-sense RNA genome with 5′- and 3′-untranslated regions flanking a polyprotein that encodes three structural (C, prM/M and E) and seven non-structural (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5) proteins. prM and E structural proteins are the primary antigenic targets of the humoural immune response in humansCitation9–12. DENV NS3 is a multifunctional protein, which contains protease, helicase and triphosphatase domain. The N-terminal amino acids (residue 1–184) of NS3 is responsible for protease activity. NS2B serves as a cofactor of NS3 protease and forms complex with NS3; the central amino acid hydrophilic domain (residue 49–92) of NS2B is critical for cofactor activityCitation13. DENV replication also requires the non-structural protein 5 (NS5) which is the essential RNA-dependent RNA polymerase (RdRp) activityCitation14. DENV NS5 RdRp has proven to be a promising target for direct-acting antiviral (DAA) drug development, because it is structurally conserved among the four DENV serotypes, and NS5 RdRp has no enzymatic counterpart in mammalian cellsCitation15.
Currently, no licensed antiviral drugs are available to block DENV infection, and while vector control efforts remain the only means to stop the spread of the infection, they have not successfully inhibited annual epidemic outbreaks throughout the tropicsCitation16. Recently, a live-attenuated DENV vaccineCitation17 based on the yellow fever virus 17 D backbone was licensed for use in the Philippines, Brazil and Mexico. However, the serotype-specific efficacy of the DENV vaccine is varying, and the long-term protection and safety of this vaccine still need more investigationCitation18. In the present study, we have focused on the identification of potential anti-DENV inhibitors by targeting the enzymatic activities of the NS5 RdRp polymerase and NS3 protease in vitro and in vivoCitation19–21. As part of a continuation of our studiesCitation22–24, we developed new pyrazole derivativesCitation25 as potential DENV NS5 RdRp inhibitors (). In addition, we carried out virtual screening (VS) studies on the NS2B/NS3 protease to design and synthesise new DENV NS3 protease inhibitors (). In sum, we identified five compounds that exhibited anti-DENV replication activity without cytotoxicity; two compounds exhibited anti-DENV activity in DENV-infected ICR-suckling mouse model. Interestingly, combination treatment with compounds, respectively, targeting NS5 RdRp polymerase and NS3 protease, demonstrated a synergistic inhibitory effect on DENV replication.
Table 1. Activity of Compounds 1–3 against DENV-2 Replication and DENV-2 NS5 RdRp.
Table 2. Activity of DENV inhibitors 4 and 5 against DENV-2 replication.
Materials and methods
Chemistry
Microwave (MW)-assisted reactions were performed on a CEM Discover SP single-mode reactor equipped with Explorer 72 autosampler, controlling the instrument settings by PC-running CEM Synergy 1.60 software (Matthews, NY, USA). Closed vessel experiments were carried out in capped MW-dedicated vials (10 ml) with cylindrical stirring bar (length 8 mm, diameter 3 mm). Stirring, temperature, irradiation power, maximum pressure (Pmax), PowerMAX (simultaneous cooling while heating), ActiVent (simultaneous venting while heating), and ramp and hold times were set as indicated. Temperature of the reaction was monitored by an external fibre optic temperature sensor. After completion of the reaction, the mixture was cooled to 25 °C via air jet cooling. Organic solutions were dried over anhydrous sodium sulphate. Evaporation of the solvents was carried out on a Bu¨chi Rotavapor R-210 equipped with a Bu¨chi V-850 vacuum controller and a Bu¨chi V-700 vacuum pump. Column chromatography was performed on columns packed with silica gel from Macherey-Nagel (70 − 230 mesh). Silica gel thin-layer chromatography (TLC) cards from Macherey-Nagel (silica gel pre-coated aluminium cards with fluorescent indicator visualisable at 254 nm) were used for TLC. Developed plates were visualised with a Spectroline ENF 260 C/FE UV apparatus. Melting points (mp) were determined on a Stuart Scientific SMP1 apparatus and are uncorrected. Infrared spectra (IR) were run on a PerkinElmer Spectrum 100 FT-IR spectrophotometer equipped with universal attenuated total reflectance (ATR) accessory and IR data acquired and processed by PerkinElmer Spectrum 10.03.00.0069 software. Band position and absorption ranges are given in cm−1. Proton nuclear magnetic resonance (1H NMR) spectra were recorded a Bruker Avance (400 MHz) spectrometer in the indicated solvent and corresponding fid files were processed by MestreLab Research SL MestreReNova 6.2.1–769 software (Santiago de Compostela, Spain). Chemical shifts are expressed in δ units (ppm) from tetramethylsilane. Elemental analyses of biologically evaluated compounds were found to be within ±0.4% of the theoretical values and their purity was found to be >95% by high-pressure liquid chromatography (HPLC). The HPLC system used Thermo Fisher Scientific Dionex UltiMate 3000, consisted of a SR-3000 solvent rack, a LPG-3400SD quaternary analytical pump, a TCC-3000SD column compartment, a DAD-3000 diode array detector and an analytical manual injection valve with a 20 µL loop. Samples were dissolved in acetonitrile (1 mg/mL). HPLC analysis was performed by using a Thermo Fisher Scientific Acclaim 120 C18 column (5 µm, 4.6 mm × 250 mm) at 25 ± 1 °C with an appropriate solvent gradient (acetonitrile/water), flow rate of 1.0 mL/min and signal detector at 206, 230, 254 and 365 nm. Chromatographic data were acquired and processed by Thermo Fisher Scientific Chromeleon 6.80 SR15 Build 4656 software (Waltham, MA, USA).
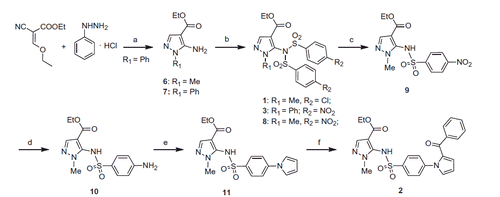
General procedure for preparation of compounds 1, 3 and 8. Ethyl 5–(4-chloro-N-((4-chlorophenyl)sulphonyl)phenylsulphonamido)-1-methyl-1H-pyrazole-4-carboxylate (1). 4-Chlorobenzenesulphonyl chloride (0.15 g, 0.72 mmol) was added to solution of commercially available 6 (0.10 g, 0.6 mmol) in pyridine (2 mL) while stirring. The reaction was stirred at 60 °C overnight. The mixture was neutralised with 1 N HCl and extracted with ethyl acetate. The organic layer was washed with brine, dried and filtered. Removal of the solvent gave a residue that was purified by column chromatography (silica gel, n-hexane:ethyl acetate = 2:1 as eluent) to furnish 1 (0.10 g, 33%), mp 132–135 °C (from ethanol). 1H NMR (DMSO-d6): δ 0.96 (t, J = 6.2 Hz, 3H), 3.52 (s, 3H), 3.71–3.77 (m, 2H), 7.81 (d, J = 8.0 Hz, 4H), 7.94 (d, J = 7.9 Hz, 4H), 8.06 ppm (s, 1H). FT-IR (ATR): ν 1176, 1391, 1709 cm−1. Anal. calcd. for C19H17Cl2N3O6S2 (518.38): C, 44.02; H, 3.31; N, 8.11; Cl, 13.68; S, 12.37. Found: C, 43.78; H, 3.26; N, 7.84; Cl, 13.36; S, 12.02.
Ethyl 5–(4-nitro-N-((4-nitrophenyl)sulphonyl)phenylsulphonamido)-1-phenyl-1H-pyrazole-4-carboxylate (3). Was synthesised as 1 starting from 7Citation26 and 4-nitrobenzensulphonyl chloride. Yield 79%, mp 230–232 °C (from ethanol). 1H NMR (DMSO-d6): δ 1.01 (t, J = 7.0, 3H), 3.70–3.76 (m, 2H), 7.32–7.38 (m, 3H), 7.48 (d, J = 6.9 Hz, 2H), 8.04 (d, J = 8.9 Hz, 4H), 8.33–8.36 ppm (m, 5H). FT-IR (ATR): ν 1167, 1336, 1597, 1706 cm−1. Anal. calcd. for C24H19N5O10S2 (601.06): C, 47.92; H, 3.18; N, 11.64; S, 10.66. Found: C, 47.78; H, 3.11; N, 11.39; S, 10.42.
Ethyl 1-methyl-5–(4-nitro-N-((4-nitrophenyl)sulphonyl)phenylsulphonamido)-1H-pyrazole-4-carboxylate (8). Was synthesised as 1 starting from 6 and 4-nitrobenzenesulphonyl chloride. Yield 33%, mp 220–222 °C (from ethanol). 1H NMR (DMSO-d6): δ 0.93 (t, J = 7.1 Hz, 3H), 3.54 (s, 3H), 3.69–3.74 (m, 2H), 8.11 (s, 1H), 8.23 (d, J = 8.9 Hz, 4H), 8.52 ppm (d, J = 8.9 Hz, 4H). FT-IR (ATR): ν 1173, 1349, 1528, 1717 cm−1. Anal. calcd. for C19H17N5O10S2 (539.49): C, 42.30; H, 3.18; N, 12.98; S, 11.89. Found: C, 42.12; H, 3.14; N, 12.60; S, 11.70.
Ethyl 1-methyl-5–(4-nitrophenylsulphonamido)-1H-pyrazole-4-carboxylate (9). A mixture of 8 (0.032 g, 0.06 mmol) and lithium hydroxide monohydrate (0.01 g, 0.24 mmol) in THF/H2O (1:1, 5 mL) was heated at 50 °C for 15 min, cooled, made acid with 1 N HCl aqueous solution (pH ≈ 5) and extracted with ethyl acetate. The organic layer was washed with brine, dried and filtered. Removal of the solvent gave 9 (0.02 g, 95%), mp 150–152 °C (from ethanol). 1H NMR (DMSO-d6): δ 0.98 (t, J = 7.08, 3H), 3.71–3.76 (m, 5H), 7.78 (s, 1H), 7.87 (d, J = 8.9 Hz, 2H), 8.40 (d, J = 8.9 Hz, 2H), 11.01 ppm (br s, disappeared on treatment with D2O, 1H). FT-IR (ATR): ν 1170, 1347, 1532, 1709 cm−1. Anal. calcd. for C13H14N4O6S (354.34): C, 44.07; H, 3.98; N, 15.81; S, 9.05. Found: C, 43.84; H, 3.92; N, 15.62; S, 8.91.
Ethyl 5–(4-aminophenylsulphonamido)-1-methyl-1H-pyrazole-4-carboxylate (10). A mixture of 9 (0.14 g, 0.4 mmol) and tin(II) chloride dihydrate (0.27 g, 1.2 mmol) in ethyl acetate (15 ml) was heated at reflux for 3 h. After cooling, the mixture was made basic (pH ≈ 8) with a saturated aqueous solution of NaHCO3 and filtered. The layers were separated and the organic one was washed with brine, dried and filtered. Removal of the solvent gave a residue that was purified by column chromatography (silica gel, dichloromethane:ethanol = 9.8:0.2 as eluent) to furnish 10 (0.06 g, 50%), mp 118–120 °C (from ethanol). 1H NMR (DMSO-d6): δ 1.09 (t, J = 7.1 Hz, 3H), 3.66 (s, 3H), 3.83–3.88 (m, 2H), 6.02 (s, 2H), 6.52 (d, J = 8.7 Hz, 2H), 7.20 (d, J = 8.7 Hz, 2H), 7.72 (s, 1H), 9.71 ppm (br s, disappeared on treatment with D2O, 1H). FT-IR (ATR): ν 1152, 1378, 1698, 3397 cm−1.
Ethyl 5–(4-(1H-pyrrol-1-yl)phenylsulphonamido)-1-methyl-1H-pyrazole-4-carboxylate (11). A mixture of 10 (0.05 g, 0.15 mmol) and 2,5-dimethoxytetrahydrofuran (0.02 g, 0.02 mL, 0.15 mmol) in glacial acetic acid (1 mL) was heated at reflux for 30 min. The solvent was removed in vacuo, and after cooling water and ethyl acetate were added. Layers were separated and the organic one was washed with a saturated aqueous solution of NaHCO3, brine and dried. Removal of the solvent gave a residue that was purified by column chromatography (silica gel, n-hexane:ethyl acetate = 1:1 as eluent) to furnish 11 (0.035 g, 66%), mp 200–203 °C (from ethanol). 1H NMR (DMSO-d6): δ 0.97 (t, J = 7.1 Hz, 3H), 3.73–3.77 (m, 5H), 6.32 (t, J = 2.2 Hz, 2H), 7.49 (t, J = 2.2 Hz, 2H), 7.62 (d, J = 8.8 Hz, 2H), 7.76–7.78 (m, 3H), 10.48 ppm (br s, disappeared on treatment with D2O, 1H). FT-IR (ATR): ν 1132, 1336, 1702, 3139 cm−1. Anal. calcd. for C17H18N4O4S (374.41): C, 54.53; H, 4.85; N, 14.96; S, 8.56. Found: C, 54.28; H, 4.79; N, 14.69; S, 8.22.
Ethyl 5–(4-(2-benzoyl-1H-pyrrol-1-yl)phenylsulphonamido)-1-methyl-1H-pyrazole-4-carboxylate (2). A mixture of 11 (0.06 g, 0.14 mmol), benzoyl chloride (0.02 g, 0.016 mL, 0.14 mmol) and anhydrous aluminium chloride (0.02 g; 0.14 mol) in anhydrous 1,2-dichloroethane (3 mL) was placed into the MW cavity (closed vessel mode, Pmax = 250 psi). A starting MW irradiation of 70 W was used, the temperature being ramped from 25 to 110 °C, while stirring. Once 110 °C was reached, taking about 1 min, the reaction mixture was held at this temperature for 4 min. The reaction mixture was quenched on 1 N HCl aqueous solution/crushed ice and extracted with dichloromethane. The organic layer was washed with brine, dried and filtered. Removal of the solvent gave a residue that was purified by column chromatography (silica gel, n-hexane:acetone = 2:1 as eluent) to furnish 2 (0.04 g, 71%), mp 136–140 °C (from ethanol). 1H NMR (DMSO-d6): δ 1.07 (t, J = 7.1 Hz, 3H), 3.67 (s, 3H), 3.87–3.93 (m, 2H), 6.43–6.45 (m, 1H), 6.87–6.89 (m, 1H), 7.45–7.46 (m, 1H), 7.52–7.56 (m, 4H), 7.63–7.69 (m, 3H), 7.78 (s, 1H), 7.82–7.84 (m, 2H) 10.60 ppm (br s, disappeared on treatment with D2O, 1H). FT-IR (ATR): ν 1166, 1347, 1595, 1713, 3102 cm−1. Anal. calcd. for C24H22N4O5S (478.52): C, 60.24; H, 4.63; N, 11.71; S, 6.70. Found: C, 59.92; H, 4.58; N, 11.46; S, 6.46. Further elution with the same eluent gave ethyl 5–(4-(3-benzoyl-1H-pyrrol-1-yl)phenylsulphonamido)-1-methyl-1H-pyrazole-4-carboxylate (0.02 g, 28%), mp 216–218 °C (from ethanol). 1H NMR (DMSO-d6): δ 0.97 (t, J = 7.0 Hz, 3H), 3.73–3.79 (m, 5H), 6.80–6.82 (m, 1H), 7.53–7.57 (m, 2H), 7.62–7.68 (m, 4H), 7.76 (s, 1H), 7.84–7.86 (d, J = 8.4 Hz, 2H), 7.93–7.96 (d, J = 9.0 Hz, 2H), 8.04 (s, 1H), 10.58 ppm (br s, disappeared on treatment with D2O, 1H). FT-IR (ATR): ν 1166, 1280, 1642, 1706, 3132 cm−1. Anal. calcd. C24H22N4O5S (478.52): C, 60.24; H, 4.63; N, 11.71; S, 6.70. Found: C, 59.88; H, 4.55; N, 11.65; S, 6.60.
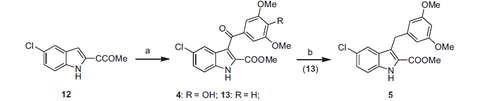
Methyl 5-chloro-3–(4-hydroxy-3,5-dimethoxybenzoyl)-1H-indole-2-carboxylate (4). Was synthesised as 2 starting from 12 and 4-hydroxy-3,5-dimethoxybenzoyl chloride. Yield 11%, mp 118–120 °C (from ethanol). 1H NMR (CDCl3): δ 3.70 (s, 3H), 3.86 (s, 6H), 6.01 (br s, disappeared on treatment with D2O, 1H), 7.17 (s, 2H), 7.32–7.35 (m, 1H), 7.41–7.43 (m, 1H), 7.60–7.61 (m, 1H), 9.31 ppm (br s, disappeared on treatment with D2O, 1H). FT-IR (ATR): ν 1715, 3312 cm−1. Anal. calcd. for C19H16ClNO6 (389.79): C, 58.55; H, 4.14; N, 3.59; Cl, 9.09. Found: C, 58.36; H, 4.09; N, 3.23; Cl, 8.81.
Methyl 5-chloro-3–(3,5-dimethoxybenzoyl)-1H-indole-2-carboxylate (13). Was synthesised as 2 starting from 12 and 3,5-dimethoxybenzoyl chloride. Yield 21%, mp 185–188 °C (from ethanol). 1H NMR (CDCl3): δ 3.70 (s, 3H), 3.82 (s, 6H), 6.07 (t, J = 2.3 Hz, 1H), 7.0 (d, J = 4.7 Hz, 2H), 7.33–7.36 (m, 1H), 7.41–7.44 (m, 1H), 7.65–7.66 (m, 1H), 9.31 ppm (br s, disappeared on treatment with D2O, 1H). FT-IR (ATR): ν 1698, 3287 cm−1.
Methyl 5-chloro-3–(3,5-dimethoxybenzyl)-1H-indole-2-carboxylate (5). A mixture of 13 (0.1 g, 0.3 mmol), triethylsilane (0.072 g, 0.1 mL, 0.6 mmol) and trifluoroacetic acid (0.31 g, 0.21 mL, 3 mmol) was stirred at 25 °C for 48 h. The mixture was diluted with a saturated aqueous solution of NaHCO3 and extracted with ethyl acetate. The organic layer was washed with brine, dried and filtered. Removal of the solvent gave a residue that was purified by column chromatography (silica gel, n-hexane/ethyl acetate = 3:1 as eluent) to furnish 5 (0.05 g, 50%), mp 182–185 °C (from ethanol). 1H NMR (CDCl3): δ 3.71 (s, 6H), 3.92 (s, 3H), 4.43 (s, 2H), 6.30–6.31 (m, 1H), 6.41–6.44 (m, 2H), 7.25–7.35 (m, 2H), 7.58–7.60 (m, 1H), 8.80 ppm (br s, disappeared on treatment with D2O, 1H). FT-IR (ATR): ν 1690, 3323 cm−1. Anal. calcd. for C19H18ClNO4 (359.81): C, 63.43; H, 5.04; Cl, 9.85; N, 3.89. Found: C, 63.27; H, 4.97; Cl, 9.59; N, 3.70.
Molecular modelling studies
All molecular modelling studies were performed on a MacPro dual 2.66 GHz Xeon running Ubuntu 12. The DENV protease structure (PDB ID: 2FOM)Citation27 was downloaded from the protein data bank. An in-house compound library matching Lipinski’s rule of fiveCitation28 was used as a training set. No pre-filter was applied. The docking simulations were performed with PLANTSCitation29 using a 13 Å radius grid sphere. The centre of the binding site was settled according to TamiriCitation30. Molecules were scored by ChemPLP scoring function.Citation29 was generated by PyMOLCitation31.
Ethics statement and experimental animals
Six-day-old ICR strain-suckling mice were obtained from BioLasco Taiwan Co. Ltd (Taipei, Taiwan). All animal studies were conducted in specific pathogen-free conditions and carried out in accordance with the Guide for the Care and Use of Laboratory Animals. The experimental protocol was approved by the Animal Research Committee of Kaohsiung Medical University of Taiwan (IACUC, protocol number 102177) under the guidance of the Public Health Service (PHS) Policy on Humane Care and Use of Laboratory Animals.
Cells and virus
Human hepatoma Huh-7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% foetal bovine serum, 1% non-essential amino acids, and 1% antibiotic–antimycotic within 5% CO2 supplement at 37 °C. Aedes albopictus C6/36 mosquito cells were cultured in RPMI1640 medium containing 10% fetal bovine serum, 1% non-essential amino acids, 1% antibiotic–antimycotic and 1% sodium pyruvate within 5% CO2 supplement at 37 °C. DENV-2 (DENV type-2 strain 16681) was amplified in C6/36 mosquito cells. Spodoptera frugiperda (Sf9) insect cells were cultured in Grace’s medium with 10% hear-inactivated foetal bovine serum (FBS) and 1% antibiotic–antimycotic at 26 °C.
Evaluation of anti-DENV RNA activity
Huh-7 cells were seeded in 24-well plate and infected DENV at an MOI of 0.2 for 2 h and followed by test compound treatment at concentration of 0, 1, 5, 10, 25, 50 and 100 µM for 3 days. The total cellular RNA was harvested by RNA extraction kitCitation32 following manufacturer’s instrument. DENV RNA and cellular mRNA levels were determined by quantitative real-time reverse-transcription polymerase chain reaction (RT-qPCR) with specific primersCitation33. The DENV RNA level was normalised by cellular glyceraldehydes-3-phosphate dehydrogenase (GAPDH) mRNA level. The relative DENV RNA level was calculated by StepOne™ Software v2.2.2 (Applied Biosystems, Foster City, CA, USA) following normalisation of cellular glyceraldehydes-3-phosphate dehydrogenase (GAPDH) mRNA level. The zero dose was defined as 100%. The EC50 values were calculated from non-linear regression curve fitting using GraphPad Prism7 software (San Diego, CA, USA). Results were obtained from three independent experiments.
Cell cytotoxicity assay
The Huh-7 cells were seeded in 96-well and treated with test compound at concentration of 0, 10, 25, 50, 100, 150 and 200 µM. After 3 days incubation, the cell viability was determined by CellTiter 96 AQueous One Solution Cell Proliferation Assay (MTS assay) as previously reportedCitation34. Briefly, the MTS assay buffer was added to the cultured plate following removal of the culture medium. After 2 h of incubation at 37 °C, the absorbance at 492 nm was measured. The cell viability was calculated from non-linear regression curve fitting using GraphPad Prism7. The zero dose was defined as 100%. Results were obtained from three independent experiments.
Purification of DENV NS5 protein
The DENV-2 NS5 protein purification was performed as previously describedCitation33. Briefly, Sf9 cells were infected by the recombinant baculovirus vAc-DENV-NS5 at an MOI of 10 for 3 days. The cells were pelleted by centrifugation at 1000×g for 5 min at 4 °C and washed twice with phosphate-buffered saline (PBS). The cells were suspended in 5 mL of binding buffer (20 mM sodium phosphate, pH 7.5, 300 mM NaCl, 20 mM imidazole) containing protease inhibitor cocktail, and then disrupted by sonication. After centrifugation, the supernatant containing His-tagged DENV-2 NS5 protein was subjected to metal affinity chromatography employing the AKTA prime protein purification systemCitation35. The eluted DENV-2 NS5 protein was concentrated by an Amicon Ultra-15 30 k centrifugal filter deviceCitation36 and then dialysed against PBS overnight at 4 °C.
Evaluation of anti-DENV RdRp activity
The cell-based experiments were performed as previously describedCitation33. Briefly, the Huh-7 cells were transfected with 0.5 µg of p(+)RLuc-(–)DV-UTRΔC-FLuc and DENV NS5 expression vector pcDNA-NS5-Myc followed by compound treatment for 3 days. The RLuc and FLuc activities were analysed by Dual-Glo Luciferase Assay SystemCitation37. The enzyme-based fluorescence-based alkaline phosphatase-coupled polymerase assay (FAPA) was performed as previously describedCitation33. Briefly, the template was amplified from the cDNA of DENV-2 minus strand 3′-UTR and its RNA was synthesised by the T7 Megascript kitCitation38. The 100 nM DENV NS5 protein was incubated with 1, 5 or 10 µM test compound, 100 nM (−) 3′-UTR RNA, 20 µM CTP, GTP, UTP and 20 µM BBT-ATP (Jena Bioscience) in a total volume of 30 µL in assay buffer (50 mM Tris-HCl, pH 7.5, 10 mM KCl, 1 mM MgCl2, 0.3 mM MnCl2, 0.001% Triton X-100 and 10 µM cysteine) at 25 °C for 60 min. The 30 µL of 2.5× stop buffer (200 mM NaCl, 25 mM MgCl2, 1.5 M diethanolamine, pH 10) with 25 nM calf intestinal alkaline phosphatase (Promega) was added to the reaction after 60-min incubation. The fluorescence signal was measured at excitation wavelength of 422 nm and emission wavelength of 566 nm, respectively.
Purification of DENV NS2B/NS3pro protein
The DENV NS2B/NS3pro purification was performed as previously describedCitation39. Briefly, the pET24b-NS2B-NS3pro plasmid was transformed into competent Escherichia coli BL21 (DE3) and grew in kanamycin (50 µg/mL)-containing LB at 26 °C until OD600 nm reached 0.6. The isopropyl-β-D-thiogalactopyranose was added into the bacterial culture at 0.5 mM for protein induction. After 12-h incubation, cells were collected and subjected to protein purification following previous description.
Evaluation of anti-DENV protease activity
For cell-based activity assay, the Huh-7 cells were cotransfected with 0.5 µg of pEG(MITA)SEAP reporter vector and DENV protease expression vector pcDNA-NS2B-GSG-NS3-Myc followed by compound treatment for 3 days. The SEAP activity was analysed by Phospha-Light SEAP Reporter Gene Assay SystemCitation40. The enzyme-based experiment was performed as previously describedCitation39. Briefly, the 7-amino-4-methylcoumarin (AMC) fluorophore-linked peptide substrate Boc-GRR-AMCCitation41 and purified DENV NS2B/NS3pro protein were used to analyse DENV NS2B/NS3 protease activity. The 100 nM DENV NS2B/NS3pro protein was incubated with 1, 5 or 10 µM tested compound and 10 µM Boc-GRR-AMC in cleavage buffer (200 mM Tris [pH 9.5], 20% glycerol) for 30 min at 25 °C. The fluorescence signal was detected at excitation wavelength of 380 nm and emission wavelength of 465 nm, respectively.
Evaluation of anti-DENV activity by ICR-suckling mice
Breeder ICR mice were purchased from BioLasco Taiwan Co. Ltd. The experimental protocol was approved by the Animal Research Committee of Kaohsiung Medical University of Taiwan (IACUC, protocol number 102177). All mice received humane care and fed with standard rodent chew and water ad libitum. Mice were kept under a standard laboratory condition following the Animal Use Protocol of Kaohsiung Medical University. The 6-day-old ICR-suckling mice were intracerebrally injected with DENV-2 followed by intracerebrally injecting saline or test compound at 1, 3, 5 days postinfection (dpi). Mice intracerebrally injected with 65 °C heat-inactive DENV-2 followed with saline treatment served as healthy control. The body weight, clinical scores and survival rate were recorded every day. The mice were sacrificed at 6 dpi.
Results and discussion
Chemistry
Compounds 1–3 were synthesised as shown in Scheme 1. Ethyl 5-amino-1-methyl-1H-pyrazole-4-carboxylate (6) was purchased from Maybridge. Treatment of ethyl 2-cyano-3-ethoxyacrylate with phenylhydrazine hydrochloride yielded ethyl 5-amino-1-phenylpyrazole-4-carboxylate (7)Citation26. Pyrazoles 6 and 7 were converted into final derivatives 1, 3 or intermediate 8 by reaction with 4-chloro- or 4-nitrobenzenesulphonyl chloride, respectively, in pyridine. Treatment of 8 with lithium hydroxide in aqueous tetrahydrofuran for 15 min gave the sulphonamide 9. Tin (II) chloride dihydrate reduction of 9 for 3 h in boiling ethyl acetate afforded amino derivative 10 that was converted into pyrrole derivative 11 with 2,5-dimethoxytetrahydrofuran in glacial acetic acid at 80 °C for 1 h. Derivative 2 was prepared from 11 by MW-assisted reaction with benzoyl chloride in the presence of anhydrous aluminium chloride in 1,2-dichloroethane at 110 °C (70 W) for 3 min.
Scheme 1. Synthesis of the final compounds 1–3. (a) TEA, reflux, 7 h. (b) 4-Cl- or 4-NO2-C6H4SO2Cl, pyridine, 60 °C, overnight. (c) LiOH, 50 °C, 15 min. (d) SnCl2, 80 °C, 3 h. (e) 2,5-(OMe)2-THF, 100 °C, 1 h. (f) benzoyl chloride, AlCl3, closed vessel, 110 °C, 70 W, 4 min.
Indole derivatives 4 and 5 were synthesised as shown in Scheme 2. Final compound 4 and the intermediate 13 were prepared by MW-assisted aroylation of methyl 5-chloro-1H-indole-2-carboxylate (12) with 4-hydroxy-3,5-dimethoxybenzoyl chloride or 3,5-dimethoxybenzoyl chloride, respectively, in the presence of anhydrous aluminium chloride in 1,2-dichloroethane at 110 °C (70 W) for 2 min. Treatment of 13 with triethylsilane in trifluoroacetic acid for 48 h afforded the corresponding methylene compound 5.
Virtual screening studies
In search for allosteric inhibitors of the NS2B/NS3 protease, we carried out virtual screening (VS) and docking studies. We focused our VS studies against the allosteric site, of the enzymeCitation42,Citation43. To our knowledge, only few protease allosteric inhibitors have been reportedCitation30, and many of them are natural compounds that do not possess drug-like propertiesCitation30. We performed structure-based screening studies on an open conformation of the NS2B/NS3 protease structureCitation44 (pdb code: 2FOM)Citation27 available at the protein data bank webserverCitation45. This structure of the NS2B/NS3 protease was previously used in VS campaign in search for new allosteric inhibitorsCitation43.
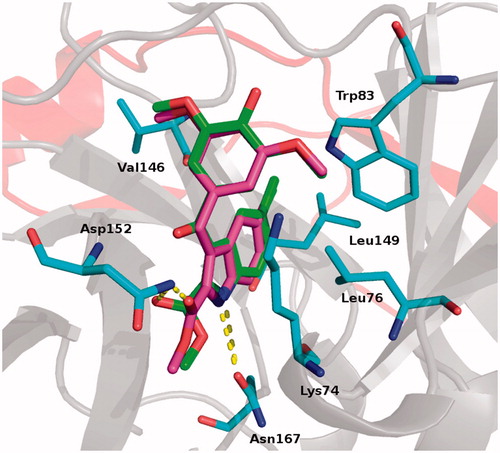
Compounds 4 and 5 shared virtually superimposable binding modes: (a) the 2-methoxycarbonyl ester occupied the inner part of the binding pocket and was stabilised by an H-bond with Asp152 side chain; (b) the indole NH formed an H-bond with Asn167; (c) the indole ring arranged a series of hydrophobic contacts with Val146, Leu149 and Leu76; (d) the methoxylphenyl moiety of both inhibitors formed hydrophobic interactions with Trp83 and a pi-cation interaction with Lys74. ().
Biological activity
DENV NS5 RdRp inhibitors
Compounds 1–3 were evaluated in DENV-2 infected Huh-7 cells by determining DENV RNA levels using specific qRT-PCR primer targeting viral NS5 levels, normalised to cellular GADPH mRNA levels. Compound 1 inhibited the DENV-2 with EC50 of 11.09 ± 0.21 µM and selectivity index (SI = CC50/EC50 ratio) of 17.7. Compound 2 bearing a benzoyl tail at position 2 of the pyrrole potently inhibited DENV-2 with EC50 of 7.61 ± 0.36 µM and SI >26.3. 1-Phenylpyrazole derivative compound 3 with the 4-nitrophenyl group showed potent anti-DENV-2 activity with EC50 of 5.66 ± 0.25 µM and SI > 35.3. Collectively, compounds 1–3 strongly reduced DENV RNA level ( and , panel A).
Figure 2. Inhibition of DENV RNA replication and RdRp activity by compounds 1–3. Panel A. Huh-7 cells were infected with DENV-2 at a multiplicity of infection (M.O.I) of 0.2 and followed by treatment with the DENV polymerase inhibitor for 3 days. The DENV RNA level was analysed by RT-qPCR with specific primer targeting viral NS5 gene, and relative viral RNA levels were normalised against cellular GADPH mRNA levels. Treatment of 50 μM 2′-C-methylcytidine (2CMC) direct against DENV RdRp served as positive control. 0.1% DMSO (Mock) served as negative control. Panel B. Huh-7 cells were transfected with pEG(MITA)SEAP and pcDNA-NS2B-GSG-NS3-Myc followed by incubation with each compound. The luciferase activity was analysed after 3 days treatment. Error bars denote the means ± SD of three independent experiments. *p < .05; **p < .01.
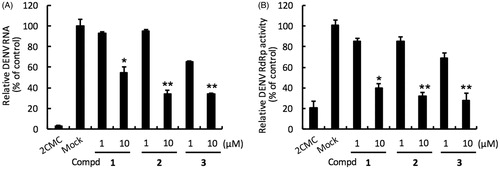
To confirm the anti-RdRp activity of 1–3, we performed a cell-based RdRp reporter assay. The Huh-7 cells were transiently expressed p(+)RLuc-(−)DV-UTRΔC-FLuc and DENV NS5 expression vector pcDNA-NS5-Myc and then treated with the tested compound. Our data showed that compounds 1–3 efficiently inhibited DENV NS5 RdRp activity with EC50 of 8.07 ± 0.26, 7.22 ± 0.38 and 5.99 ± 0.30 µM ( and , panel B). To further confirm the inhibitory specificity of compounds 1–3 against NS5 RdRp activity, we performed an enzyme-based FAPA assay. Our data showed that compounds 1–3 exhibited direct-action against DENV RdRp activity () with EC50 of 7.78 ± 0.31, 5.34 ± 0.17 and 4.87 ± 0.24 µM, respectively, whose values correlated with results of cell-based assay. Taken together, these results demonstrated that compounds 1–3 attenuate DENV replication by directly inhibiting DENV RdRp activity.
DENV NS3 protease inhibitors
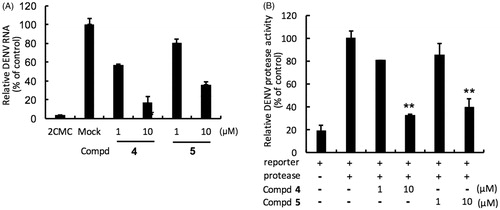
Huh-7 cells were infected with DENV-2 and then treated with protease inhibitors for 3 days and DENV RNA synthesis was evaluated with specific qRT-PCR primers targeting viral NS5 gene. Compounds 4 and 5 strongly inhibited the DENV-2 DNA replication with EC50s of 4.60 ± 0.03 µM and 7.28 ± 0.13 µM, respectively ( and , panel A). In addition, a cell-based DENV protease reporter assay was used to characterise the protease specificity of compounds 4 and 5. Huh-7 cells were transfected with pEG(MITA)SEAP and DENV protease expression vector pcDNA-NS2B-GSG-NS3-Myc and then treated with 4 or 5. Both compounds showed strong inhibitory activity against the DENV protease with EC50 of 6.71 ± 0.20 µM and 7.92 ± 0.62 µM, respectively ( and , panel B). We further performed an enzyme-based assay to evaluate the inhibitory specificity of compounds 4–5. The data showed that compounds 4–5 specifically suppressed DENV protease activity with EC50 of 4.72 ± 0.3 µM and 6.90 ± 0.11 µM, respectively (). In conclusion, the results indicated that both compound 4 and 5 suppress DENV replication by directly inhibiting DENV protease activity.
Figure. 3. Inhibition of DENV RNA and NS3-protease activity by compounds 4 and 5. Panel A. Huh-7 cells were infected with DENV-2 at a multiplicity of infection (M.O.I) of 0.2 and followed by the treatment of each DENV protease inhibitors for 3 days. The DENV RNA level was analysed by RT-qPCR with specific primer targeting viral NS5 gene, and relative viral RNA levels were normalised against cellular GADPH mRNA levels. Treatment of 50 μM 2′-C-methylcytidine (2CMC) direct against DENV RdRp served as positive control. 0.1% DMSO (Mock) served as negative control. Panel B. Huh-7 cells were transfected with pEG(MITA)SEAP and pcDNA-NS2B-GSG-NS3-Myc followed by incubation of each compounds, and the luciferase activity was analysed after 3 days treatment. Error bars denote the means ± SD of three independent experiments. *p < .05; **p < .01.
Evaluation of 3 and 4 in DENV-infected ICR-suckling mouse model
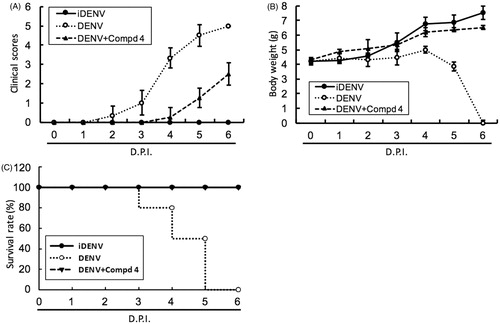
To access the antiviral activity of compound 3 and 4 in vivo, 6-day-old ICR-suckling mice were inoculated with 2.5 × 105 pfu of DENV-2 by intracerebral injection and then were administered with compound 3 (10 mg/kg) or 4 (1 mg/kg) by intracerebral injection at 1-, 3- and 5-day postinfection (dpi); inoculation of heat-inactivated DENV-2 (iDENV) served as a mock control. The results of clinical scores () and body weight () showed that compound 3 or compound 4 treatment reduced DENV-induced pathology, including ruffled fur, anorexia, severe paralysis, lethargy, on DENV-infected mice within 4 to 6 dpi. Furthermore, compound 3 or 4 treatment significantly protected the mice against life-threatening DENV infection, as compared to the untreated mice (). Collectively, these results demonstrated that both classes of small molecules, targeting polymerase and protease activities, inhibited DENV replication and represent prototypical DAA molecules for further development through compound modification.
Figure 4. DENV RdRp inhibitor 3 protected ICR-suckling mice from DENV infection. 6-day-old ICR-suckling mice were intracerebrally injected with heat-inactive DENV (iDENV, n = 5) or active DENV (DENV, n = 4). Mice-receiving DENV were treated with 10 mg/kg of compound 3 (n = 5) at 1, 3, 5 dpi. Panel A, clinical scores; Panel B, body weight and Panel C, survival rate were recorded every day. Disease severity was scored as follow: 0: healthy, 1: slightly sick (reduced mobility), 2: inappetance, 3: weight loss and difficult to move, 4: paralysis, 5: death. Each group included 6 mice. Error bars denote the means ± SD.
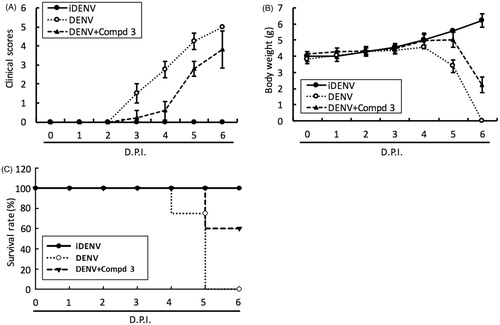
Figure 5. DENV protease inhibitor 4 protected ICR-suckling mice from DENV infection. Six-day-old ICR-suckling mice were intracerebrally injected with heat-inactive DENV (iDENV, n = 6) or active DENV (DENV, n = 6). Mice-receiving DENV were treated with 1 mg/kg of compound 4 (n = 6) at 1, 3, 5 dpi. Panel A, clinical scores, Panel B, body weight, and Panel C, survival rate were recorded every day. Disease severity was scored as follow: 0: healthy, 1: slightly sick (reduced mobility), 2: inappetance, 3: weight loss and difficult to move, 4: paralysis, 5: death. Each group included six mice. Error bars denote the means ± SD.
Synergistic effect of NS5 RdRp inhibitor 3 and NS3 protease inhibitor 4 against DENV replication
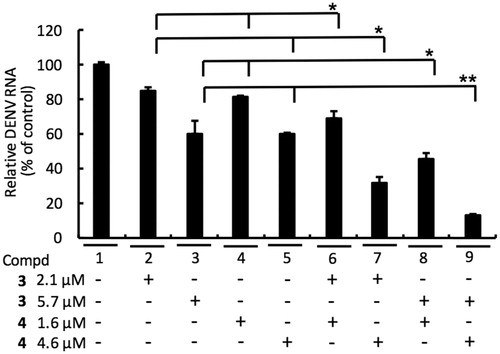
To examine whether combinational treatment of NS5 RdRp inhibitor 3 and NS3 protease inhibitor 4 could synergistically inhibit DENV replication, DENV-2-infected Huh-7 cells were cotreated with 3 and 4 at the indicated concentration for 3 days. As shown in , a synergistic inhibition of DENV replication was observed with the 3 and 4 combination (lines 6–9), as compared to single-compound treatment (lines 2–5).
Figure 6. Combination assay of RdRp (3) and NS3 protease (4) inhibitors. Huh-7 cells were infected with DENV-2 at a multiplicity of infection (M.O.I) of 0.2 and followed by treatment of 3 and 4 with indicated concentration for 3 days. The DENV RNA level was analysed by RT-qPCR with specific primer targeting viral NS5 gene, and relative viral RNA levels were normalised against cellular GADPH mRNA levels. Error bars denote the means ± SD of three independent experiments. *p < .05; **p < .01.
Conclusions
We sought to design and characterise potential anti-DENV inhibitors by targeting the viral enzymatic, NS5 RNA-dependent RNA polymerase and the NS3 protease activities. We identified five potential compounds demonstrating anti-DENV replication activity without cytotoxicity. Two compounds targeting DENV NS3 protease and NS5 RdRp also exhibited anti-DENV activity in ICR-suckling mouse model of DENV infection. Combination treatment exhibited enhanced inhibition of DENV replication. Taken together, these results demonstrate that both classes of small molecules inhibited DENV replication and represent prototypical DAA molecules for further development through compound modification.
Acknowledgements
The authors would like to thank Dr. Huey-Nan Wu (Institute of Molecular Biology, Academia Sinica, Taipei, Taiwan) for giving DENV-2 (type-2 strain 16681) and Dr. Charles Rice (Rockefeller University and Apath, LCC, USA) for kindly providing human hepatoma Huh-7 cells.
Disclosure statement
The authors report no declarations of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Rothman AL. Immunity to dengue virus: a tale of original antigenic sin and tropical cytokine storms. Nat Rev Immunol 2011;11:532–43.
- Halstead SH. Dengue virus-mosquito interactions. Annu Rev Entomol 2008;53:273–91.
- Wilder-Smith A, Ooi EE, Vasudevan SG, Gubler DJ. Update on dengue: epidemiology, virus evolution, antiviral drugs, and vaccine development. Curr Infect Dis Rep 2010;12:157–64.
- Roche C, Cassar O, Laille M, Murgue B. Dengue-3 virus genomic differences that correlate with in vitro phenotype on a human cell line but not with disease severity. Microbes Infect 2007;9:63–9.
- Shurtleff AC, Beasley DW, Chen JJ, et al. Genetic variation in the 3' non-coding region of dengue viruses. Virology 2001;281:75–87.
- Silva RLA, de Silva AM, Harris E, MacDonald GH. Genetic analysis of dengue 3 virus subtype III 5′ and 3′ non-coding regions. Virus Res 2008;135:320–5.
- Vasilakis N, Weaver SC. The history and evolution of human dengue emergence. Adv Virus Res 2008;72:1–76.
- Zhou Y, Mammen MP, Klungthong C, et al. Comparative analysis reveals no consistent association between the secondary structure of the 3’-untranslated region of dengue viruses and disease syndrome. J Gen Virol 2006;87:2595–603.
- Dejnirattisai W, Jumnainsong A, Onsirisakul N, et al. Cross-reacting antibodies enhance dengue virus infection in humans. Science 2010;328:745–8.
- Beltramello M, Williams KL, Simmons CP, et al. The human immune response to dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 2010;8:271–83.
- de Alwis R, Beltramello M, Messer WB, et al. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis 2011;6:e1188.
- Smith SA, de Alwis AR, Kose N, et al. Isolation of dengue virus-specific memory B cells with live virus antigen from human subjects following natural infection reveals the presence of diverse novel functional groups of antibody clones. J Virol 2014;88:12233–41.
- Yotmanee P, Rungrotmongkol T, Wichapong K, et al. Binding specificity of polypeptide substrates in NS2B/NS3pro serine protease of dengue virus type 2: a molecular dynamics study. J Mol Graph Model 2015;60:24–33.
- Khromykh AA, Kenney MT, Westaway EG. Trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol 1988;72:7270–9.
- Rawlinson SM, Pryor MJ, Wright PJ, Jans DA. Dengue virus RNA polymerase NS5: a potential therapeutic target? Curr Drug Targets 2006;7:1623–38.
- Tatem AJ, Hay SI, Rogers DJ. Global traffic and disease vector dispersal. Proc Natl Acad Sci USA 2006;103:6242–7.
- Halstead SB. Licensed Dengue vaccine: public health conundrum and scientific challenge. Am J Trop Med Hyg 2016;95:741–5.
- Guy E, Lang J, Saville M, Jackson N. Vaccination against Dengue: challenges and current developments. Ann Rev Med 2016;67:387–404.
- Noble CG, Chen YG, Dong H, et al. Strategies for development of dengue virus inhibitors. Antivir Res 2010;8:5450–62.
- Lim SP, Christian G, Noble CG, Shi PY. The dengue virus NS5 protein as a target for drug discovery. Antiviral Res 2015;119:57–67.
- Tomlinsona SM, Malmstroma RD, Russoa A, et al. Structure-based discovery of dengue virus protease inhibitors. Antiviral Res 2009;82:110–14.
- Silvestri R, Cascio MG, La Regina G, et al. Synthesis, cannabinoid receptor affinity and molecular modeling studies of substituted 1-aryl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides. J Med Chem 2008;51:1560–76.
- La Pietra V, La Regina G, Coluccia A, et al. Design, synthesis, and biological evaluatio of 1-phenylpyrazolo[3,4-e]pyrrolo[3,4-g]indolizine-4,6(1H,5H)-diones as new glycogen synthase kinase-3β inhibitors. J Med Chem 2013;56:10066–78.
- Manvar D, Pelliccia S, La Regina G, et al. New 1-phenyl-5-(1H-pyrrol-1-yl)-1H-pyrazole-3-carboxamides inhibit hepatitis C virus replication via suppression of cyclooxygenase-2. Eur J Med Chem 2015;90:497–506.
- Khan MF, Alam MM, Verma G, et al. The therapeutic voyage of pyrazole and its analogs: a review. Eur J Med Chem 2016;120:170–201.
- Corelli F, Massa S, Stefancich G, et al. Agenti antiinfiammatori non steroidei. Nota V. - Sntesi di acidi 1-aril-5-(1-pirril)pirazolil-4-acetici a potenziale attività antiinfiammatoria. Farmaco 1988;43:251–65.
- Erbel P, Schiering N, D'Arcy A, et al. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol 2006;13:372–3.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3–26.
- Korb O, Stutzle T, Exner TE, PLANTS: Application of ant colony optimization to structure-based drug design. In: Dorigo M, Gambardella LM, Birattari M, Martinoli A, Poli R, Stutzle T, eds. Ant colony optimization and swarm intelligence. Proceedings of the 5th International Workshop, ANTS, Lecture Notes in Computer Science, Series 4150, Berlin: Springer; 2006:47–258.
- Timiri AK, Sinha BN, Jayaprakash V. Progress and prospects on DENV protease inhibitors. Eur J Med Chem 2016;117:125–43.
- PyMOL, v1.8.6.0, release March 9, 2017. Available from: https://www.pymol.org.
- Lin YT, Wu YH, Tseng CK, et al. Green tea phenolic epicatechins inhibit hepatitis C virus replication via cycloxygenase-2 and attenuate virus-induced inflammation. PLoS One 2013;8:e54466.
- Lee JC, Tseng CK, Wu YH, et al. Characterization of the activity of 2'-C-methylcytidine against dengue virus replication. Antiviral Res 2015;116:1–9.
- Wu SF, Lin CK, Chuang YS, et al. Anti-hepatitis C virus activity of 3-hydroxy caruilignan C from Swietenia macrophylla stems. J Viral Hepat 2012;19:364–70.
- Mayani M, Filipe CDM, Ghosh R. Cascade ultrafiltration systems integrated processes for purification and concentration of lysozyme. J Membrane Sci 2010;347:150–8.
- Hartwig S, Raschke S, Knebel B, et al. Secretome profiling of primary human skeletal muscle cells. Biochim Biophys Acta 2014;1844:1011–17.
- Chao WW, Hong YH, Chen ML, Lin BF. Inhibitory effects of Angelica sinensis ethyl acetate extract and major compounds on NF-κB trans-activation activity and LPS-induced inflammation. J Ethnopharmacol 2010;129:244–9.
- Hu G, Lou Z, Gupta M. The long non-coding RNA GAS5 cooperates with the eukaryotic translation initiation factor 4E to regulate c-Myc translation. PLoS One 2014;9:e107016.
- Tseng CK, Lin CK, Wu YH, et al. Human heme oxygenase 1 is a potential host cell factor against dengue virus replication. Sci Rep 2016;6:32176.
- Tannous BA. Gaussia luciferase reporter assay for monitoring biological processes in culture and in vivo. Nat Protoc 2009;4:582–91.
- Tomlinson SM, Watowich SJ. Anthracene-based inhibitors of dengue virus NS2B-NS3 protease. Antiviral Res 2011;89:127–35.
- 7-Amino-4-methylcoumarin (AMC) fluorophore-linked peptide substrate Boc-GRR-AMC. Bachem, CA, USA; 2016. Available from: http://www.bachem.com/ [last accessed 29 Nov 2016].
- Yildiz M, Ghosh S, Bell AJ, et al. Allosteric inhibition of the NS2B-NS3 protease from dengue virus. ACS Chem Biol 2013;8:2744–52.
- Mukhametov A, Newhouse EI, Ab Aziz N, et al. Allosteric pocket of the dengue virus (serotype 2) NS2B/NS3 protease: in-silico ligand screening and molecular dynamics studies of inhibitors. J Mol Graph Model 2014;52:103–13.
- Chandramouli S, Joseph JS, Daudenarde S, et al. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J Virol 2010;84:3059–67.
- Protein data bank. Available from: http://www.rcsb.org/pdb/home/home.do [last accessed 6 Dec 2016].