Abstract
A new series of pyrido[2,3-d]pyrimidin-4(3H)-one derivatives having the essential pharmacophoric features of EGFR inhibitors has been designed and synthesised. Cell viability screening was performed for these compounds against A-549, PC-3, HCT-116, and MCF-7 cell lines at a dose of 100 μM. The highest active derivatives (8a, 8 b, 8d, 9a, and 12b) were selected for IC50 screening. Compounds 8a, 8 b, and 9a showed the highest cytotoxic activities and were further investigated for wild EGFRWT and mutant EGFRT790M inhibitory activities. Compound 8a showed the highest inhibitory activities against EGFRWT and EGFRT790M with IC50 values of 0.099 and 0.123 µM, respectively. In addition, it arrested the cell cycle at pre-G1 phase and induced a significant apoptotic effect in PC-3 cells. Furthermore, compound 8a induced a 5.3-fold increase in the level of caspase-3 in PC-3 cells. Finally, docking studies were carried out to examine the binding mode of the synthesised compounds against both EGFRWT and EGFRT790M.
1. Introduction
According to WHO, cancer was the direct cause of 10 million deaths in 2020 and the cost of cancer treatment globally was US$1.16 trillion in 2010Citation1. Several internal and external factors can cause cancer. The most well-known factors are hormonal disorders, genetic mutations, radiations, smoking tobacco, metals, polluted food, chemicals, and infectious organismsCitation2–4. Resistance against anticancer drugs is considered one of the most serious problems in cancer managementCitation5. Due to the high residence of many cancer types, the discovery of new anticancer agents with high effect, less resistance, and fewer side effects is an urgent need.
Protein kinases (PKs) are a group of enzymes that are responsible for the transference of phosphate from ATP molecule to tyrosine, serine and/or threonine amino acids in protein substratesCitation6,Citation7. Furthermore, PKs promote cellular signalling processes such as cell growth regulation, differentiation, migration, and metabolismCitation8. PKs have been found to be overexpressed in a variety of human malignanciesCitation9. Accordingly, the inhibition of PKs has emerged as a selective method for killing cancer cellsCitation10. Receptor tyrosine kinases (RTKs) are vital category protein kinases. About 20 different RTKs have been discovered that have similar structuresCitation11.
The epidermal growth factor receptor (EGFR) belongs to the RTKs family that stimulates differentiation and proliferation of cells after the binding of its specific active ligandCitation12. EGFR structure has an extracellular part (at the surface of the cells) and an intracellular part. The activation of the outer part leads to an activation of the intracellular region of the receptor and a phosphorylation of the intracellular substratesCitation13. This step facilitates cell growth, synthesis of DNA, and the expression of oncogenesCitation14. It was reported that EGFR is over-expressed and implicated in the pathogenesis and progression of various human carcinomasCitation15. In many patients, resistance against cancer therapy arises from an acquired mutation in the EGFR kinase domain (T790M). Such mutant EGFR is called EGFRT790MCitation16. Thus, EGFRs (wild and mutant types) are interesting biological targets for the discovery of new anticancer agentsCitation17,Citation18.
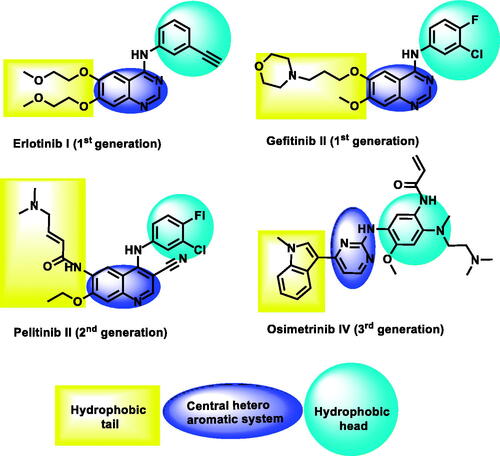
The ATP binding site of EGFR consists of five regions; an adenine-binding pocket, a sugar region (ribose binding pocket), a hydrophobic region I, a hydrophobic region II, and a phosphate-binding regionCitation19–21. Most of the reported EGFR inhibitors are ATP-competitive inhibitor small molecules that have specific moieties to occupy the adenine-binding pocket, the hydrophobic region I, and the hydrophobic region IICitation10 ().
Figure 1. The essential pharmacophoric features of erlotinib as an EGFR inhibitor occupying three pockets in the ATP binding site based on ReferenceCitation22.
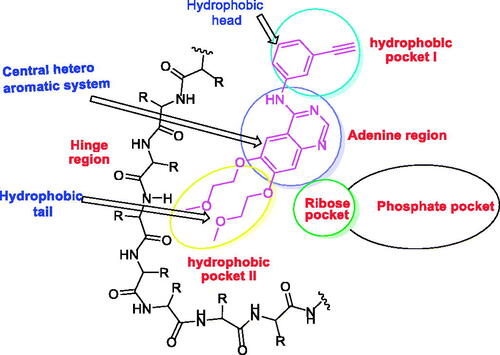
EGFR inhibitors have a specific Y-shaped structureCitation23. In addition, the structure of EGFR inhibitors should comprise many essential pharmacophoric featuresCitation24. Each feature binds at a specific region in the ATP binding site. For example, a flat hetero aromatic system is an essential feature of EGFR inhibitor to occupy the adenine binding pocket of the ATP binding site. Such hetero structure can form hydrogen bonds with some amino acids as Met769, Thr790, and Thr854Citation25. Also, a terminal hydrophobic head of the EGFR inhibitor can occupy the hydrophobic region I forming many hydrophobic interactionsCitation24. Finally, a hydrophobic tail be buried in the hydrophobic region II producing high affinityCitation19,Citation26.
Till now, three generations of EGFR inhibitors were approved by the FDA (). Erlotinib ICitation27 and gefitinib IICitation28 are examples of the first generation. The generated mutation in EGFR led to the acquired drug resistance and reduced efficacy in cancer treatmentCitation29. The mutant form of protein (EGFRT790M) resists the affinity of ATP-competitive inhibitorsCitation30. The second-generation of EGFR inhibitors was approved to overcome the drug resistance that was induced by EGFRT790M. These inhibitors can form covalent interactions with Cys797 at the ATP binding siteCitation31–33. Pelitinib IIICitation34 is a well-known example of this class. Unfortunately, low maximal-tolerated-dose, the major drawback of this class, led to poor clinical outcomesCitation35,Citation36. Osimertinib 5Citation37, an example of the third-generation EGFR inhibitors, exhibited greater activities against mutant form (EGFRT790M) than the wild form (EGFRWT). Recently, toxic epidermal necrolysis was reported upon the administration of olmutinibCitation38. Hence, many efforts are still required to reach more potent and less toxic EGFR inhibitors.
Pyrido[2,3-d]pyrimidin-4(3H)-one moiety was utilised before for the synthesis of various anticancer agentsCitation39–42, and EGFR inhibitorsCitation43. Interestingly it was included in the discovery of highly specific inhibitors against the mutant EGFRT790MCitation44.
As an extension of our previous efforts in the design and synthesis of new anticancer agentsCitation45–51, especially that target RTKsCitation52,Citation53 and EGFRCitation22 Citation54–58, we used the pyrido[2,3-d]pyrimidin-4(3H)-one moiety as a building block for the design and synthesis of new anticancer agents targeting the wild EGFR (EGFRWT) as well as the mutant EGFR (EGFRT790M).
1.1. Rationale of molecular design
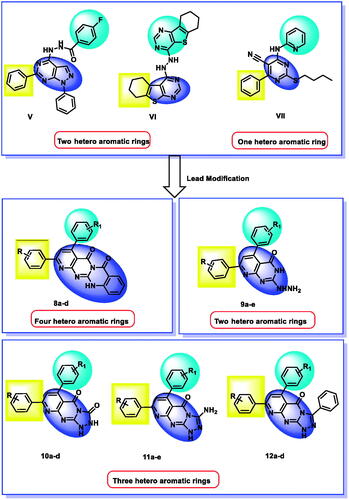
For years, our team synthesised several EGFR inhibitors which showed promising anticancer activities. In 2018, a series of 1H-pyrazolo[3,4-d]pyrimidine derivatives were synthesised and evaluated for their inhibitory activities against EGFRWT and EGFRT790M. compound V potently inhibited the two EGFR types with a good apoptotic effect and arrested the cell cycle at the G2/M phase. Such compounds comprise two hetero-aromatic rings (1H-pyrazolo[3,4-d]pyrimidine) to occupy the adenine binding pocketCitation22.
In 2019, we designed and synthesised a series of thieno[2,3-d]pyrimidine derivatives as EGFR and HER2 tyrosine kinase inhibitors. Compound VI was the most active member producing significant apoptosis. This compound contains two hetero-aromatic rings (thieno[3,2-d]pyrimidine) to occupy the adenine binding pocketCitation54.
In 2020, our team designed and synthesised a new series of pyrimidine-5-carbonitrile derivatives as EGFR inhibitors. Compound VII showed high inhibitory activities against EGFRWT and EGFRT790M. In addition, it arrested the cell cycle at the G2/M phase and induced a significant apoptotic effect in HCT-116, HepG-2, MCF-7cells. This compound contains one hetero-aromatic ring (pyrimidine) to occupy the adenine binding pocketCitation55.
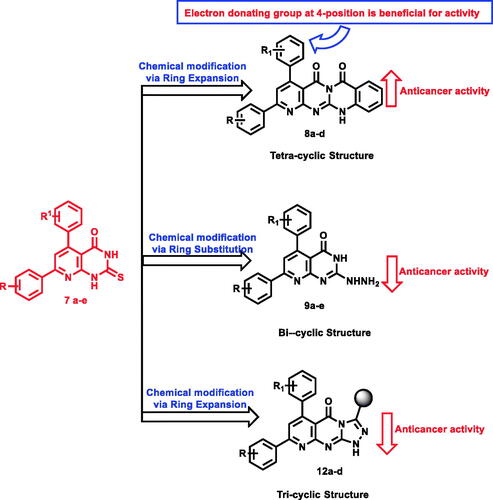
In the current work, we used the previously reported active candidates (V, VI, and VII)Citation22,Citation54,Citation55 as lead compounds in the design of the new derivatives. The rationale of our molecular design depended on the modification of such compounds to get new EGFR inhibitors. The modification was carried out at three features following the essential features of EGFR inhibitors. Concerning the terminal hydrophobic head and the hydrophobic tail, different substituted benzene rings were used to study the SAR of the synthesised compounds. Regarding the flat hetero-aromatic system, we used three different systems. The first one is pyrido[2,3-d]pyrimidin-4(3H)-one moiety which comprises two hetero-aromatic rings (compounds 9a–e). The second one is pyrido[2,3-d][1,2,4]triazolo[4,3-a]pyrimidin-5(1H)-one moiety which composes three heteroaromatic rings (compounds 10a–d, 11a–e, and 12a–d). The third one is 5H-pyrido[2′,3′:4,5]pyrimido[2,1-b]quinazoline-5,7(12H)-dione moiety which constitutes four hetero-aromatic rings (compounds 8a–d; ).
2. Results and discussion
2.1. Chemistry
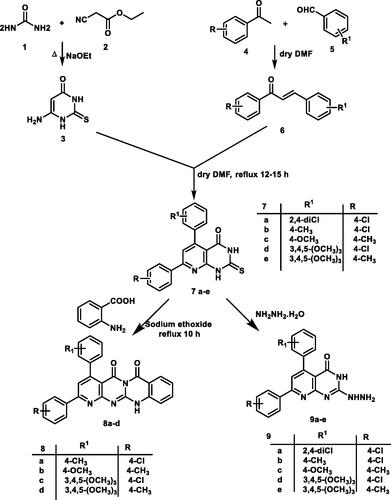
In continuation of the previous workCitation59, the starting precursor 2-thioxo-2,3-dihydropyrido[2,3-d]pyrimidin-4(1H)-one derivatives 7a–e were afforded via the reaction of the appropriate chalcones 6a–e with 6-aminothiouracil 3. The target compounds were synthesised in acceptable yield as reportedCitation59. Here in, the structure of the new 2-thioxo-2,3-dihydropyrido[2,3-d]pyrimidin-4(1H)-one derivative 7a was proved by elemental and spectral analyses. 1H NMR spectrum showed two D2O exchangeable singlet signals at δ 12.51, 13.23 ppm correspond to the two protons of each NH groups. Also, a singlet signal was recorded at δ 7.97 ppm, corresponding to the proton at C6 of pyridopyrimidine ring. The 13C NMR spectrum of 7a analogue displayed two characteristic signals at δ 162.29, 175.61 corresponding to carbons of C=O and C=S groups, respectively.
The 5H-pyrido[2′,3′:4,5]pyrimido[2,1-b]quinazoline-5,7(12H)-dione analogues 8a–d were synthesised through the reaction of compounds 7b–e with anthranilic acid in the presence of catalytic amount of sodium ethoxide under reflux conditionCitation60. Their chemical structures were confirmed by elemental and spectral data for example the 1H NMR of compound 8d revealed an increase in the integration of aromatic region at δ 6.76–8.15 ppm, and the presence of D2O exchangeable singlet signal assigned for one proton of NH group at δ 11.64 ppm. The 13 C NMR spectrum showed the characteristic two signals for the two carbons of C=O signals at δ 161.45 and 169.46 ppm. The mass spectrum for 8d revealed the expected molecular ion peak at m/z of 520. Finally, IR spectrum of 8 b displayed absorption bands at 1693, 1750 and 3410 cm−1 corresponding to two C=O and one NH groups, respectively.
The 2-hydrazinopyrido[2,3-d]pyrimidin-4(3H)-one derivatives 9a–e were depicted through the nucleophilic attack of hydrazine hydrate upon the key derivatives 7a–e following the reported methodCitation60. The newly hydrazinyl derivative 9a was proved by spectral data. The 1H NMR spectrum showed two singlet signals at δ 8.23, 9.12 ppm assigned for three protons of hydrazinyl group NHNH2.
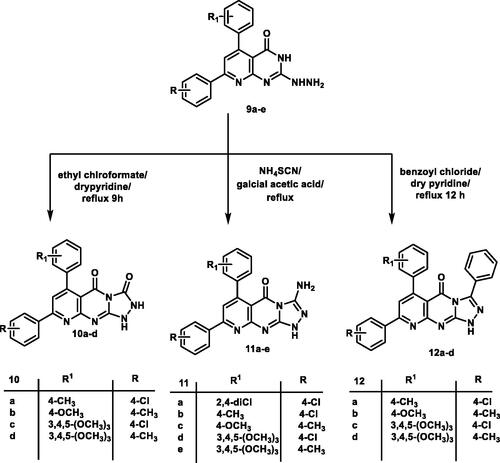
Cyclo-condensation of the 2-hydrazinyl derivative 9a–e with ethyl chloroformate in dry pyridine produced pyrido[2,3-d][1,2,4]triazolo[4,3-a]pyrimidine-3,5-dione derivatives 10a–d. The IR spectrum of compound 10d revealed the presence of three absorption bands at 1708, 3437, and 3425 cm−1 assigned for two carbonyl and two NH groups, respectively. The 1H NMR spectrum for the same compound showed two D2O exchangeable signals at δ 9.26, 11.07 ppm assigned for two NH groups. Mass spectrum of compound 10c showed molecular ion peak at m/z of 479 and its isotope at m/z of 481.
Reaction of hydrazinyl derivatives 9a–e with ammonium thiocyanate in glacial acetic acid under reflux afforded 3-aminopyrido[2,3-d][1,2,4]triazolo[4,3-a]pyrimidin-5(1H)-one derivatives 11a–e. The 1H NMR of compound 11a revealed the presence of two exchangeable singlet signals at 7.09 and 7.33 ppm assigned for NH and NH2 groups. Mass spectra of compound 11c illustrated the expected molecular ion peak at m/z of 400.5.
The 3-phenylpyrido[2,3-d][1,2,4]triazolo[4,3-a]pyrimidin-5(1H)-one analogues 12a–e were obtained via the reaction of hydrazinyl derivatives 9a–e with benzoyl chloride in pyridine under reflux conditions. Analytical and spectroscopic measurements confirmed the structures of compounds 12a–d. The IR spectrum of 12b displayed two absorption bands at 1720, 3414 cm−1 corresponds to C=O and NH groups, respectively. The 1H NMR spectrum of the same series gave an increase in aromatic integration due to the presence of an extra phenyl ring. The mass spectrum of 12b revealed a molecular ion peak at m/z of 459 (Schemes 1 and 2).
2.2. Biological evaluation
2.2.1. In vitro antiproliferative activities
All the final synthesised (19) compounds were tested for their anticancer activities against four tumour cell lines namely, lung cancer (A-549), prostate cancer (PC-3), colon cancer (HCT-116), and breast cancer (MCF-7) using standard MTT methodCitation61–63. Preliminary screening against the cancer cell lines was performed, using doxorubicin as a reference drug at doses of 100 μM. Variable results were recorded for the screened compounds as depicted in . The pyrido[2,3-d]pyrimidin-4(3H)-one derivatives (8a, 8 b, 8d, and 9a) that exhibited inhibitory activity ≥70% were selected for IC50 screening comparing erlotinib.
Table 1. Percentage of growth inhibition activity of compounds 7a, 8a–d, 9a, 10a–e against A549, PC-3, HCT-116 and MCF-7 at a concentration of 100 μM.
All compounds were barely active against breast cancer (MCF-7) cell line at 100 μM (% of inhibition ranging from 5 to 68% (). By focussing on the prostatic cell line (PC-3), the anticancer profile of the tested compounds was significantly improved especially the tetracyclic derivatives 8a (IC50 = 7.98 µM) and 8d (IC50 = 7.12 µM) that exhibited about 1.5 times more active than erlotinib (11.05 µM). In addition, compound 9a showed a strong activity against PC-3 line with an IC50 value of 9.26 µM. For compound 8d, it showed a strong anti-proliferative activity against A-549 with an IC50 value of 7.23 µM which is comparable to erlotinib (IC50 = 6.53 µM). Compound 8b revealed a moderate inhibitory activity against PC-3 cell line with an IC50 value of 18.01 µM. Generally, no cytotoxic activity was observed against the colon cancer cell line (HCT-116), but compounds 8a, 8b, 8d, 12b revealed mild cytotoxic activity.
2.2.2. Structural–activity relationship
The synthetic pathway of the target compounds was depicted in two schemes starting with thioxo-precursors 7a–e to afford tetracyclic derivatives 8a–d, hydrazinyl derivatives 9a–e, and triazolyl derivatives 10a–d, 11a–e, and 12a–d ().
Expansion of pyrido[2,3-d]pyrimidin-4(3H)-one core to give tetracyclic 5H-pyrido [2′,3′:4,5]pyrimido[2,1-b]quinazoline-5,7(12H)-dione derivatives 8a–d showed the preferred impact on the evaluated anticancer activity. Compounds 8a, b, d exhibited the most potent cytotoxic activity against A-549 cell line with IC50 values of 16.2, 16, and 7.23 μM, respectively, and the later was equipotent to erlotinib (IC50 = 6.53 μM).
Concerning prostate cancer cell line (PC-3), both compounds 8a (IC50 = 7.98 μM), and 8d (IC50 = 7.12 μM), were two-fold more potent than the reference molecule (IC50 = 11.05 μM). It was noticed that the electronic factor greatly influences the anticancer activity of the same series against lung and prostate cancer cells. For example, the existence of electron-donating (OCH3) group at 4-position of compounds 8a and 8d was beneficial for activity. The modification of tetracyclic derivatives 8a (IC50 = 7.98 μM) into hydrazinyl derivatives 9a (IC50 = 9.26 μM) decreased the anticancer activity against prostate cancer cell line (IC50 = 9.26 μM). In addition, the expansion of pyrido[2,3-d]pyrimidin-4(3H)-one scaffold into triazolyl analogues caused a remarkable drop in the activity with an inhibition range from 2 to 52% ().
Table 2. IC50 values of compounds 8a, 8b, 8d, 9a and 12b against A-549, PC-3, HCT-116 and MCF-7.
2.2.3. EGFRWT kinase inhibitory assay
The promising antiproliferative compounds (8a, 8d, and 9a) were further examined for their EGFRWT kinase inhibitory activities using Homogeneous time resolved fluorescence (HTRF) assayCitation64. Erlotinib was used as a reference molecule ().
Table 3. In vitro enzymatic inhibitory activities against EGFRL858R and EGFR790M.
The tested derivatives 8a, 8d, and 9a showed promising inhibitory activities against EGFRWT with IC50 values of 0.099, 0.419, and 0.594 µM, respectively. compounds 8a showed a good activity compared to erlotinib (IC50 = 0.043 µM). Whereas compounds 8d and 9a showed moderate act nlp0m kinase inhibitory assay
To evaluate the potential activity of the synthesised compounds against the mutant form of EGFR, the most active cytotoxic compounds (8a, 8d and 9a) were tested for their inhibitory effect against EGFRT790M. Erlotinib was used as a positive control.
The tested compounds 8a, 8d and 9a showed inhibitory effects against EGFRT790M with IC50 values of 0.123, 0.290, and 0.571 µM, respectively. Compound 8a exhibited the highest inhibitory effect but less than erlotinib (IC50 = 0.071 µM). While compounds 8d and 9a showed moderate inhibitory activities ().
2.2.4. Cell cycle analysis
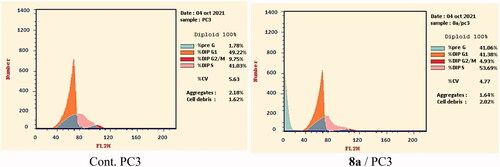
Based on the above-mentioned biological testing, the most promising candidate 8a was subjected to flow cytometry analysis to investigate its effect on the cell cycle distribution in the most sensitive cell line (PC-3). The reported protocol described by Wand et al.Citation65 was applied in this test. PC-3 cells were incubated with compound 8a for 24 h in a concentration equal to its IC50 against such cell line (7.98 µM). After that, the different phases of the cell cycle were analysed.
Compound 8a showed different effects on the cell cycle distribution. Compared to the control cells (Cont. (PC-3)), the cell population increased at the phases of pre-G1 and %S by 22 and 1.3 folds, respectively. For the Pre-G1phase, the cell increased from 1.78% (in cont. cells) to 41.06% (at the treated cells). In the S phase, the cell increased from 41.03% (in cont. cells) to 53.69% (at the treated cells). On the other hand, the cell population decreased in both the G0–G1 and the G2-M phases. Such results obviously reveal that compound 8a can arrest the PC-3 cell line at pre-G1 of the cell cycle ( and Supplementary data).
2.2.5. Annexin V-FITC apoptosis assay
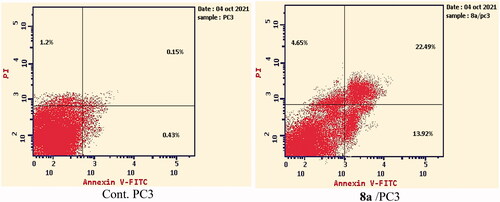
To analyse the apoptotic effect of the most active compound 8a, Annexin V and PI double staining assay with FITC was appliedCitation66. In this test, PC-3 cells were incubated with compound 8a at a concentration of 7.98 µM for 24 h. The results were depicted in ( and Supplementary data).
Investigating the results of Annexin V and PI double staining assay, revealed that compound 8a produced a significant increase in the early apoptosis ratio from 0.43 to 13.92% (32-fold). Also, it exerted an increase in the late apoptosis ratio from 0.15 to 22.49% (150-fold). Such findings indicate that compound 8a has a significant apoptotic effect against PC-3 cells.
2.2.6. Caspase-3 determination
The ability of a drug to induce apoptosis determines the sensitivity of the cancer cells against it. There are many signalling pathways that control apoptosis induction. Caspases family are considered as one of the most apoptotic regulatorsCitation67. Activation of caspases especially (caspase-3) produces cell deathCitation68. In addition, it was reported that EGFR inhibitors exhibit significant apoptotic effects through the caspase pathwayCitation69,Citation70. Here, the effect of the most active EGFR inhibitor 8a on caspase-3 was examined in PC-3 cells. Compound 8a was applied on PC-3 cells at a concentration of 3.04 µM for 24 h. The results revealed that such a compound generated a marked increase in the level of caspase-3 (452.3 pg/mL, 5.3-fold) compared to the control cells (84.24 pg/mL). In addition, the tested compound showed a comparable effect with the reference compound; staurosporine (413.1 pg/mL; and Supplementary data).
Table 4. Effect of compound 8a on active caspase-3 in PC-3 cells after 24 h treatment.
2.3. Docking studies
To confirm our rationale of design, the binding modes of the synthesised compounds were investigated against the proposed targets using a docking approach. The used biological targets in docking studies were EGFR-TK Wild-type (EGFRWT, PDB: 4HJO)Citation71 and EGFR-TK mutant type (EGFRT790M, PDB: 3W2O)Citation72 using MOE 14.0 software. The co-crystallised ligands were used as reference molecules. The output of docking studies showed a high affinity of the synthesised compounds against the two tested targets compared to the reference molecules ().
Table 5. The docking binding free energies of the synthesised compounds against EGFRWT and EGFRT790M.
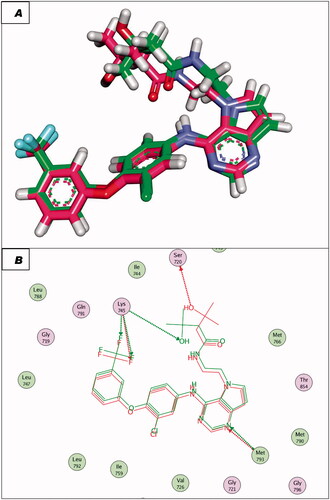
To validate the docking procedures, the co-crystallised ligands (Erlotinib and TAK-285) were re-docked against EGFRWT and EGFRT790M, respectively. The RMSD of docked and original ligands of erlotinib and TAK-285 were 0.88 and 1.05 Å, respectively. These values indicate the validity of the docking protocol ( and ).
Figure 7. (A and B) 3D and 2D superimposition of the docked ligand (erlotinib; pink) and the original ligand (green) with RMSD value of 0.88 Å.
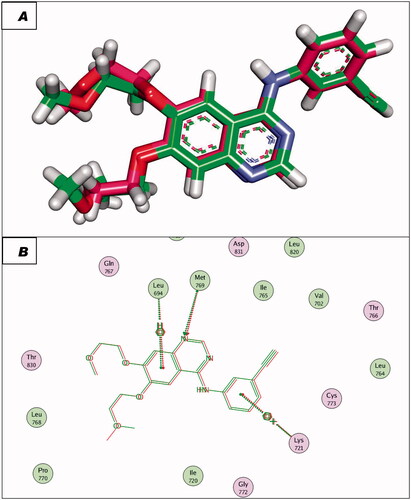
Figure 8. (A and B) 3D and 2D superimposition of the docked ligand of mutant EGFR (TAK-285; Pink) and the original ligand (green) with RMSD value of 1.06 Å.
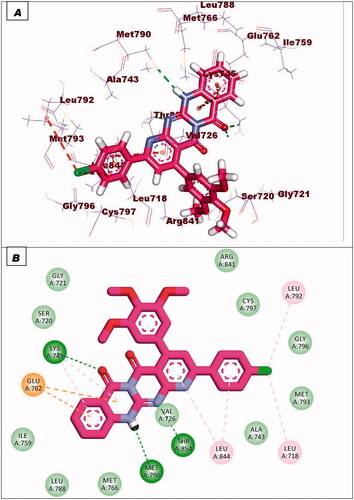
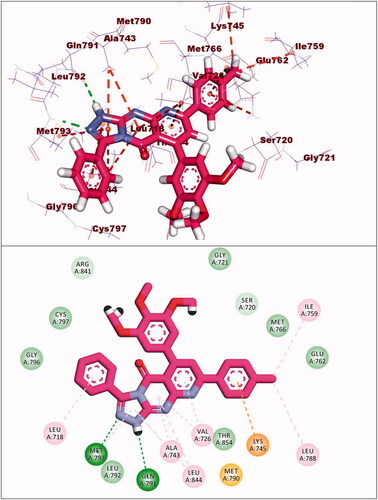
The co-crystallised ligand (erlotinib) of EGFRWT showed a binding energy of −22.12 kcal/mol. The heterocyclic system (quinazoline moiety) was buried in the adenine pocket forming a hydrogen bond with Met769. Also, it formed four hydrophobic interactions with Lue694, Ala719, and Leu820. The ethynylphenyl moiety was oriented into the hydrophobic pocket I forming three hydrophobic interactions with Ala719, Val702, and Lys721. The 2-methoxyethoxy groups occupied the hydrophobic region II forming a hydrogen bond with Cys773 ().
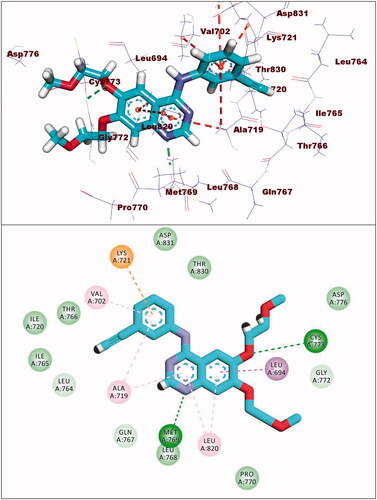
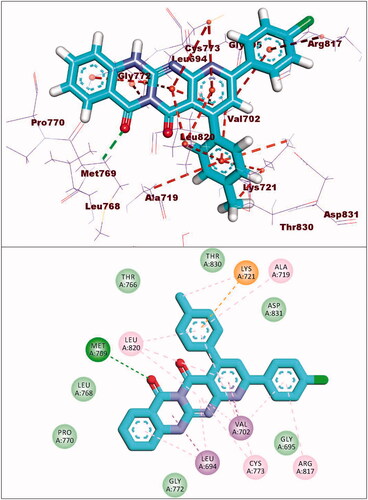
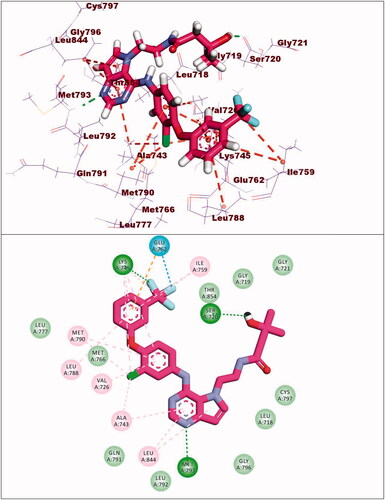
Compound 8a showed a binding mode like that of erlotinib with a binding energy of −19.29 kcal/mol. The 5H-pyrido[2′,3′:4,5]pyrimido[2,1-b]quinazoline-5,7(12H)-dione moiety occupied the adenine pocket of the EGFRWT forming one hydrogen bond with the crucial amino acid Met769. In addition, it formed nine hydrophobic interactions with Val702, Leu694, and Leu820. The tolyl moiety occupied the hydrophobic pocket I forming four hydrophobic interactions with Leu890, Ala719, and Lys721. Moreover, the 4-chlorophenyl moiety occupied the hydrophobic region II forming two hydrophobic interactions with Val702 and Arg817 ().
Compound 8b showed a binding energy of −19.06 kcal/mol. The 5H-pyrido[2′,3′:4,5]pyrimido[2,1-b]quinazoline-5,7(12H)-dione moiety was buried in the adenine pocket forming two hydrogen bonds with acid Met769 and Cys773. In addition, it formed six hydrophobic interactions with Val702, Leu694, and Leu820. The 4-methoxyphenyl moiety occupied pocket I forming two hydrophobic interactions with val702, and Cys773. Moreover, the 4-chlorophenyl moiety occupied the hydrophobic II forming three hydrophobic interactions with Val721 and Ala719 ().
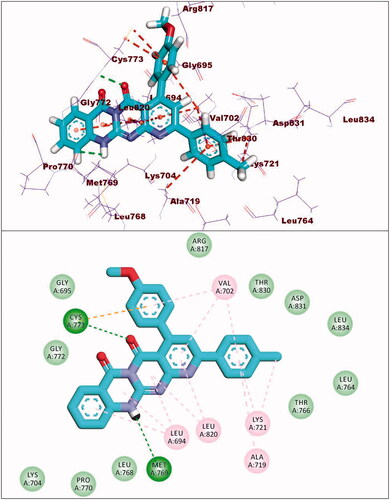
With regard to compound 8d, it showed a binding mode similar to the refrence molecules with a binding energy of −21.92 kcal/mol. The 5H-pyrido[2′,3′:4,5]pyrimido[2,1-b]quinazoline-5,7(12H)-dione moiety was involved in two hydrogen bonds with the amino acids Met769 and Cys773 in the adenine pocket. In addition, it formed eight hydrophobic interactions with Val702, Leu694, Gly772, and Leu820. The 3,4,5-trimethoxyphenyl moiety occupied pocket I forming two hydrophobic interactions with Ala719 and Leu820. It formed two hydrogen bonds with Thr766 and Thr830. Moreover, the tolyl moiety occupied the hydrophobic II forming three hydrophobic interactions with Val702, Arg817 and Cys773 ().
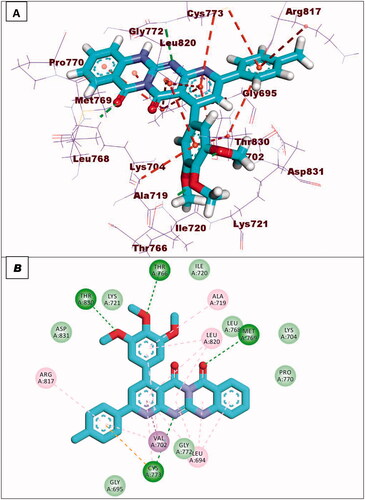
Compound 9a showed a binding energy of −15.80 kcal/mol. The 2-hydrazinylpyrido[2,3-d]pyrimidin-4(3H)-one moiety was inserted in the adenine pocket forming a hydrogen bond with the amino acid Met769. Further, it formed five hydrophobic interactions with Val702, Leu694, and Leu820. The 4-chlorophenyl moiety occupied pocket I forming three hydrophobic interactions with Leu764, Lys721, and Ala719. Moreover, the 2,4-dichlorophenyl moiety occupied the hydrophobic II forming five hydrophobic interactions with Val702, Arg817, Leu694, and Cys773 ().
The synthesised compounds showed good binding affinities against EGFRT790M with binding free energies ranging from −11.59 to −22.39 kcal/mol (). The co-crystallised ligand (TAK-285) exhibited a binding energy of −18.70 kcal/mol. The pyrrolo[3,2-d]pyrimidine moiety was buried in the adenine pocket forming a hydrogen bond with Met793 and three hydrophobic bonds with Leu844 and Ala743. The terminal 3-(trifluoromethyl)phenoxy group occupied the hydrophobic pocket I forming a hydrogen bond with Lys745. Also, it formed seven hydrophobic interactions with Lys745, Glu762, Leu788, and Ile759. In addition, the N-ethyl-3-hydroxy-3-methylbutanamide moiety occupied the hydrophobic region II forming hydrogen bond with Ser720. The phenyl moiety formed hydrophobic interactions with Met790, Val726, and Ala743 ().
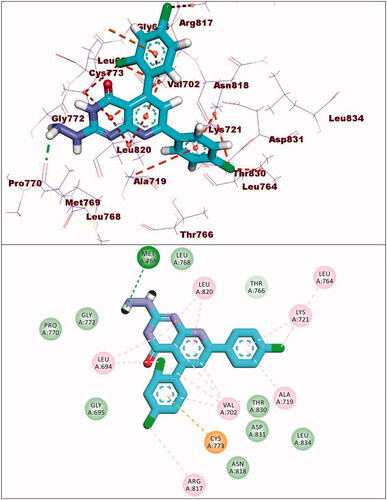
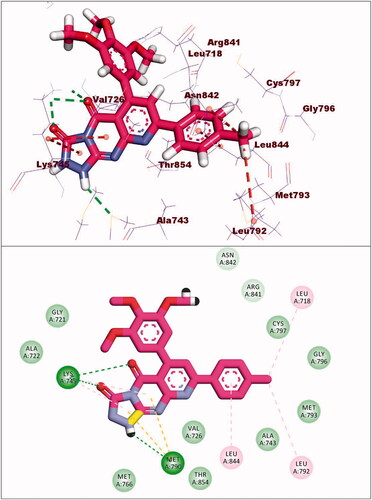
Compound 8c exhibited a binding mode similar to that of TAK-285 with an affinity value of −19.40 kcal/mol. The 5H-pyrido[2′,3′:4,5]pyrimido[2,1-b]quinazoline-5,7(12H)-dione moiety occupied the adenine pocket of forming five hydrophobic interaction with Lys745, Glu762, and Leu844. Also, it formed three hydrogen bonds with Thr854, Met790, and Lys745. The 3,4,5-trimethoxyphenyl moiety occupied the hydrophobic pocket I and 4-chloro phenyl moiety occupied the hydrophobic region II forming three hydrophobic bonds with Leu718, Leu844, and Leu792 ().
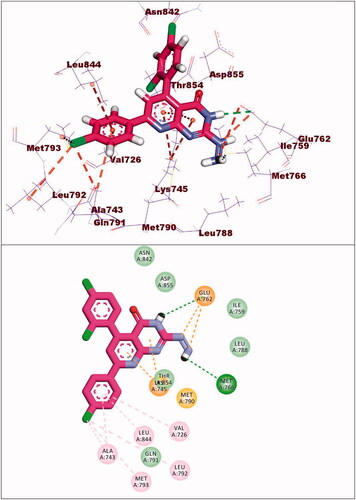
Compound 9a exhibited a binding energy of −12.15 kcal/mol. The 2-hydrazinylpyrido[2,3-d]pyrimidin-4(3H)-one moiety occupied the adenine pocket of forming four hydrophobic interaction with Lys745,Glu762, and Thr854. Also, it formed two hydrogen bonds with Glu762 and Mey766. The 2,4-dichlorophenyl moiety occupied the hydrophobic pocket I and 4-chloro phenyl moiety occupied the hydrophobic region II forming six hydrophobic bonds with Val726, Met793, Ala743, Leu844 and Leu792 ().
Compound 10d exhibited a binding energy of −19.86 kcal/mol. The 1,2-dihydropyrido[2,3-d][1,2,4]triazolo[4,3-a]pyrimidine-3,5-dione moiety occupied the adenine pocket of forming three hydrogen bonds with Lys745, and Met790. Also, it formed four hydrophobic interactions with Lys745, and Met790. The 3,4,5-trimethoxyphenyl moiety occupied the hydrophobic pocket I and tolyl moiety occupied the hydrophobic region II forming three hydrophobic bonds with Leu718, Leu844 and Leu792 ().
Compound 12d exhibited a binding energy of −22.39 kcal/mol. The pyrido[2,3-d][1,2,4]triazolo[4,3-a]pyrimidin-5(1H)-one moiety occupied the adenine pocket of forming two hydrogen bonds with Gln791 and Met793. Also, it formed six hydrophobic interactions withVal726, Leu844, Ala743, and Met793. The 3,4,5-trimethoxyphenyl moiety occupied the hydrophobic pocket I and tolyl moiety occupied the hydrophobic region II forming three hydrophobic bonds with Leu788, Ile759 and Lys745 ().
3. Conclusion
New nineteen pyrido[2,3-d]pyrimidin-4(3H)-one derivatives have been designed and synthesised as EGFR inhibitors. These compounds were evaluated for antiproliferative activities against A-549, PC-3, HCT-116, and MCF-7 cell lines. Compounds 8a, 8 b, 8d, 9a, and 12b exhibited the highest activities. Compound 8a showed promising activities against A-549, PC-3, and HCT-116 cell lines with IC50 values of 16.2, 7.98, and 25.61 µM, respectively. Compounds 8a, 8b, and 9a showed promising inhibitory activities against EGFRWT with IC50 values of 0.099, 0.419, and 0.594 µM, respectively. In addition, such derivatives showed good inhibitory effects against EGFRT790M with IC50 values of 0.123, 0.290, and 0.571 µM, respectively. The most promising candidate 8a induced a significant apoptotic effect in PC-3 cells and arrested the cell cycle at the pre-G1 phase. Structure-activity relationship studies revealed that tetracyclic 5H-pyrido [2′,3′:4,5]pyrimido[2,1-b]quinazoline-5,7(12H)-dione derivatives 8a–d have the preferred impact on the anticancer activity. In addition, the existence of an electron-donating (OCH3) group at 4-position of compounds 8a and 8d is beneficial for activity. To give an additional comprehensive investigation about the mechanism of action of the synthesised compounds, docking studies were performed against EGFRWT and EGFRT790M. Docking studies revealed that the synthesised compounds have similar binding modes against the prospective biological targets. This work introduces compounds 8a as a potential promising EGFR inhibitor.
4. Experimental
4.1. Chemistry
4.1.1. General
All details of chemicals and different apparatus for analyses were provided in Supplementary data.
4.1.2. General procedure for synthesis of thioxopyridopyrimidinone 7a–e
A mixture of the appropriate α, β-unsaturated ketones 6a–e (0.01 mol) and 6-amino-2,3-dihydro-2-thioxopyrimidin-4(1H)-one (3) (1.43 g, 0.01 mol) was heated in dry DMF (20 ml) under reflux for 10–15 h. After cooling, the precipitates were filtered and crystallised from DMF to afford compounds 7a–e. All spectral data of thioxo derivatives 7b–e was reported in our previous workCitation73. Herein, we described our newly synthesised thioxo precursor 7a.
4.1.2.1. 7–(4-Chlorophenyl)-5–(2,4-dichlorophenyl)-2-thioxo-2,3-dihydropyrido [2,3-d]pyrimidin-4(1H)-one (7a)
Yield (50%); m.p. 318–320 °C. IR (KBr) (cm−1): 3387 (NH), 1701 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 7.40–7.76(m, 5H, Ar-H), 7.97 (s, 1H, pyridine-H6), 8.26 (d, J = 8 Hz, 2H, chlorophenyl-H2,H6), 12.50 (brs, 1H, NH, D2O exchangeable); 13.23 (brs, 1H, NH, D2O exchangeable); 13CNMR (DMSO-d6) δ (ppm): 108.8, 118.5, 126.5, 127.7, 128.7, 129.1, 130.1, 132.4, 133.1, 134.7, 136.7, 149.3, 152.4, 158.3, 158.6, 162.2, 175.6; MS (m/z) 434; Anal. Calc. for: (C19H10Cl3N3OS): C, 52.50; H, 2.32; N, 9.67; Found: C, 52.57; H, 2.36; N, 9.73%.
4.1.3. General procedure for synthesis of 2,4-diaryl-5Hpyrido [2',3':4,5] pyrimido[2,1-b]quinazoline-5,7(12H)-dione(8a–d)
A mixture of 2-thioxopyrido[2,3-d]pyrimidine derivatives 7a–e (0.01 mol) and anthranilic acid (1.37 g, 0.01 mol) was heated under reflux for 20 h in the presence of 2% sodium ethoxide (20 ml) The reaction mixture was cooled, poured into ice cold water and acidified by diluted hydrochloric acid. The formed precipitate was filtered, washed several times with water, dried and washed with hot ethanol to give the compounds 8a–d.
4.1.3.1. 2–(4-Chlorophenyl)-4-(p-tolyl)-5H-pyrido[2',3':4,5]pyrimido [2,1-b] quinazoline-5,7(12H)-dione (8a)
Yield (68%); m.p. >300 °C. IR(KBr) (cm−1): 3479 (NH), 1750 (C=O), 1685 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 2.36 (s, 3H, CH3), 6.45 (m, 1H, Ar-H), 6.66 (d, J = 8.4 Hz, 2H, Ar-H), 7.10 (t, J = 8 Hz, 1H, Ar-H), 7.20 (d, J = 8 Hz, 2H, Ar-H), 7.31(d, J = 8 Hz, 2H, Ar-H), 7.50 (s, 1H, C6-pyridine), 7.56 (d, J = 8.4 Hz, 2H, Ar-H), 8.42 (d, J = 8.4 Hz, 2H, Ar-H), 11.19 (brs, 1H, NH, D2O exchangeable); 13CNMR (DMSO-d6) δ (ppm): 21.3, 106.3, 114.5, 116.1, 118.4, 127.7, 128.6, 129.0, 129.3, 129.36, 129.6, 131.8, 132.6, 135.5, 136.0, 136.2, 137.4, 150.1, 153.6, 154.2, 157.9, 169.7, 169.8; MS (m/z): 466 (M + 2), 464 (M+); Anal. Calc. for: (C27H17ClN4O2): C, 69.75; H, 3.69; N, 12.05; % Found: C, 69.82; H, 3.74; N, 12.11%.
4.1.3.2. 4–(4-Methoxyphenyl)-2-(p-tolyl)-5H-pyrido[2',3':4,5] pyrimido[2,1-b] quinazoline-5,7(12H)-dione (8b)
Yield (52%); m.p. >300 °C. IR (KBr) (cm−1): 3410 (NH), 1693 (C=O), 1750 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 2.36 (s, 3H, CH3), 3.79 (s, 3H, OCH3), 6.92–6.95 (m, 2H, Ar-H), 7.29–7.44 (m, 6H, Ar-H), 7.49 (s, 1H, C-6 pyridine), 8.07–8.10 (m, 4H, Ar-H), 11.11 (brs, 1H, NH, D2O exchangeable); 13CNMR (DMSO-d6) δ (ppm): 21.3, 55.6, 106.3, 114.5, 116.2, 118.7, 127.3, 128.1, 129.3, 129.3, 129.6, 131.4, 132.2, 135.4, 136.8, 136.2, 137.8, 150.2, 153.7, 154.8, 154.5, 157.6, 169.7, 169.8; MS (m/z): 460; Anal. Calc, for: (C28H20N4O3); C, 73.03; H, 4.38; N, 12.17%; Found: C, 73.07; H, 4.44; N, 12.23%.
4.1.3.3. 2–(4-Chlorophenyl)-4–(3,4,5-trimethoxyphenyl)-5H-pyrido [2',3':4,5] pyrimido[2,1-b]quinazoline-5,7(12H)-dione (8c)
Yield (87%); m. p. <300 °C. IR (KBr) (cm−1): 3468 (NH), 1708 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 3.74 (s, 3H, OCH3), 3.80 (s, 6H, 2OCH3), 6.51 (t, 1H, Ar-H,), 6.74 (d, J = 8 Hz, 2H, Ar-H), 7.23 (m, 1H, Ar-H,), 7.62–7.68 (m, 2H, Ar-H,), 7.76 (s, 1H, C-pyridine), 8.32 (d, J = 8 Hz, 4H, Ar-H), 11.24 (brs, 1H, NH, D2O exchangeable); 13CNMR (DMSO-d6) δ (ppm): 56.6, 60.6, 106.3, 106.7, 109.0, 115.0, 119.6, 126.9, 129.3, 129.6, 129.9, 130.4, 131.5, 133.9, 134.2, 135.6, 137.9, 150.5, 152.5, 153.4, 157.8, 159.1, 161.4, 170.6; MS (m/z): 542 (M + 2), 540 (M+); Anal. Calc. for: (C29H21ClN4O5): C, 64.39; H, 3.91; N, 10.36%; Found: C, 64.44; H, 3.95; N, 10.41%.
4.1.3.4. 2-(p-Tolyl)-4–(3,4,5-trimethoxyphenyl)-5H-pyrido[2',3':4,5] pyrimido[2,1-b]quinazoline-5,7(12H)-dione (8d)
Yield (57%); m.p. >300 °C. IR (KBr) (cm−1): 3421 (NH), 1697(C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 2.39 (s, 3H, CH3), 3.74 (s, 3H, OCH3), 3.80 (s, 6H, 2OCH3), 6.75 (s, 2H, Ar-H), 7.21–6.76 (m, 2H, Ar-H,), 7.35 (d, J = 8 Hz, 2H, Ar-H), 7.46 (s, 1H, C6-pyridine), 7.57 (m, 2H, Ar-H), 7.99 (d, J = 8 Hz, 1H, Ar-H), 8.14 (d, J = 8 Hz, 1H, Ar-H), 11.65 (brs, 1H, NH, D2O exchangeable); 13 C NMR (DMSO-d6) δ (ppm): 21.6, 56.2, 60.2, 106.0, 106.3, 106.7, 118.0, 127.9, 128.6, 129.3, 129.3, 129.6, 129.9, 134.2, 134.6, 136.5, 137.9, 140.9, 144.2, 150.1, 152.2, 153.4, 154.1, 159.1, 161.4, 169.4; MS (m/z): 520; Anal. Calc. for: (C30H24N4O5); C, 69.22; H, 4.65; N, 10.76%; Found: C, 69.25; H, 4.71; N, 10.82%.
4.1.4. General procedure for synthesis of 2-Hydrazinyl -5,7-diarylpyrido[2,3-d]pyrimidin-4(3H)-one (9a–e)
A mixture of 2-thioxopyrido[2,3-d]pyrimidine derivatives 7a–e (0.004 mol) and hydrazine hydrate (99%, 3 ml, 0.006 mol,) was heated under reflux in absolute ethanol (20 ml) for 10–15 h. After cooling, the precipitate was filtered and washed with hot ethanol to give compounds 9a–e. All spectral data of hydrazino derivatives 9b–e was reported in our previous workCitation73. Herein we described our newly synthesised hydrazine precursor 9a.
4.1.4.1. 7-(4-Chlorophenyl)-5–(2,4-dichlorophenyl)-2-hydrazineylpyrido[2,3-d]pyrimidin-4(3H)-one 9a
Yield (45%); m.p. 265–267 °C. IR (KBr) (cm−1): 3398, 3367 (NH) and (NH2), 1685 (C=O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 7.59 (d, J = 8 Hz, 2H, Ar-H), 7.61–7.62 (m, 3H, Ar-H), 8.09 (s, 1H, H6-pyridine), 8.23 (brs, 2H, NH2, D2O exchangeable), 8.47 (d, J = 8 Hz, 2H, Ar-H), 9.18 (brs, 1H, NH, D2O exchangeable), 12.83, (brs, 1H, NH, D2O exchangeable); 13 C NMR (DMSO-d6) δ (ppm): 106.5, 112.8, 120.3, 124.7, 129.0, 129.2, 130.1, 131.6, 132.9, 136.14, 136.19, 144.7, 147.8, 148.6, 157.9, 160.1, 174.6; MS (m/z): 438 (M + 6), 436 (M + 4), 434 (M + 2), 432 (M+). Anal. Calc. for: (C19H12Cl3N5O): C, 52.74; H, 2.80; N, 16.19%; Found: C, 52.78; H, 2.86; N, 16.25.
4.1.5. General procedure for synthesis of 6.8-diaryl-pyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidine-3,5-dione (10a–d)
A mixture of 2-hydrazinylpyrido[2,3-d]pyrimidine 9 b–e (1 mmol) and ethyl chloroformate (0.22 g, 2 mmol) in dry pyridine (10 ml) was heated under reflux for 9 hCitation60. The reaction mixture was cooled and the obtained solid was filtered, washed with ethanol, dried, and crystallised from DMF: EtOH (1:2).
4.1.5.1. 8-(4-Chlorophenyl)-6-(p-tolyl)-1,2-dihydropyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidine-3,5-dione (10a)
Yield (80%); m.p. 347–349 °C. IR (KBr) (cm−1): 3383 (NH), 1685 (C=O), 1647 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 2.37 (s, 3H, CH3), 7.24–7.26 (m, 3H, Ar-H), 7.37 (d, J = 8 Hz, 1H, Ar-H), 7.48–7.62 (m, 4H, 2Ar-H + 2NH, D2O exchangeable), 7.92 (s, 1H, C-6pyridine,), 8.48 (d, J = 8 Hz, 1H, Ar-H), 8.67 (d, J = 8 Hz, 1H, Ar-H); 13 C NMR (DMSO-d6) δ (ppm): 21.2, 108.6, 121.5, 128.1, 128.8, 129.4, 129.8, 134.5, 134.8, 135.7, 137.8, 145.7, 147.4, 148.0, 155.7, 159.4, 169.9; MS (m/z): 405 (M + 2), 403 (M+); Anal. Calc. for: (C21H14ClN5O2): C, 62.46; H, 3.49; N, 17.34%; Found: C, 62.54; H, 3.55; N, 17.40%.
4.1.5.2. 6-(4-Methoxyphenyl)-8-(p-tolyl)-1,2-dihydropyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidine-3,5-dione (10b)
Yield (54%); m.p. 366–368 °C. IR(KBr) (cm−1): 3398 (NH), 3375 (NH), 1697 (C=O), 1654 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 2.38 (s, 3H, CH3), 3.83 (s, 3H, OCH3), 6.96 (d, J = 8 Hz, 2H, Ar-H), 7.32–7.40 (m, 5H, Ar-H), 7.79 (s, 1H, C6-pyridine), 8.06 (brs, 2H, 2NH, D2O exchangeable), 8.57 (d, J = 8 Hz, 1H, Ar-H); MS (m/z): 399; Anal. Calc. for: (C22H17N5O3): C, 66.16; H, 4.29; N, 17.53%; Found: C; 66.24, H, 4.35; N, 17.60%.
4.1.5.3. 8-(4-Chlorophenyl)-6–(3,4,5-trimethoxyphenyl)-1,2-dihydropyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidine-3,5-dione (10c)
Yield (67%); m.p. 367–369 °C. IR(KBr) (cm−1): 3160 (2NH), 1759, 1697(2 C=O) 1HNMR (400 MHz, DMSO-d6) δ (ppm): 3.73 (s, 3H, OCH3), 3.78 (s, 6H, 2OCH3), 6.79 (s, 2H, Ar-H), 7.53–7.61 (m, 2H, Ar-H), 7.89 (s, 1H, C6-pyridine), 8.22–8.77 (m, 2H, Ar-H), 9.46 (s, 2H, 2NH, D2O exchangeable); 13C NMR (DMSO-d6) δ (ppm); 56.1, 59.7, 105.7, 106.8, 116.4, 117.5, 119.1, 128.0, 129.6, 137.7, 140.5, 148.8, 149.4, 152.3, 155.8, 158.4, 161.9, 169.6; MS (m/z): 481 (M + 2), 479 (M+). Anal. Calc. for: (C23H18ClN5O5): C, 57.57; H, 3.78; N, 14.59%; Found: C; 57.64, H, 3.85; N, 14.64%.
4.1.5.4. 8-(p-Tolyl)-6–(3,4,5-trimethoxyphenyl)-1,2-dihydropyrido[2,3-d][1, 2, 4]triazolo [4,3-a]pyrimidine-3,5-dione (10d)
Yield (54%); m.p. 391–393 °C. IR (KBr) (cm−1): 3494 (NH), 3383 (NH), 1685 (C=O), 1660 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 2.37 (s, 3H, CH3), 3.73 (s, 6H, 2 OCH3), 6.73 (s, 2H, Ar-H), 7.34–7.45 (m, 2H, Ar-H), 7.95 (s, 1H, C6-pyridine), 8.07–8.11 (m, 2H, Ar-H), 9.26 (s, 1H, NH, D2O exchangeable), 11.07 (s, 1H, NH, D2O exchangeable); MS (m/z): 459. Anal. Calc. for: (C24H21N5O5): C, 62.74; H, 4.61; N, 15.24%; Found: C; 62.81, H, 4.67; N, 15.31%.
4.1.6. General procedure for 3-amino-6,8-diaryl-2,3-dihydropyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidin-5(1H)-one 11(a–e)
A mixture of 2-hydrazinylpyrido[2,3-d]pyrimidines 9a–e (0.002 mol) and ammonium thiocyanate (2.38 g, 0.3 mol) in glacial acetic acid (15 ml) was heated under reflux for 10 h. The reaction mixture was cooled, poured onto iced water and the precipitate was filtered, dried and washed with hot ethanolCitation60.
4.1.6.1. 3-Amino-8–(4-chlorophenyl)-6–(2,4-dichlorophenyl)-10,10a-dihydropyrido [2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidin-5(1H)-one (11a)
Yield (33%); m.p. 381–383 °C. IR (KBr) (cm−1): 3421, 3356 (NH, NH2), 1681 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 7.09 (brs, 1H, NH, D2O exchangeable), 7.33 (brs, 2H, NH2, D2O exchangeable), 7.52–7.63 (m, 3H, Ar-H), 7.70 (d, J = 8 Hz, 1H, Ar-H), 7.85 (s, 1H, C6-pyridine), 8.04 (s, 1H, Ar-H), 8.28 (d, J = 12 Hz, 2H, Ar-H); 13C NMR (DMSO-d6) δ (ppm): 119.5, 127.4, 128.4, 129.1, 129.8, 131.0, 132.1, 132.4, 134.4, 134.7, 136.4, 136.7, 138.0, 141.1, 147.6, 148.5, 154.5, 167.2; MS (m/z): 463 (M + 6), 461 (M + 4), 459 (M + 2), 457 (M+). Anal. Calc. for: (C20H11Cl3N6O): C, 52.48; H, 2.42; N, 18.36%; Found: C, 52.55; H, 2.47; N, 18.42%.
4.1.6.2. 3-Amino-8–(4-chlorophenyl)-6-(p-tolyl)-2,3-dihydropyrido[2,3-d][1, 2, 4] triazolo[4,3-a]pyrimidin-5(1H)-one (11b)
Yield (33%); m.p. 383–385 °C. IR (KBr) (cm−1): 3422, 3394 (NH, NH2), 1697 (C=O); 1HNMR (400 MHz, DMSO-d6) δ (ppm): 2.34 (s, 3H, CH3), 7.16–7.22 (m, 2H, Ar-H), 7.33 (d, J = 8 Hz, 2H, Ar-H), 7.53–7.62 (d, J = 8 Hz, 2H), 7.88 (s, 1H, C-6pyridine), 8.28 (d, J = 8 Hz, 2H), 11.66 (brs, 1H, NH, D2O exchangeable), 11.65, (brs, 2H, NH2, D2O exchangeable); 13C NMR (DMSO-d6) δ (ppm): 20.6, 106.5, 109.3, 119.7, 122.7, 127.1, 128.3, 129.4, 134.7, 135.5, 137.4, 145.8, 147.3, 150.2, 153.7, 155.6, 167.4; MS (m/z): 404 (M + 2), 402 (M+); Anal. Calc. for: (C21H15ClN6O); C, 62.61; H, 3.75; N, 20.86%; Found: C, 62.66; H, 3.84; N, 20.91%.
4.1.6.3. 3-Amino-6–(4-methoxyphenyl)-8-(p-tolyl)-2,3-dihydropyrido[2,3-d][1, 2, 4] triazolo[4,3-a]pyrimidin-5(1H)-one (11c)
Yield (47%); m.p. 381–383 °C. IR (KBr) (cm−1): 3425, 4332 (NH, NH2), 1697 (C=O);1H NMR (400 MHz, DMSO-d6) δ (ppm): 2.35 (s, 3H, CH3), 3.81 (s, 3H, OCH3), 6.91–6.96 (m, 2H, Ar-H), 7.31–7.40 (m, 4H, Ar-H), 7.81 (s, 1H, C6-pyridine), 8.14 (d, J = 8 Hz, 2H, Ar-H), 11.11 (brs, 2H, NH2, D2O exchangeable), 11.57 (brs, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ (ppm): 21.2, 54.7, 112.8, 127.7, 129.4, 130.1, 133.4, 139.7, 140.7, 145.4, 145.7, 149.6, 150.0, 153.3, 153.6, 159.3, 162.6, 167.9; MS (m/z): 398. Anal. Calc. for: (C22H18N6O2): C, 66.32; H, 4.55; N, 21.09%; Found: C; 66.36, H, 4.60; N, 21.14%.
4.1.6.4. 3-Amino-8–(4-chlorophenyl)-6–(3,4,5-trimethoxyphenyl) -2,3dihydropyrido[2,3-d][1, 2, 4]triazolo[4,3-a] pyrimidin-5(1H)-one(11d)
Yield (85%); m.p. 365–367 °C. IR (KBr) (cm−1): 3437, 3425 (NH, NH2), 1708 (C=O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 3.71 (s, 3H, OCH3), 3.78 (s, 6H, 2OCH3), 7.17 (s, 2H, Ar-H), 7.61–8 (m, 3H, Ar-H + C6-pyridine), 8.23–8.31 (m, 2H, Ar-H), 11.18 (brs, 2H, NH2, D2O exchangeable), 11.71 (brs, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ (ppm): 56.0, 60.0, 106.4, 106.2, 128.8, 129.1, 129.4, 129.8, 134.1, 135.7, 140.7, 150.0, 151.7, 152.9, 153.6, 157.3, 160.9, 167.3; MS (m/z): 480 (M + 2), 478 (M+) Anal. Calc. for: (C23H19ClN6O4); C, 57.69; H, 4.00; N, 17.55%; Found: C, 57.74; H, 4.07; N, 17.59%.
4.1.6.5. 3-Amino-8-(p-Tolyl)-6–(3,4,5-trimethoxyphenyl)-2,3-dihydropyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidin-5(1H)-one (11e)
Yield (85%); m.p. 358–360 °C. IR (KBr) (cm−1): 3433, 3367 (NH, NH2), 1701 (C=O) 1H NMR (400 MHz, DMSO-d6) δ (ppm): 2.37 (s, 3H, CH3), 3.77 (s, 3H, OCH3), 3.78 (s, 6H, 2OCH3), 6.73–6.78 (m, 2H, Ar-H), 7.32–7.39 (m, 2H, Ar-H), 7.92 (s, 1H, C6-pyridine,), 8.19 (d, J = 8 Hz, 2H, Ar-H), 11.15 (brs, 2H, NH2, D2O exchangeable), 11.61 (s, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ (ppm): 20.5, 55.7, 59.7, 106.5, 107.9, 114.8, 117.5, 127.5, 129.4, 134.1, 137.1, 140.1, 150.0, 152.0, 153.4, 153.7, 158.6, 161.3, 167.7; MS (m/z): 458; Anal. Calc. for: (C24H22N6O4); C, 62.87; H, 4.84; N, 18.33%; Found: C, 62.94; H, 4.91; N, 18.39%.
4.1.6.6. 3-Phenyl-6,8-disubistitutedphenylpyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidin-5(1H)-one (12a–e)
To a solution of hydrazine derivative 9b–e (0.001 mol) in dry pyridine (20 ml) benzoyl chloride was added (0.001 mol), and the resulting mixture was heated under reflux for 10–15 h. After cooling, the formed precipitate was filtered washed with hot ethanol to afford 12a–d, respectively.
4.1.6.7. 8–(4-Chlorophenyl)-3-phenyl-6-(p-tolyl)pyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidin-5(1H)-one (12a)
Yield (67%); m.p. 315–317 °C. IR(KBr) (cm−1): 3414(NH) 1750 (C=O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 2.40 (s, 3H, CH3), 7.27–8.30 (m, 14H, Ar-H), 11.21(brs, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ (ppm): 21.3, 106.3, 118.0, 128.2, 128.2, 128.6, 129.3, 135.5, 136.6, 136.9, 138.5, 142.2, 150.8, 151.5, 153.8, 154.1, 155.2, 158.1, 162.8, 166,1, 188.4; MS m/z (%): 465(M + 2), 463 (M+); Anal. Calc. for: (C27H18ClN5O); C, 69.90; H, 3.91; N, 15.10%; Found: C, 69.97; H, 3.98; N, 15.14%.
4.1.6.8. 6-(4-Methoxyphenyl)-3-phenyl-8-(p-tolyl)pyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidin-5(1H)-one (12b)
Yield (33%); m.p. 325–327 °C. IR (KBr) (cm−1): 3414, NH, 1720 (C=O);1H NMR (400 MHz, DMSO-d6) δ (ppm); 2.39 (s, 3H, CH3), 3.85 (s, 3H, OCH3), 7.03–8.32 (m, 15H 14Ar-H + NH- D2O exchangeable); 13C NMR (DMSO-d6) δ (ppm): 21.6, 55.5, 113.2, 127.1, 127.6, 128.5, 129.0, 129.5, 129.8, 129.9, 130.3, 130.9, 131.5, 132.7, 135.4, 143.6, 145.2, 150.4, 154.6, 158.4, 160.3, 170.6; MS (m/z): 459; Anal. Calc. for: (C28H21N5O2); C, 73.19; H, 4.61; N, 15.24%; Found: C, 73.25; H, 4.66; N, 15.27%.
4.1.6.9. 8-(4-Chlorophenyl)-3-phenyl-6–(3,4,5-trimethoxyphenyl)pyrido[2,3-d][1, 2, 4]triazolo[4,3-a]pyrimidin-5(1H)-one (12c)
Yield (33%); m.p. 313–315 °C. IR (KBr) (cm−1): 3417 (NH), 1693 (C=O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 3.7 (s, 3H, OCH3), 3.8 (s, 6H, 2OCH3), 7.47–59 (m, 3H, Ar-H), 7.59–7.63 (m, 4H, Ar-H), 7.93–8.06 (m, 3H, 2Ar-H + 1NH-D2O exchangeable), 8.55–8.59 (m, 1H, C6-pyridine), 8.91–8.92 (m, 2H, Ar-H); 13CNMR (DMSO-d6) δ (ppm): 55.6, 59.6, 113.3, 123.9, 127.2, 128.9, 129.3, 129.5, 129.7, 129.8, 129.9, 130.3, 130.9, 132.6, 143.3, 146.5, 152.7, 160.2, 169.2; MS (m/z): 541(M + 2), 539 (M+); Anal. Calc. for: (C29H22ClN5O4): C, 64.51; H, 4.11; N, 12.97%; Found: C, 64.57; H, 4.16; N, 13.03%.
4.1.6.10. 3-Phenyl-8-(p-tolyl)-6–(3,4,5-trimethoxyphenyl)pyrido[2,3-d][1, 2, 4]triazolo [4,3-a]pyrimidin-5(1H)-one(12d)
Yield (33%); m.p. 322–324 °C. IR (KBr) (cm−1): 3464 (NH), 1701 (C=O); 1H NMR (400 MHz, DMSO-d6) δ (ppm): 2.37 (s, 3H, CH3), 3.75 (s, 3H, OCH3), 3.9 (s, 6H, 2OCH3), 6.76–7.07 (m, 2H, Ar-H), 7.09–7.87 (m, 8H, Ar-H), 8.10–8.22 (m, 2H, Ar-H), 11.16 (brs, 1H, NH, D2O exchangeable); 13C NMR (DMSO-d6) δ (ppm): 20.8, 56.1, 60.1, 105.5, 106.5, 107.9, 108.9, 117.5, 118.8, 120.4, 127.4, 128.4, 129.1, 130.5, 132.3, 133.8, 134.4, 137.1, 140.1, 150.0, 151.7, 153.3, 158.6,162.6; MS (m/z): 519; Anal. Calc. for: (C30H25N5O4); C, 69.35; H, 4.85; N, 13.48%; Found: C, 69.41; H, 4.91; N, 13.52%.
4.2. Biological evaluation
4.2.1. In vitro cytotoxic activity
In vitro cytotoxicity was carried out using MTT assay protocolCitation63 as described in Supplementary data.
4.2.2. In vitro EGFR kinase assay
In vitro EGFR inhibitory activity was assessed using Homogeneous time-resolved fluorescence (HTRF) assayCitation64 as described in Supplementary data.
4.2.3. Cell cycle analysis
The effect of compound 8a on cell cycle distribution was performed using propidium iodide (PI) staining technique as described in Supplementary dataCitation65,Citation74,Citation75.
4.2.4. Apoptosis analysis
The effect of compound 8a on cell apoptosis was investigated as described in Supplementary dataCitation76–78.
4.3. Docking studies
Molecular docking studies of the synthesised compounds were carried out against EGFRWT (PDB ID: 4HJO, resolution 2.75 Å and EGFRT790M (PDB ID: 3W2O, resolution 2.35 Å) as described in Supplementary dataCitation22.
Supplemental Material
Download PDF (2.6 MB)Acknowledgement
This paper is based upon work supported by Science, Technology & Innovation Funding Authority (STIFA) under grant number 43327.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- WHO. Cancer. Available from: https://www.who.int/news-room/fact-sheets/detail/cancer. (last accessed 13 Jan 2022).
- Boyle P, Ferlay JJ. Cancer incidence and mortality in Europe, 2004. Annals Oncol 2005;16:481–8.
- Gavalas NG, Karadimou A, Dimopoulos MA, et al. Immune response in ovarian cancer: how is the immune system involved in prognosis and therapy: potential for treatment utilization. J Immunol Res 2010;2010:1–15.
- Li M, Huo X, Davuljigari CB, et al. MicroRNAs and their role in environmental chemical carcinogenesis. Environ Geochem Health 2019;41:225–47.
- Chorawala M, Oza P, Shah GJ. Mechanisms of anticancer drugs resistance: an overview. J Pharm Technol Drug Res 2012;4:1–9.
- Lavogina D, Enkvist E, Uri A. Bisubstrate inhibitors of protein kinases: from principle to practical applications. ChemMedChem 2010;5:23–34.
- Luković E, González-Vera JA, Imperiali B. Recognition-domain focused chemosensors: versatile and efficient reporters of protein kinase activity. J Am Soc 2008;130:12821–7.
- Braun T, Gautel MJ. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol cell Biol 2011;12:349–61.
- Meulenbeld HJ, Mathijssen RH, Verweij J, et al. Danusertib, an aurora kinase inhibitor. Expert Opin Investig Drugs 2012;21:383–93.
- Abdellatif KR, Bakr RB. Pyrimidine and fused pyrimidine derivatives as promising protein kinase inhibitors for cancer treatment. Med Chem J 2021;30:31–49.
- Ullrich A, Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell 1990;61:203–12.
- Voldborg BR, Damstrup L, Spang-Thomsen M, Poulsen HS. Epidermal growth factor receptor (EGFR) and EGFR mutations, function and possible role in clinical trials. Ann Oncol 1997;8:1197–206.
- Meierjohann S, Mueller T, Schartl M, Buehner MJZ. A structural model of the extracellular domain of the oncogenic EGFR variant Xmrk. Zebrafish 2006;3:359–69.
- Ren S, Chen X, Kuang P, et al. Association of EGFR mutation or ALK rearrangement with expression of DNA repair and synthesis genes in never-smoker women with pulmonary adenocarcinoma. Cancer 2012;118:5588–94.
- Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006;366:2–16.
- Godin-Heymann N, Ulkus L, Brannigan BW, et al. The T790M “gatekeeper” mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther 2008;7:874–9.
- Abdelsalam EA, Zaghary WA, Amin KM, et al. Synthesis and in vitro anticancer evaluation of some fused indazoles, quinazolines and quinolines as potential EGFR inhibitors. Bioorg Chem 2019;89:102985.
- Ismail RS, Abou-Seri SM, Eldehna WM, et al. Novel series of 6-(2-substitutedacetamido)-4-anilinoquinazolines as EGFR-ERK signal transduction inhibitors in MCF-7 breast cancer cells. Eur J Med Chem 2018;155:782–96.
- Gandin V, Ferrarese A, Dalla Via M, et al. Targeting kinases with anilinopyrimidines: discovery of N-phenyl-N'-[4-(pyrimidin-4-ylamino)phenyl]urea derivatives as selective inhibitors of class III receptor tyrosine kinase subfamily. Sci Rep 2015;5:16750.
- Traxler P, Furet P. Strategies toward the design of novel and selective protein tyrosine kinase inhibitors. Pharmacol Therapeutics 1999;82:195–206.
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 2009;9:28–39.
- Gaber AA, Bayoumi AH, El-Morsy AM, et al. Design, synthesis and anticancer evaluation of 1H-pyrazolo[3,4-d]pyrimidine derivatives as potent EGFRWT and EGFRT790M inhibitors and apoptosis inducers. Bioorg Chem 2018;80:375–95.
- Sharma VK, Nandekar PP, Sangamwar A, et al. Structure guided design and binding analysis of EGFR inhibiting analogues of erlotinib and AEE788 using ensemble docking, molecular dynamics and MM-GBSA. RSC Adv 2016;6:65725–35.
- Mowafy S, Galanis A, Doctor ZM, et al. Toward discovery of mutant EGFR inhibitors; design, synthesis and in vitro biological evaluation of potent 4-arylamino-6-ureido and thioureido-quinazoline derivatives. Biorg Med Chem 2016;24:3501–12.
- Zhao Z, Wu H, Wang L, et al. Exploration of type II binding mode: a privileged approach for kinase inhibitor focused drug discovery? ACS Chem Biol 2014;9:1230–41.
- Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol 2006;2:358–64.
- Bonomi P. Erlotinib: a new therapeutic approach for non-small cell lung cancer. Expert Opin Invest Drugs 2003;12:1395–401.
- Pao W, Wang TY, Riely GJ, et al. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS Med 2005;2:e17.
- Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLOS Med 2005;2:e73.
- Kobayashi S, Boggon TJ, Dayaram T, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med 2005;352:786–92.
- Kwak EL, Sordella R, Bell DW, et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. Proc Natl Acad Sci USA 2005;102:7665–70.
- Engelman JA, Zejnullahu K, Gale C-M, et al. PF00299804, an irreversible pan-ERBB inhibitor, is effective in lung cancer models with EGFR and ERBB2 mutations that are resistant to gefitinib. Cancer Res 2007;67:11924–32.
- Li D, Ambrogio L, Shimamura T, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene 2008;27:4702–11.
- Sohn SH, Sul HJ, Kim B, et al. RNF43 and PWWP2B inhibit cancer cell proliferation and are predictive or prognostic biomarker for FDA-approved drugs in patients with advanced gastric cancer. J Cancer. 2021;12:4616–25.
- Sequist LV, Besse B, Lynch TJ, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non–small-cell lung cancer. J Clin Oncol 2010;28:3076–83.
- Kim Y, Ko J, Cui Z, et al. The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor. Mol Cancer Therapeutics 2012;11:784–91.
- Jänne PA, Yang JC-H, Kim D-W, et al. AZD9291 in EGFR inhibitor-resistant non-small-cell lung cancer. N Engl J Med 2015;372:1689–99.
- J. Carroll. Following lethal tox report, Boehringer scraps plans for high-speed development, kills $730M Hanmi deal; 2016. Available from: https://endpts.com/following-lethal-tox-report-boehringer-scraps-plans-for-high-speed-development-kills-730m-hanmi-deal/. (last accessed May 2019).
- P, Callery G, Peter Cancer and cancer chemotherapy. In: David, AW and Thomas, LL, eds. Foye’s principles of medical chemistry. Philadelphia: Lippincott, Williams and Wilkins; 2002.
- Abdel-Mohsen HT, Ragab FA, Ramla MM, El Diwani HI. Novel benzimidazole-pyrimidine conjugates as potent antitumor agents. Eur J Med Chem 2010;45:2336–44.
- Shao H, Shi S, Foley DW, et al. Synthesis, structure-activity relationship and biological evaluation of 2,4,5-trisubstituted pyrimidine CDK inhibitors as potential anti-tumour agents. Eur J Med Chem 2013;70:447–55.
- Fathalla O, Zeid I, Haiba M, et al. Synthesis, antibacterial and anticancer evaluation of some pyrimidine derivatives. World J Chem 2009;4:127–32.
- Yu L, Huang M, Xu T, et al. A structure-guided optimization of pyrido[2,3-d]pyrimidin-7-ones as selective inhibitors of EGFRL858R/T790M mutant with improved pharmacokinetic properties. Eur J Med Chem 2017;126:1107–17.
- Xu T, Peng T, Ren X, et al. C5-substituted pyrido [2, 3-d] pyrimidin-7-ones as highly specific kinase inhibitors targeting the clinical resistance-related EGFR T790M mutant. MedChemComm 2015;6:1693–7.
- El-Naggar AM, Abou-El-Regal MM, El-Metwally SA, et al. Synthesis, characterization and molecular docking studies of thiouracil derivatives as potent thymidylate synthase inhibitors and potential anticancer agents. Mol Div 2017;21:967–83.
- Eldehna WM, Abo-Ashour MF, Nocentini A, et al. Novel 4/3-((4-oxo-5-(2-oxoindolin-3-ylidene)thiazolidin-2-ylidene)amino) benzenesulfonamides: synthesis, carbonic anhydrase inhibitory activity, anticancer activity and molecular modelling studies. Eur J Med Chem 2017;139:250–62.
- El-Naggar AM, Eissa IH, Belal A, El-Sayed AA. Design, eco-friendly synthesis, molecular modeling and anticancer evaluation of thiazol-5 (4 H)-ones as potential tubulin polymerization inhibitors targeting the colchicine binding site. RSC Adv 2020;10:2791–811.
- Eissa IH, El-Naggar AM, El-Hashash MA. Design, synthesis, molecular modeling and biological evaluation of novel 1H-pyrazolo[3,4-b]pyridine derivatives as potential anticancer agents. Bioorg Chem 2016;67:43–56.
- Eissa IH, El-Naggar AM, Abd El-Sattar NE, Youssef AS. Design and discovery of novel quinoxaline derivatives as dual DNA intercalators and topoisomerase II inhibitors. Anti-Cancer Agents Med Chem 2018;18:195–209.
- Ibrahim M, Taghour M, Metwaly A, et al. Design, synthesis, molecular modeling and anti-proliferative evaluation of novel quinoxaline derivatives as potential DNA intercalators and topoisomerase II inhibitors. Eur J Med Chem 2018;155:117–34.
- Eissa IH, Metwaly AM, Belal A, et al. Discovery and antiproliferative evaluation of new quinoxalines as potential DNA intercalators and topoisomerase II inhibitors. Archiv der Pharmazie 2019;352:1900123.
- Mahdy HA, Ibrahim MK, Metwaly AM, et al. Design, synthesis, molecular modeling, in vivo studies and anticancer evaluation of quinazolin-4(3H)-one derivatives as potential VEGFR-2 inhibitors and apoptosis inducers. Bioorg Chem 2020;94:103422.
- El‐Helby AGA, Sakr H, Eissa IH, et al. Benzoxazole/benzothiazole‐derived VEGFR‐2 inhibitors: design, synthesis, molecular docking, and anticancer evaluations. Archiv der Pharmazie 2019;352:1900178.
- Elmetwally SA, Saied KF, Eissa IH, Elkaeed EB. Design, synthesis and anticancer evaluation of thieno[2,3-d]pyrimidine derivatives as dual EGFR/HER2 inhibitors and apoptosis inducers. Bioorg Chem 2019;88:102944.
- Nasser AA, Eissa IH, Oun MR, et al. Discovery of new pyrimidine-5-carbonitrile derivatives as anticancer agents targeting EGFRWT and EGFRT790M. Org Biomol Chem 2020;18:7608–34.
- Othman IM, Alamshany ZM, Tashkandi NY, et al. New pyrimidine and pyrazole-based compounds as potential EGFR inhibitors: synthesis, anticancer, antimicrobial evaluation and computational studies. Bioorg Chem 2021;114:105078.
- Abd El-Meguid EA, Moustafa GO, Awad HM, et al. Novel benzothiazole hybrids targeting EGFR: design, synthesis, biological evaluation and molecular docking studies. Anticancer Agents Med Chem 2021;1240:130595.
- Khattab RR, Alshamari AK, Hassan AA, et al. Click chemistry based synthesis, cytotoxic activity and molecular docking of novel triazole-thienopyrimidine hybrid glycosides targeting EGFR. J Enzyme Inhib Med Chem 2021;36:504–16.
- Elzahabi HS, Nossier ES, Khalifa NM, et al. Anticancer evaluation and molecular modeling of multi-targeted kinase inhibitors based pyrido[2,3-d]pyrimidine scaffold. J Enzyme Inhib Med Chem 2018;33:546–57.
- Khalifa NM, Adel A-H, Abd-Elmoez SI, et al. A convenient synthesis of some new fused pyridine and pyrimidine derivatives of antimicrobial profiles. Res Chem Intermed 2015;41:2295–305.
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 1983;65:55–63.
- Denizot F, Lang RJJoim. Rapid colorimetric assay for cell growth and survival: modifications to the tetrazolium dye procedure giving improved sensitivity and reliability. J Immunol Methods 1986;89:271–7.
- Thabrew MI, Hughes RD, McFarlane IGJJop. Screening of hepatoprotective plant components using a HepG2 cell cytotoxicity assay. J Pharm Pharmacol 2011;49:1132–5.
- Jia Y, Quinn CM, Gagnon AI, Talanian R. Homogeneous time-resolved fluorescence and its applications for kinase assays in drug discovery. Anal Biochem 2006;356:273–81.
- Wang J, Lenardo MJ. Roles of caspases in apoptosis, development, and cytokine maturation revealed by homozygous gene deficiencies. J Cell Sci 2000;113:753–7.
- Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. A novel assay for apoptosis flow cytometric detection of phosphatidylserine expression on early apoptotic cells using fluorescein labelled annexin V. J Immunol Methods 1995;184:39–51.
- Earnshaw WC, Martins LM, Kaufmann SH. Mammalian caspases: structure, activation, substrates, and functions during apoptosis. Ann Rev Biochem 1999;68:383–424.
- Bortner CD, Cidlowski JA. Apoptotic volume decrease and the incredible shrinking cell. Cell Death Different 2002;9:1307–10.
- Bae SS, Choi JH, Oh YS, et al. Proteolytic cleavage of epidermal growth factor receptor by caspases. FEBS Lett 2001;491:16–20.
- He Y, Huang J, Chignell C. Cleavage of epidermal growth factor receptor by caspase during apoptosis is independent of its internalization. Oncogene 2006;25:1521–31.
- Park JH, Liu Y, Lemmon MA, Radhakrishnan R. Erlotinib binds both inactive and active conformations of the EGFR tyrosine kinase domain. Biochem J 2012;448:417–23.
- Sogabe S, Kawakita Y, Igaki S, et al. Structure-based approach for the discovery of pyrrolo[3,2-d]pyrimidine-based EGFR T790M/L858R mutant inhibitors. ACS Med Chem Lett 2013;4:201–5.
- Taylor EC, Cheng C. Studies in purine chemistry. VI. A convenient one-step synthesis of hypoxanthine. J Am Chem Soc 1959;1:9–11.
- Eldehna WM, Hassan GS, Al-Rashood ST, et al. Synthesis and in vitro anticancer activity of certain novel 1-(2-methyl-6-arylpyridin-3-yl)-3-phenylureas as apoptosis-inducing agents. J Enzyme Inhib Med Chem 2019;34:322–32.
- Al-Warhi T, Abo-Ashour MF, Almahli H, et al. Novel [(N-alkyl-3-indolylmethylene)hydrazono]oxindoles arrest cell cycle and induce cell apoptosis by inhibiting CDK2 and Bcl-2: synthesis, biological evaluation and in silico studies. J Enzyme Inhib Med Chem 2020;35:1300–9.
- Lo KK-W, Lee TK-M, Lau JS-Y, et al. Luminescent biological probes derived from ruthenium(II) estradiol polypyridine complexes. Inorg Chem 2008;47:200–8.
- Sabt A, Abdelhafez OM, El-Haggar RS, et al. Novel coumarin-6-sulfonamides as apoptotic anti-proliferative agents: synthesis, in vitro biological evaluation, and QSAR studies. J Enzyme Inhib Med Chem 2018;33:1095–107.
- Al-Sanea MM, Al-Ansary GH, Elsayed ZM, et al. Development of 3-methyl/3-(morpholinomethyl)benzofuran derivatives as novel antitumor agents towards non-small cell lung cancer cells. J Enzyme Inhib Med Chem 2021;36:987–99.