?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Oestrogen related receptor α participated in the regulation of oxidative metabolism and mitochondrial biogenesis, and was overexpressed in many cancers including triple-negative breast cancer. A set of new ERRα inverse agonists based on p-nitrobenzenesulfonamide template were discovered and compound 11 with high potent activity (IC50 = 0.80 μM) could significantly inhibit the transcription of ERRα-regulated target genes. By regulating the downstream signalling pathway, compound 11 could suppress the migration and invasion of the ER-negative MDA-MB-231 cell line. Furthermore, compound 11 demonstrated a significant growth suppression of breast cancer xenograft tumours in vivo (inhibition rate 23.58%). The docking results showed that compound 11 could form hydrogen bonds with Glu331 and Arg372 in addition to its hydrophobic interaction with ligand-binding domain. Our data implied that compound 11 represented a novel and effective ERRα inverse agonist, which had broad application prospects in the treatment of triple-negative breast cancer.
1. Introduction
Breast cancer is one of the most common malignancies in women worldwide, with more than 2 million newly diagnosed female breast cancer cases in 2018, accounting for almost 25% of the cancer cases among womenCitation1. Although the existing endocrine and targeted therapy had significant effects on breast cancer, triple-negative breast cancer still had a high metastasis recurrence rate and poor prognosisCitation2–5. The overexpression of ERRα was proved to be related to the poor prognosis of TNBC in former studiesCitation6. By interacting with PGC-1α and RIP-140Citation7,Citation8, ERRα could adjust the expression of oxidative metabolism genes and mitochondrial biosynthesis, which caused the hypoxia of tumour cellsCitation9. Through the protein kinase ERK signalling pathway, reducing the expression of ERRα demonstrated the anti-proliferate activity on vascular smooth muscle cellsCitation10. Downregulation of ERRα could prevent cancer angiogenesis through PI3K/Akt/STAT3 signalling pathway and reduce the production of vascular endothelial growth factors (VEGF)Citation11. Research on the MDA-MB-231 cell line with malignant metastasis revealed that turning down the expression level of ERRα could effectively inhibit the migration of tumour cellsCitation12,Citation13. Knocking out the ERRα gene could slow down the growth rate of tumour cells without affecting the health of miceCitation14. Therefore, ERRα might be a promising target to treat triple-negative breast cancer. Studies had shown that the development of effective ERRα inverse agonists was considered as a potential therapy for non-hormone-dependent breast cancer, especially triple-negative breast cancerCitation15.
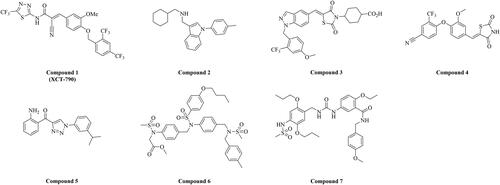
Few ERRα inverse agonists (compounds 1–7) were discovered as shown in . The inverse agonists with thiadiazole acrylamide template were the first reported series, and XCT-790 (compound 1) was the most active compound with strong selectivity and moderate activity (IC50 = 0.37 μM) in vitroCitation16. The N-arylindole analogue compound 2 was the ligand of the first reported inverse agonist crystalline complex 2PJL, demonstrating moderate activity (IC50 = 0.19 μM) in FRET assayCitation17. The crystalline complex (PDB code: 3K6P) with covalent binding between residue Ser325 and ligand was constituted of ERRα and inverse agonist compound 4 based on thiazolidinedione scaffold, and further optimisation gave the most active compound 3 (IC50 = 0.008 μM)Citation18,Citation19. Compound 5 based on 1-Phenyl-4-benzoyl-triazole template demonstrated high potent activity on diabetes (IC50 = 0.021 μM) and oral availabilityCitation20. Compound 6 (IC50 = 1.47 μM) was considered as a potential ERRα inverse agonist, which was proved to suppress the growth of human xenografts (43.7%) in nude miceCitation21. Compound 7 with 1–(2,5-diethoxy-benzyl)-3-phenylurea temple revealed moderate activity in TR-FRET assay (IC50 = 1.46 μM), and demonstrated notably growth inhibition (40.9%) on the ER-negative human breast cancer xenografts in vivoCitation22. However, very few ERRα inverse agonists had been described, especially in triple-negative breast cancer. In this paper, a new type of p-nitrobenzenesulfonamide analogues was identified as high potent ERRα inverse agonists in vitro and in vivo against triple-negative breast cancer.
2. Design
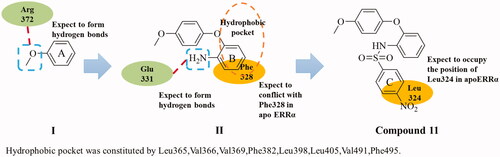
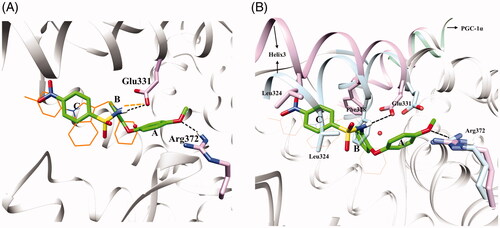
Comparison of the ERRα structure from the crystalline complex with inverse agonist compound 2 (PDB code: 2PJL)Citation17 and the crystalline structure of apoERRα (PDB code: 1XB7)Citation23 demonstrated that the conformational inversions of Leu324 and Phe328 were the key to the inverse agonistic activity of ERRα. Crystalline complex 2PJL revealed that the ligand-binding domain was mainly composed of hydrophobic residues and the hydrogen bond interaction with Glu331 might increase the activity. The hydrogen bond between compound 4 and Arg372 in crystalline complex 3K6PCitation18 indicated that Arg372 might be a key residue to activity. All these features provide the guidance for the design of novel ERRα inverse agonists.
As shown in , the design of novel high effective ERRα inverse agonists was started from fragment I with the oxygen atom for forming a hydrogen bond with Arg372. To occupy the position of Phe328 in apoERRα, the ring B was introduced at the para position of the methoxy group, and then fragment II was obtained. The ortho amino on the ring B was intended to interact with the Glu331. To occupy the position where Leu324 in apoERRα was, p-nitrosulfonyl group was designed to fragment II through sulfonylation with amino on the ring B and compound 11 was obtained.
To explore the structure-active relationships on the ring A, C and nitrogen atom of sulphonamide, compounds 8–18 were designed and synthesised for the test of biological activity.
3. Molecular docking results
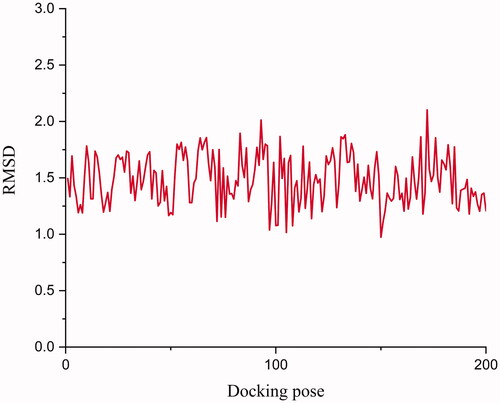
In order to gain insight into the hypothetical binding mode of compound 11 with ERRα, the docking simulation was performed by the Sybyl X 2.0 with Surflex-dock package. In the docking simulation, each ligand was generated 200 poses by Surflex-dock package, and the Surflex-dock GeomX was selected as the docking mode. As shown in , the RMSD between co-crystallized ligand (compound 2) and docking poses were relatively stable, which could validate the docking parameters as reliable parameters. The binding site could be regarded as a sphere with a radius of 8 Å.
Figure 3. RMSD between co-crystallized ligand (compound 2) and docking pose. The docking pose was generated by Surflex-dock package in Sybyl X 2.0. The RMSD was calculated by Discovery studio 2019.
The docking result of ERRα with compound 11 was shown in . The oxygen atom of methoxy on ring A and the nitrogen atom of sulphonamide on ring B formed hydrogen bonds with Arg372 and Glu331 respectively (), which could enhance the stability of compound 11 and ERRα. The positions Phe328 and Leu324 in apoERRα (PDB code: 1XB7) were occupied by ring B and ring C of compound 11 respectively in reverse-excited ERRα (PDB code: 2pjl) (), which transferred the apoERRα into in-active conformation. All these transformations made compound 11 act as an inverse agonist.
Figure 4. (A) Superimposition of docking result of ERRα with compound 11 (green) and crystal structure of ERRα with inverse agonist (compound 2, orange, PDB code 2PJL). The dotted black lines in the figure represent the hydrogen bonding interactions between ligand and protein. (B) The superimposed apoERRα crystal complex (1XB7) and compound 11 (green) docked with ERRα inverse agonist crystal complex crystals (2PJL). Plum: 2PJL, Sky blue: 1XB7. The dotted black lines in the figure represent the hydrogen bonding interactions between ligand and protein. These photographs were obtained by the chimaera 1.15rc program.
4. Chemistry
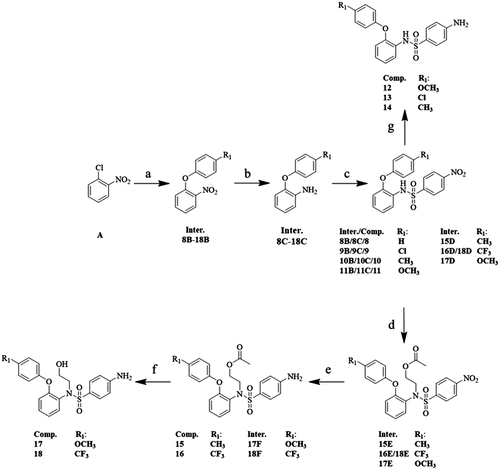
The synthetic steps of p-nitrobenzenesulfonamide analogues 8–18 were shown in Scheme 1. Intermediates 8B–18B were obtained from raw material A through arylation reaction with K2CO3 in dry DMF at 120 °C. After nitro reduction, intermediates 8C–18C were produced from intermediates 8B–18BCitation24. Through sulfonylation reaction in DCM at 0 °C, the intermediates 8C–18C were transformed into intermediates 12D–18DCitation25. After nitro reduction with NaBH4 in MeOH, intermediates 12D–18D were converted to products 12–14Citation24. Intermediates 15D–18D reacted with 2-bromoethyl acetate and produced intermediates 15E–18E, which were sequentially subjected to nitro reduction to give products 15–16 and intermediates 17F–18FCitation24. Products 17–18 were prepared through hydrolysation reaction with K2CO3 in MeOH at 60 °C.
5. Results and discussion
5.1. ERRα transcriptional activity and structure-activity relationships
The transcriptional activity of compounds 8–18 at cells level was shown in . When methyl group on 4-position of ring A and R2 without substituent, the inhibiting activity of nitro-substituted compound 10 (IC50 = 1.23 μM) is 1.5 times that of amino-substituted compound 14 (IC50 = 1.88 μM). Replacing the methyl group on ring A of compound 10 by methoxy group gave compound 11 (IC50 = 0.80 μM) with 1.5-fold more active than compound 10. After reducing the nitro of compound 11, the corresponding amino compound 12 was inactive. It was suggested that nitro-substituted compounds 10 and 11 with methyl or methoxy on ring A were more beneficial to activity than corresponding amino-substituted compounds 14 and 12. By introducing ethyl acetate to the R2 group of compound 14, a slightly better analogue 15 (IC50 = 1.78 μM) was obtained. Replacing methyl on R1 with trifluoromethyl to optimise compound 15, more potent compound 16 (IC50 = 1.66 μM) was discovered. After introducing ethanol to R2 of inactive compound 12, the obtained compound 17 (IC50 = 1.29 μM) regained its activity. Compound 17 (IC50 = 1.29 μM) with methoxy group exhibited more potent ERRα transrepression activity than compound 18 (IC50 = 1.78 μM) possessing a trifluoromethyl group. The trend observed for compounds 8–11 was that substituted methoxy group (compound 11, IC50 = 0.80 μM) took precedence over methyl group (compound 10, IC50 = 1.23 μM), the hydrogen atom (in compound 8, IC50 = 1.76 μM) and the chlorine atom (in compound 9, IC50 = 2.02 μM). Amino compound 13 (IC50 = 1.61 μM) with chlorine on ring A exhibited more inhibitory than corresponding nitro compound 9 (IC50 = 2.02 μM). The most active compound 11 with methoxy and nitro groups was selected for further study of its biological activity.
Table 1. The inhibition of compounds 8–18 on ERRα transcriptional activity.
5.2. Compound 11 inhibited the proliferation of human breast cancer cells
Base on the previous results, cell proliferation assay and colony formation assay was performed to detect the anticancer effect of the most potent ERRα inverse agonist (compound 11) in human breast cancer cells. The results of the present study showed that compound 11 demonstrated excellent anti-proliferation activity in breast cancer cells by CCK-8 assay. Treatment with 10 μM compound 11 suppressed cell proliferation by 23.5%, 38.9% and 31.0% at 24 h and 37.4%, 44.3% and 38.0% at 48 h for MCF-7, MDA-MB-231 and HCC-1937, respectively (p < 0.05 for all) ((A)). And with an IC50 value at 15.44, 9.15 and 14.67 μM for MCF-7, MDA-MB-231 and HCC-1937, respectively ().
Figure 5. Compound 11 inhibits growth and colony formation of breast cancer cells. (A) Influence of compound 11 on the viability of breast cancer cells MDA-MB-231, MCF-7 and HCC-1937. Cells were treated with vehicle or compound 11 (1, 3, 10 μM) for 24 h and 48 h, the viability was detected with the CCK-8 kit. (B) Breast carcinoma cells were treated with various concentrations of compound 11 for 24 h. Cell proliferation was determined by the CCK-8 assay. IC50 value of compound 11 in breast cancer cell lines was then determined. (C) Efficacy of compound 11 on colony formation of breast cancer cells. (D) Breast cancer cells were treated with compound 11 (0.3, 1 μM) for 7 days, and then fixed, stained and counted. Values are the means ± SEM (n = 3). *p < 0.05 or **p < 0.01 indicates significant differences from the vehicle group as assessed by a two-tailed unpaired Student’s t-test.
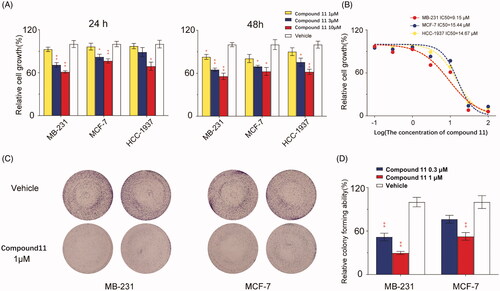
When evaluated by the colony formation assay, high-dose compound 11 (1 μM) significantly reduced the number of colonies by 47.5% and 70.3% for MCF-7 and MDA-MB-231, respectively (p < 0.01 for both). The number of colonies decreased significantly by low-dose compound 11 (0.3 μM) only in MDA-MB-231 cells (). The results showed that the killing influence of compound 11 on ER-negative MDA-MB-231 cells was higher than that on ER-positive MCF-7 cells. The consequences mentioned above indicated that compound 11 was a more effective and sensitive breast cancer cell proliferation inhibitor, which was regardless of whether the ER-positive.
5.3. Compound 11 inhibited human breast cancer cells migration and invasion
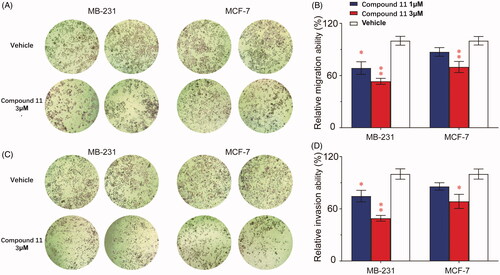
The increase in motility and invasiveness of cancer cells is related to the progression of breast cancer, which was the main cause of incidence rate and mortality of cancer patients. Experiments were performed to evaluate the inhibition rate of compound 11 on breast tumour cells migration and invasion. Transwell migration assay showed that compound 11 (1 μM) had moderate inhibition of breast cancer cells migration, and high-dose compound 11 (3 μM) had a much more potently inhibitory effect (). In analogy with the results above, low-dose compound 11 had a weak effect on MB-231cells invasion, but high-dose compound 11 treatment led to 31.5% and 50.9% reduction in MCF-7 and MDA-MB-231 cell invasion, respectively (). Taken together, these results showed that compound 11 markedly improved the antimetastatic activity against human breast cancer cells.
Figure 6. Compound 11 inhibits breast cancer cells migration and invasion. (A, B) MCF-7 and MDA-MB-231 cells were treated with compound 11 (1, 3 μM) for 24 h. Transwell migration assays were used to determine the migratory ability of MCF-7 and MDA-MB-231 cells. The experiments were performed in quadruplicate, and data represent mean ± SEM, *p < 0.05 and **p < 0.01. (C, D) MCF-7 and MDA-MB-231 cells were treated with compound 11 (1, 3 μM) for 24 h. Transwell invasion assays were performed in Matrigel to determine the invasion ability of MCF-7 and MDA-MB-231 cells. Values are the means ± SEM (n = 4). *p < 0.05 or **p < 0.01 indicates significant differences from the vehicle group as assessed by a two-tailed unpaired Student’s t-test.
5.4. Compound 11 is an inverse agonist of ERRα and mediates anti-proliferation effect through ERRα in breast cancer cell
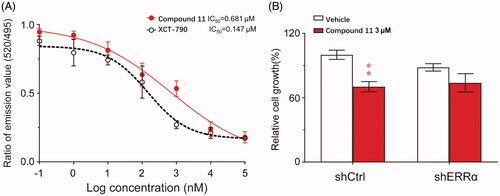
Through physical binding with co-activator PGC-1α, the transcriptional function of ERRα was activated. TR-FRET (time-resolved fluorescence energy transfer) analysis was used to determine the interaction between a terbium chelate labelled ERRα LBD fragment and the PGC-1α co-activator peptide with fluorescein. Therefore, the receptor affinity with ERRα of compound 11 was measured by assessing the ability to interfere with the interaction of ERRα with PGC-1α in a TR-FRET assay. Similar to the positive control XCT-790, an effective ERRα specific inverse agonist, compound 11 also dose-dependently decreased the FRET signal between ERRα-LBD and PGC-1α with an IC50 value of 0.681 μM (). Thence, compound 11 could bind directly with ERRα and disturb the interaction with its co-factors, which was supposed to act as an ERRα particular inverse agonist. To explore the relationship between the ERRα and the anti-proliferation effect of compound 11, we knocked down ERRα in MDA-MB-231 cells by siRNA. The results suggested knock-down of ERRα reduced compound 11 induced anti-proliferation in breast cancer cells ().
Figure 7. Compound 11 is an inverse agonist of ERRα. (A) TR-FRET assay was used to detect whether compound 11 could bind to ERRα LBD to suppress the interaction with PGC-1α peptide. (B) Effect of compound 11 on the viability of MB-231 cells after treatment with shERRα or shCtrl. Values are the means ± SEM (n = 3). *p < 0.05 or **p < 0.01 indicates significant differences from the vehicle group as assessed by a two-tailed unpaired Student’s t-test.
5.5. Derivative 11 inhibits the expression of target genes regulated by ERRα
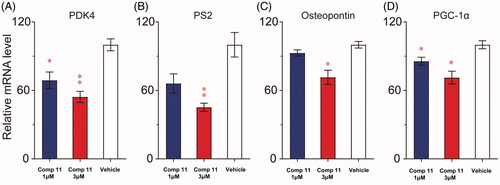
Then, quantitative RT-PCR was used to assess the mRNA levels of ERRα-regulated target genes including peroxisome proliferator-activated receptor coactivator 1α (PGC-1α), pyruvate dehydrogenase kinase 4 (PDK4), osteopontin (SPP1) and pS2 (TFF1) in breast cancer cells hatched with derivative 11. The results demonstrated that PGC-1α, PDK4, osteopontin and pS2 mRNA expression in high-dose derivative 11 (3 μM) treated MDA-MB-231 cells were all notably reduced (. All the consequences indicated that derivative 11 reduced the transcription of target genes adjusted by ERRα.
Figure 8. Compound 11 down-regulates the expression of ERRα-regulated target genes. (A–C) MDA-MB-231 cells were incubated with vehicle or compound 11 (1, 3 μM) for 24 h, and the mRNA levels of ERRα target genes (PDK4, pS2 and osteopontin) were determined by RT-PCR analysis. (D) Effect of compound 11 on mRNA levels of PGC-1α. Values are the means ± SEM (n = 3). *p < 0.05 or **p < 0.01 indicates significant differences from the vehicle group as assessed by a two-tailed unpaired Student’s t-test.
5.6. Compound 11 suppresses the growth of tumour cells in xenograft mice
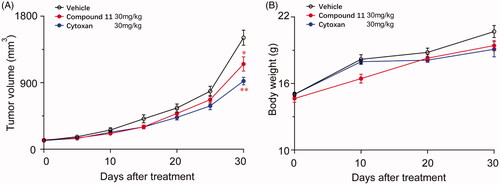
The results of in vitro study suggested that compound 11 might demonstrate inhibition of cancer in vivo. To confirm this surmise, we established a breast tumour xenograft in nude mice. As shown in , compound 11 showed an effective inhibitory influence on the growth of xenograft tumour of human breast cancer. At the end of this experiment, the tumour volume of mice treated with compound 11 dose of 30 mg/kg every other day was 1152 mm3, while the tumours size of control mice had reached 1508 mm3. After treatment with compound 11, tumour growth was inhibited by 23.58%. There were no notably compound-related influences on the weight of mice or any other symptoms of overt toxicity were observed in the treatment group compared with the control group (). Overall, these evidences proved that compound 11 might be a promising lead compound to treat triple-negative breast cancer (TNBC).
Figure 9. Compound 11 suppresses the growth of breast tumours in mice xenograft models. (A) The volume of the MDA-MB-231 xenograft tumour was measured after 30 days of treatment with vehicle or compound 11. Compound 11 was intraperitoneally injected 30 mg/kg every other day. (B) The body weight of mice in different treatment groups. Values are the means ± SEM (n = 6). *p < 0.05 or **p < 0.01 indicates significant differences from the vehicle group as assessed by a two-tailed unpaired Student’s t-test.
6. Conclusion
A set of original ERRα inverse agonists (compounds 8–18) based on p-nitrobenzenesulfonamide template were discovered by design, structural optimisation and ERRα assays in vitro and in vivo. The most potential leading compound of ERRα inverse agonist was compound 11 (IC50 = 0.80 μM) demonstrating potent anti-proliferate and anti-metastatic activity on MDA-MB-231 cells. Further assay in vitro revealed the expression of ERRα and target genes regulated by ERRα was notably decreased by compound 11 (IC50 = 0.681 μM). In human breast cancer xenografts experiment, compound 11 significantly suppressed the growth of tumour in nude mice with an inhibitory rate of 23.58%. Docking results indicated that the methoxy and nitrogen atom of sulphonamide in analogue 11 constituted hydrogen binding interactions with residues Glu331 and Arg372, which could increase the activity of compound 11 by enhancing the binding stability. Overall, these series compounds broken new ground for the reasonable exploitation of potent ERRα inverse agonists aimed at triple-negative breast cancer.
7. Experimental section
7.1. Chemistry experiments
7.1.1. General synthetic methods of compounds 8–18:
Unless otherwise noted, all analytical materials, such as reagents and solvents, were purchased through the reagent supplier and used directly. All reaction processes of the synthetic experiments were tracked by using thin-layer chromatography (TLC) on silica gel plates. All of the final compounds were defined by using the Bruker AVANCE II 400 spectrometers using the CDCl3 or DMSO as solvent.
7.1.2. 4-Nitro-N-(2-phenoxyphenyl)benzenesulfonamide (8)
The mixture of phenol (670 mg, 7.62 mmol, 1.2eq), potassium carbonate (2.19 g, 15.87 mmol, 2.5eq) and potassium iodide(105 mg, 0.635 mmol, 0.01eq) in N,N-dimethylformamide (DMF) was heated to 90 °C for 10 min, then the 2-chloronitrobenzene (1g, 6.35 mmol, 1.0eq) was added to the solution followed by stirring at 120 °C for 8 h. DMF was removed by distillation and the residue was dissolved in ethyl acetate and extracted with water (3 200 ml). The combined organic phases were dried with Na2SO4 and concentrated under reduced pressure to obtain pure brown liquid (8B) 2-nitrophenyl phenyl ether 1.22 g (yield 89%).
To a solution of 8B (1 g, 4.65 mmol, 1.0eq) in MeOH (10 ml) was added nickel chloride hexahydrate(1.3g, 5.58 mmol, 1.2eq), NaBH4 (530 mg, 13.95 mmol, 3.0eq), and the mixture was stirred at 0 °C for 2 h. The mixture was made acidic with a 10% HCl solution and extracted with ethyl acetate (3 150 ml), the organic layer was washed with a saturated NaCl solution (3
100 ml), and the combined organic phases were dried with Na2SO4, filtered, concentrated, and purified by flash column chromatography to give 2-phenoxy-aniline (8C, 689 mg, yield 80%) as a light brown liquid.
Intermediate 8C (600 mg, 3.24 mmol, 1.0eq) was dissolved in dichloromethane after addition of pyridine (1.3 g, 16.2 mmol, 5.0eq). The solution was stirred at 0 °C for 10 min and p-nitrobenzenesulfonyl chloride (718 mg, 3.24 mmol, 1.0eq) was added dropwise to the solution, followed by stirring at room temperature for 8 h. Dichloromethane was then removed by distillation. The solid residue was dissolved in dichloromethane (50 ml) and washed with a saturated NaCl solution (3 100 ml). The combined organic phases were dried with Na2SO4 and concentrated under reduced pressure. The remaining solid was purified by flash column chromatography to give the pale red solid N-(2-phenoxy -Phenyl)-benzenesulfonamide (compound 8, 1.1 g, yield 92%). 1H NMR (400 MHz, CDCl3) δ 8.14 (d, J = 8.8 Hz, 2H), 7.86 (d, J = 8.8 Hz, 2H), 7.74 − 7.65 (m, 1H), 7.20 (t, J = 7.9 Hz, 2H), 7.08 (d, J = 12.3 Hz, 3H), 6.73 (d, J = 9.3 Hz, 1H), 6.56 (d, J = 7.9 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 155.19, 149.70, 147.24, 144.08, 129.42, 129.14, 127.98, 126.31, 123.71, 123.29, 120.07, 117.93, 117.34, 114.84. HRMS (ESI+), m/z calcd for C18H14N2O5S [M – H]– 369.0623, found 369.0540. Melt point: 128.6–129.4 °C.
7.1.3. N-(2–(4-chlorophenoxy)phenyl)-4-nitrobenzenesulfonamide (9)
Substitute p-chlorophenol for phenol, compound 9 (light pink solid, 950 mg, yield 86%) was obtained by a scheme closely resemble to that depicted for the synthesis of compound 8. 1H NMR (400 MHz, DMSO) δ 10.42 (s, 1H), 8.20 (d, J = 8.7 Hz, 2H), 7.88 (d, J = 8.7 Hz, 2H), 7.42 (d, J = 7.6 Hz, 1H), 7.25 − 7.16 (m, 4H), 6.88 (d, J = 7.8 Hz, 1H), 6.58 (d, J = 8.8 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 153.69, 149.77, 146.88, 144.11, 129.47, 129.06, 128.00, 126.31, 124.79, 124.13, 123.66, 122.92, 118.82, 117.70. HRMS (ESI+), m/z calcd for C18H13ClN2O5S [M– H]– 403.0234, found 403.0150. Melt point: 149.6–150.4 °C.
7.1.4. 4-Nitro-N-(2-(p-tolyloxy)phenyl)benzenesulfonamide (10)
Substitute p-methyl phenol for phenol, compound 10 (light yellow solid, 500 mg, yield 87%) was obtained by a scheme closely resemble that depicted for the synthesis of compound 8. 1H NMR (400 MHz, DMSO) δ 10.36 (s, 1H), 8.18 (d, J = 8.7 Hz, 2H), 7.88 (d, J = 8.7 Hz, 2H), 7.40 (d, J = 7.7 Hz, 1H), 7.14 (dt, J = 15.3, 7.1 Hz, 2H), 6.97 (d, J = 8.3 Hz, 2H), 6.73 (d, J = 8.0 Hz, 1H), 6.44 (d, J = 8.3 Hz, 2H), 2.19 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 152.76, 149.67, 147.71, 144.13, 133.43, 129.86, 127.98, 126.32, 126.02, 123.67–123.21, 117.40, 20.06. HRMS (ESI+), m/z calcd for C19H16N2O5S [M– H]– 383.0780, found 383.0694. Melt point: 126.1–126.7 °C.
7.1.5. N-(2–(4-methoxyphenoxy)phenyl)-4-nitrobenzenesulfonamide (11)
Substitute p-methylphenol for phenol, compound 11 (light yellow solid, 1 g, yield 90%) was obtained by a scheme closely resemble to that depicted for the synthesis of compound 8. 1H NMR (400 MHz, CDCl3) δ 8.17 (d, J = 8.7 Hz, 2H), 7.90 (d, J = 8.8 Hz, 2H), 7.66 (d, J = 9.4 Hz, 1H), 7.18 (d, J = 14.5 Hz, 1H), 7.09 − 6.99 (m, 2H), 6.73 (d, J = 8.9 Hz, 2H), 6.66 − 6.57 (m, 1H), 6.51 (d, J = 9.0 Hz, 2H), 3.75 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 155.92, 149.71, 148.47, 148.00, 144.25, 128.07, 126.26, 125.51, 123.58, 122.98, 119.29, 116.29, 114.43, 55.14. HRMS (ESI+), m/z calcd for C19H16N2O6S [M + H]+ 401.0729, found 401.0799. Melt point: 122.6–123.5 °C.
7.1.6. 4-Amino-N-(2–(4-methoxyphenoxy)phenyl)benzenesulfonamide (12)
To a solution of compound 11 (100 mg, 0.25 mmol, 1.0eq) in MeOH (10 ml) was added nickel chloride hexahydrate (71.3 mg, 0.30 mmol, 1.2eq), NaBH4 (28.4 mg, 0.75 mmol, 3.0eq), and the mixture was stirred at 0 °C for 2 h. The mixture was made acidic with a 10% HCl solution and extracted with ethyl acetate (3 150 ml), the organic layer was washed with a saturated NaCl solution (3
100 ml), and the combined organic phases were dried with Na2SO4, filtered and concentrated to give 4-amino-N-(2–(4-methoxyphenoxy)phenyl)benzenesulfonamide (compound 12, 74 mg, yield 80%) as a pure brown solid. 1H NMR (400 MHz, CDCl3) δ 8.19 (d, J = 8.5 Hz, 2H), 7.91 (d, J = 8.5 Hz, 2H), 7.49 (d, J = 7.6 Hz, 1H), 7.09 (t, J = 7.5 Hz, 1H), 6.77 (d, J = 8.7 Hz, 2H), 6.68 (d, J = 8.2 Hz, 1H), 6.52 (d, J = 8.7 Hz, 2H), 3.78 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 155.70, 150.36, 148.58, 148.09, 128.93, 127.24, 124.71, 122.61, 121.58, 119.85, 115.97, 114.37, 113.33, 55.16. HRMS (ESI+), m/z calcd for C19H18N2O4S [M + H]+ 371.0987, found 371.1056. Melt point: 153.7–154.3 °C.
7.1.7. 4-Amino-N-(2–(4-chlorophenoxy)phenyl)benzenesulfonamide (13)
Compound 13 (68.5 mg, yield 74%) was obtained from compound 9 by a scheme closely resemble to that depicted for the synthesis of compound 12.
1H NMR (400 MHz, CDCl3) δ 7.67 (d, J = 8.0 Hz, 1H), 7.49 (d, J = 8.6 Hz, 2H), 7.12 − 7.02 (m, 2H), 6.98 (d, J = 8.5 Hz, 2H), 6.68 (t, J = 8.3 Hz, 3H), 6.52 (d, J = 8.6 Hz, 2H), 4.10 (d, J = 6.6 Hz, 2H). 13 C NMR (101 MHz, CDCl3) δ 163.08, 152.75, 149.72, 147.70, 144.14, 133.47, 129.87, 127.96, 126.31, 126.06, 125.37, 123.37, 117.40, 114.04. HRMS (ESI+), m/z calcd for C18H15ClN2O3S [M –H]– 373.0492, found 373.0405. Melt point: 131.9–132.6 °C.
7.1.8. 4-Amino-N-(2-(p-tolyloxy)phenyl)benzenesulfonamide (14)
Compound 14 (77.4 mg, yield 84%) was obtained from compound 10 by a scheme closely resemble to that depicted for the synthesis of compound 12. 1H NMR (400 MHz, DMSO) δ 10.35 (s, 1H), 8.23 (d, J = 8.7 Hz, 2H), 7.94 (d, J = 8.8 Hz, 2H), 7.34 (dd, J = 15.6, 7.8 Hz, 2H), 7.17 − 6.99 (m, 4H), 6.88 (s, 1H), 6.72 (d, J = 8.2 Hz, 1H), 6.56 (d, J = 8.3 Hz, 2H), 4.09 (s, 2H), 3.69 (d, J = 7.8 Hz, 1H), 2.22 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 153.09, 150.27, 147.36, 133.10, 129.80, 128.95, 127.65, 124.63, 122.95, 121.40, 118.26, 116.74, 113.36, 20.21. HRMS (ESI+), m/z calcd for C19H18N2O3S [M + H]+ 355.1038, found 355.1110. Melt point: 128.9–129.5 °C.
7.1.9. 2-((4-Amino-N-(2-(p-tolyloxy)phenyl)phenyl)sulfonamido) ethyl acetate (15)
The mixture of compound 10 (1 g, 2.5 mmol, 1.0eq), and NaH (300 mg, 12.5 mmol, 5eq) in N,N-dimethylformamide (DMF) was heated to 90 °C under nitrogen protection for 10 min, then the 2-bromoethyl acetate (501 mg, 3.0 mmol, 1.2eq) was added dropwise to the solution followed by stirring at 90 °C for 7–8 h. DMF was removed by distillation and the residue was dissolved in ethyl acetate and extracted with water (3 200 ml). The combined organic phases dried with Na2SO4, filtered, concentrated, and purified by flash column chromatography to give pale yellow liquid (15E, 450 mg, yield 37%).
To a solution of 15E (400 mg, 0.83 mmol, 1.0eq) in MeOH (10 ml) was added nickel chloride hexahydrate(236.7, 0.996, 1.2eq), NaBH4 (94.2 mg, 2.49 mmol, 3.0eq), and the mixture was stirred at 0 °C for 2 h. The mixture was made acidic with a 10% HCl solution and extracted with ethyl acetate (3 150 ml), the organic layer was washed with a saturated NaCl solution (3
100 ml), and the combined organic phases were dried with Na2SO4, filtered and concentrated to give 2-((4-amino-N-(2-(p-tolyloxy)phenyl)phenyl)sulfonamido)ethyl acetate (compound 15, 239 mg, yield 63%) as a pure yellow solid. 1H NMR (400 MHz, CDCl3) δ 8.10 (d, J = 8.7 Hz, 2H), 7.85 (d, J = 8.7 Hz, 2H), 7.50 (d, J = 9.1 Hz, 1H), 7.09 (t, J = 7.3 Hz, 1H), 6.98 (d, J = 8.3 Hz, 3H), 6.72 (d, J = 7.9 Hz, 1H), 6.42 (d, J = 8.4 Hz, 2H), 4.20 (s, 2H), 3.76 (s, 2H), 2.26 (s, 3H), 1.98 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 170.26, 155.94, 154.30, 149.30, 147.38, 145.35, 133.32, 128.18, 122.26, 119.65, 116.47, 114.38, 61.60, 48.72, 20.24. HRMS (ESI+), m/z calcd for C23H24N2O5S [M + H]+ 441.1406, found 441.1482. Melt point: 126.3–126.8 °C.
7.1.10. 2-((4-Amino-N-(2–(4-(trifluoromethyl)phenoxy)phenyl)phenyl) sulfonamido)ethyl acetate (16)
Compound 16 (pale yellow solid, 221 mg, 67%) was obtained by a scheme closely resemble to that depicted for the synthesis of compound 15. 1H NMR (400 MHz, CDCl3) δ 7.55 (s, 1H), 7.55 − 7.31 (m, 5H), 7.12 (t, J = 7.6 Hz, 2H), 6.85 (d, J = 8.1 Hz, 3H), 6.46 (d, J = 8.4 Hz, 2H), 4.14 (t, J = 5.5 Hz, 2H), 3.85 (s, 3H), 1.90 (s, 3H). 13C NMR (101 MHz, CDCl3) δ 170.64, 158.19, 149.76, 146.01, 144.23, 127.93, 127.05–126.63, 126.41, 124.89, 123.60, 123.29, 118.87, 116.94, 59.86, 52.92, 20.48. HRMS (ESI+), m/z calcd for C23H21F3N2O5S [M + H]+ 495.1123, found 495.1184. Melt point: 128.7–129.3 °C.
7.1.11. 4-Amino-N-(hydroxymethyl)-N-(2–(4-methoxyphenoxy)phenyl) benzenesulfonamide (17)
Intermediate 17F (239 mg, yellow solid, yield 63%) was obtained by a scheme closely resemble that depicted for the synthesis of compound 15. The mixture of intermediate 17F (200 mg, 0.44 mmol) and potassium carbonate (182 mg, 1.2 mmol) in MeOH was heated to 60 °C under nitrogen protection for 5–6 h. MeOH was removed by distillation and the residue was dissolved in ethyl acetate and extracted with water (3 200 ml). The combined organic phases dried with Na2SO4, filtered, concentrated, and purified by flash column chromatography to give compound 17 (pale brown solid, 102 mg, yield 56%). 1H NMR (400 MHz, CDCl3) δ 7.53 (d, J = 8.5 Hz, 2H), 7.18 (t, J = 7.0 Hz, 2H), 6.98 (t, J = 7.5 Hz, 1H), 6.86 (s, 4H), 6.68 (d, J = 8.3 Hz, 1H), 6.60 (d, J = 8.5 Hz, 2H), 3.79 (s, 3H), 3.72 (s, 2H), 3.64 (s, 2H). 13C NMR (101 MHz, CDCl3) δ 156.09 (dd, J = 19.9, 4.7 Hz), 150.09, 148.16, 131.14, 129.51, 129.19, 128.13, 122.14, 120.88, 116.32, 114.43, 113.35, 59.93, 55.26–54.87, 53.11. HRMS (ESI+), m/z calcd for C21H22N2O5S [M + H]+ 415.1249, found 415.1320. Melt point: 154.0–154.9 °C.
7.1.12. 4-Amino-N-(hydroxymethyl)-N-(2–(4-(trifluoromethyl)phenoxy) phenyl)benzenesulfonamide (18)
Compound 18 (pale yellow solid, 103 mg, yield 57%) was obtained by a scheme closely resemble that depicted for the synthesis of compound 17. 1H NMR (400 MHz, CDCl3) δ 7.56 (d, J = 8.6 Hz, 2H), 7.45 (d, J = 8.6 Hz, 2H), 7.27 (d, J = 7.8 Hz, 2H), 7.13 (t, J = 7.6 Hz, 1H), 7.00 (d, J = 8.5 Hz, 2H), 6.89 (d, J = 8.2 Hz, 1H), 6.54 (d, J = 8.6 Hz, 2H), 3.69 (s, 2H), 3.64 (d, J = 4.7 Hz, 2H). 13C NMR (101 MHz, CDCl3) δ 158.39, 153.68, 149.87, 131.52, 129.90, 129.39, 127.26–127.14, 126.81, 124.12, 118.95, 118.30, 113.49, 59.93, 53.18. HRMS (ESI+), m/z calcd for C21H19F3N2O4S [M + H]+ 453.1018, found 453.1090. Melt point: 156.3–156.9 °C.
7.2. Biological assays
7.2.1. Cell culture and reagents
The human breast cancer cell lines MDA-MB-231, MCF-7, HCC1937, were purchased from American Type Culture Collection. MDA-MB-231 and HCC1937 were cultured in RPMI-1640 medium (Hyclone). MCF-7 was cultured in a DMEM medium (Hyclone). Both media were supplemented with 10% foetal bovine serum (FBS, Yeasen Biological Technology Co., Ltd), and 100 U/ml penicillin/streptomycin (Wisent) at 37 °C in a humidified incubator with 5% CO2 and 95% air.
The lentiviral ERRα overexpression vector and scrambled control were produced by Genechem Co., Ltd. For transduction, MDA-MB-231 cells were seeded at 60% confluence in 6-well plates, and infected with a viral suspension containing 5 μg/ml polybrene (Sigma-Aldrich) for 8 h.
7.2.1.1. Cell viability assay
Cell Counting Kit-8 (CCK-8) assay was used to detect the effects of compounds on the tumour cells proliferation. MDA-MB-231, MCF-7, HCC1937 cells in log growth phase were seeded in 96- well plates. After overnight incubation, the cells were treated with various concentrations of compound 11 (1, 3, 10 μM) for 24 h and 48 h. CCK-8 assay were performed according to the manufacturer's procedure (Proteintech Group, Inc, USA).
7.2.1.2. Colony formation assay
For colony formation assay, MDA-MB-231 and MCF-7 cells were plated in 6-well plates (1 × 103 cells/well) and cultured under standard culture conditions overnight. Then, the medium was replaced with low and high doses of compound 11 and vehicle for 2 weeks until the colonies were visible in the culture plate. The supernatants were discarded and washed with PBS twice, 4% paraformaldehyde used to fix the colonies, and stained with 0.1% crystal violet solution for 30 min. Finally, the excess staining solution was washed away with distilled water and photographed on a microscope and used Image J software to calculate the number of clones.
7.2.2. ERRα functional assay
A robust mammalian two-hybrid-based reporter gene assay was performed according to previous reports to evaluate the ERRα transcriptional activities of compoundsCitation16,Citation21. In brief, GAL4-ERRα-LBD along with a luciferase reporter 5 × UAS-Luc Cells were co-transfected into HEK 293 T cells using Lipofectamine 2000. After 6 h incubation, test compounds were added to the transfected cells for a further 48 h. Subsequently, the luciferase activity was determined by a dual-luciferase reporter assay system (Promega, Madison, WI, USA).
7.2.3. TR-FRET assay
To determine the ERRα binding affinity of compounds, a time-resolved fluorescence resonance energy transfer (TR-FRET) co-activator assay was conducted according to the manufacturer’s instruction (Invitrogen). After 1 h incubation at room temperature and dark state, the TR-FRET activity of the binding mixture was measured at 340 nm excitation and 495/520 nm dual emissionCitation21.
7.2.4. Transwell human breast cancer cell migration and invasion assays
In vitro migration and invasion, assays were performed on transwell chambers with 8-mm pore-size filters without (for migration) or with (for invasion) coated Matrigel (Corning Incorporated, Corning, NY, USA). MDA-MB-231 and MCF-7 cells were trypsinized, washed twice, and then resuspended in a serum-free medium. The transwell chambers were placed on the upper surface of the 24-well plate, and the 250 μl of cell suspension (1 × 105 cells) were seeded into the upper chamber. A culture medium (750 μl) containing 20% FBS was added to the lower chamber. After transwell inserts were cultured at 5% CO2 at 37 °C for 24 h, cells that had not migrated were removed from the top side of the membrane with moistened cotton swabs. Cells attached on the underside of the membrane were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Finally, washing with PBS, the invasive and migration cells were counted in three random fields under a microscope.
7.2.5. Quantitative real-time polymerase chain reaction
MDA-MB-231 cells were treated with DMSO (0.1%) or compound 11 (1, 3 μM) for 24 h. Total RNA was isolated using the Trizol reagent and Ultrapure RNA kit (CW Biotech, China) according to the manufacturer’s instructions. In brief, 2 mg of total mRNA was reverse-transcribed, and quantitative real-time PCR reaction was performed on ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). Primer sequences were used according to published literatureCitation22.
7.2.6. Breast cancer xenografts
Human breast cancer xenografts were generated according to a previous studyCitation21. In brief, tumour growth of 6-week-old BALB/c nude mice was monitored once per 5-days after implantation (2 × 106 MDA-MB-231 cells per mouse). When subcutaneous tumours reached an average size of >100 mm3, mice were treated intraperitoneally with compound 11, cyclophosphamide or vehicle (0.9% saline solution containing 0.5% 2-phenylethanol and 1.5% Tween 80). Mice were weighed per 10-days and tumour volumes were calculated using the formula V= length × widthCitation2×0.5Citation21. All procedures were performed in accordance with approved protocols by the Fudan University Institutional Animal Care and Use Committee. Every effort was made to minimise potential distress, pain, or discomfort to the animals throughout all experiments.
7.3. Molecular docking of compound and ERRα
The crystal structure of the ERRα with inverse agonist (compound 2) was obtained from the RCSB Protein Data Bank (PDB entry 2PJL). The ligand-binding domain was measured by Sybyl X2.0. The docking simulations mentioned in this article were performed by using surflex-dock geomx program (Sybyl X2.0) with the default parameters, which were used to explore the binding model of ERRα with the studied compound.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA 2018;68:394–424.
- Lin NU, Vanderplas A, Hughes ME, et al. Clinicopathologic features, patterns of recurrence, and survival among women with triple-negative breast cancer in the national comprehensive cancer network. Cancer 2012;118:5463–72.
- Li X, Yang J, Peng L, et al. Triple-negative breast cancer has worse overall survival and cause-specific survival than non-triple-negative breast cancer. Breast Cancer Res Treat 2017;161:279–87.
- Guney Eskiler G, Cecener G, Egeli U, Tunca B. Triple negative breast cancer: new therapeutic approaches and BRCA status. APMIS 2018;126:371–9.
- Ye S, Xu Y, Wang L, et al. Estrogen-related receptor α (ERRα) and G protein-coupled estrogen receptor (GPER) synergistically indicate poor prognosis in patients with triple-negative breast cancer . Onco Targets Ther 2020;13:8887–99.
- Manna S, Bostner J, Sun Y, et al. ERRα is a marker of tamoxifen response and survival in triple-negative breast cancer . Clin Cancer Res 2016;22:1421–31.
- Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (pgc-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within pgc-1alpha. J Biol Chem 2002;277:40265–74.
- Seth A, Steel JH, Nichol D, et al. The transcriptional corepressor rip140 regulates oxidative metabolism in skeletal muscle. Cell Metab 2007;6:236–45.
- Chang CY, McDonnell DP. Molecular pathways: the metabolic regulator estrogen-related receptor α as a therapeutic target in cancer. Clin Cancer Res 2012;18:6089–95.
- Lu Y, Li Q, Chen L, Shi X. XCT790 inhibits rat vascular smooth muscle cells proliferation through down-regulating the expression of estrogen-related receptor alpha . Acta Pharmaceutica Sinica 2014;49:190–7.
- Zhang LD, Chen L, Zhang M, et al. Downregulation of ERRα inhibits angiogenesis in human umbilical vein endothelial cells through regulating VEGF production and PI3K/Akt/STAT3 signaling pathway . Eur J Pharmacol 2015;769:167–76.
- Stein RA, Chang CY, Kazmin DA, et al. Estrogen-related receptor alpha is critical for the growth of estrogen receptor-negative breast cancer. Cancer Res 2008;68:8805–12.
- Vargas G, Bouchet M, Bouazza L, et al. ERRα promotes breast cancer cell dissemination to bone by increasing RANK expression in primary breast tumors. Oncogene 2019;38:950–64.
- Stein RA, Gaillard S, McDonnell DP. Estrogen-related receptor alpha induces the expression of vascular endothelial growth factor in breast cancer cells. J Steroid Biochem Mol Biol 2009;114:106–12.
- Bianco S, Lanvin O, Tribollet V, et al. Modulating estrogen receptor-related receptor-alpha activity inhibits cell proliferation. J Biol Chem 2009;284:23286–92.
- Busch BB, Stevens WC Jr, Martin R, et al. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor alpha. J Med Chem 2004;47:5593–6.
- Kallen J, Lattmann R, Beerli R, et al. Crystal structure of human estrogen-related receptor alpha in complex with a synthetic inverse agonist reveals its novel molecular mechanism. J Biol Chem 2007;282:23231–9.
- Patch RJ, Searle LL, Kim AJ, et al. Identification of diaryl ether-based ligands for estrogen-related receptor alpha as potential antidiabetic agents. J Med Chem 2011;54:788–808.
- Patch RJ, Huang H, Patel S, et al. Indazole-based ligands for estrogen-related receptor alpha as potential anti-diabetic agents. Eur J Med Chem 2017;138:830–53.
- Xu S, Zhuang X, Pan X, et al. 1-phenyl-4-benzoyl-1h-1,2,3-triazoles as orally bioavailable transcriptional function suppressors of estrogen-related receptor α. J Med Chem 2013;56:4631–40.
- Zhang L, Liu P, Chen H, et al. Characterization of a selective inverse agonist for estrogen related receptor α as a potential agent for breast cancer. Eur J Pharmacol 2016;789:439–48.
- Du Y, Song L, Zhang L, et al. The discovery of novel, potent err-alpha inverse agonists for the treatment of triple negative breast cancer. Eur J Med Chem 2017;136:457–67.
- Kallen J, Schlaeppi JM, Bitsch F, et al. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (erralpha): crystal structure of erralpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J Biol Chem 2004;279:49330–7.
- Takeuchi Y, Oda T, Chang M-R, et al. Synthesis and antitumor activity of fused quinoline derivatives. Iv. Novel 11-aminoindolo[3,2-b]quinolines. Chem Pharm Bull 1997;45:406–11.
- Rassadin VA, Grosheva DS, Arefeva IA, et al. Access to a wide range of sultams by cyclodialkylation of α-substituted methanesulfonanilides. Eur J Org Chem 2012; 2012:5028–37.