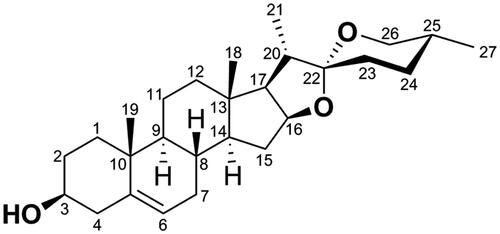
Abstract
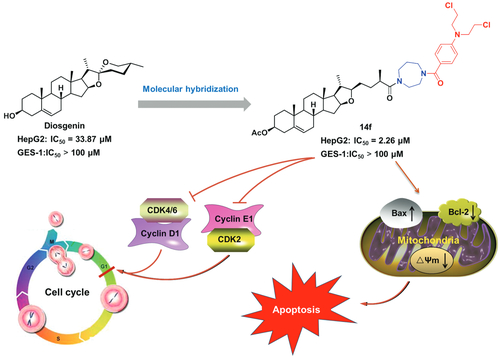
To discover new lead compounds with anti-tumour activities, in the present study, natural diosgenin was hybridised with the reported benzoic acid mustard pharmacophore. The in vitro cytotoxicity of the resulting newly synthesised hybrids (8–10, 14a–14f, and 15a–15f) was then evaluated in three tumour cells (HepG2, MCF-7, and HeLa) as well as normal GES-1 cells. Among them, 14f possessed the most potential anti-proliferative activity against HepG2 cells, with an IC50 value of 2.26 µM, which was 14.4-fold higher than that of diosgenin (IC50 = 32.63 µM). Furthermore, it showed weak cytotoxicity against GES-1 cells (IC50 > 100 µM), thus exhibiting good antiproliferative selectivity between normal and tumour cells. Moreover, 14f could induce G0/G1 arrest and apoptosis of HepG2 cells. From a mechanistic perspective, 14f regulated cell cycle-related proteins (CDK2, CDK4, CDK6, cyclin D1 and cyclin E1) as well mitochondrial apoptosis pathway-related proteins (Bax, Bcl-2, caspase 9, and caspase 3). These findings suggested that hybrid 14f serves as a promising anti-hepatoma lead compound that deserves further research.
Graphical Abstract
1. Introduction
As of September 2019, approximately 84.3% (156 out of 185) of anticancer small molecules approved by the FDA are natural products or their derivativesCitation1, which affirms the position of natural products as a focal point for finding structural and medicinal inspiration for drug discovery. Natural steroids and their synthetic derivatives are attracting increasing research interest for their promising anti-tumour activities and potential utilisation in the discovery and design of new anti-tumour agentsCitation2–4.
Diosgenin (DSG, ) is a natural steroidal sapogenin isolated from fenugreek seeds and the roots of wild yam (Dioscorea villosa)Citation5. It possesses many pharmacological functions, such as anti-oxidation, anti-inflammatory, cardiovascular protection, neuroprotection, hypolipidemic, anti-diabetic, and anti-cancer, etcCitation6.
In-depth studies on the anti-tumour effects and mechanistic patterns of DSG have revealed that it could regulate multiple genes and several signalling pathways in various types of human cancers. For instance, DSG could arrest the cell cycle at the G2/M phase by regulating the Cdc25C-Cdc2-cyclin B pathway in human breast cancer cellsCitation7, exert tumour-suppressive function by inhibiting Cdc20 in osteosarcoma cellsCitation8, induce mitochondria-mediated apoptosis in human cholangiocarcinoma cells and apoptosis via suppression of Skp2 in human breast cancer cellsCitation5,Citation9, inhibit the activation of cAMP/PKA/CREB pathway in colorectal cancer cellsCitation6, and so on. Although DSG possess extensive anti-cancer activity, the application of DSG for cancer therapy was limited by its moderate potency. Therefore, it is important to optimise the scaffold of DSG to obtain promising anti-tumour compounds with improved inhibitory effect.
Nitrogen mustards, which are a type of DNA bifunctional alkylating agents, are developed as clinically useful anti-cancer agents, and include compounds such as chlorambucil, mechlorethamine, melphalan, cyclophosphamide, and estramustine. They exert cytotoxicity by binding to DNA, cross-linking the two chains, and preventing cell replicationCitation10,Citation11.
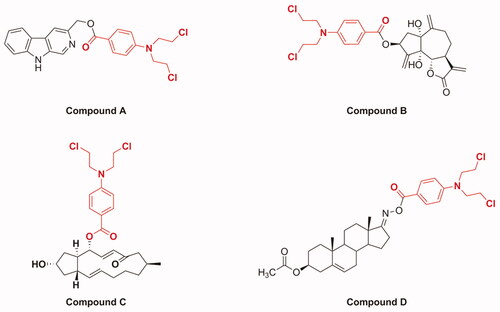
Molecular hybridisation is a widely used strategy in drug discovery, which forms new molecular entities by incorporating two or more bioactive substructures through suitable linkagesCitation12–16. These hybridised molecules possess improved or new biological properties relative to their individual componentsCitation17. Recently, it has been reported that the hybridisation of natural products with nitrogen mustards provides new strategies for discovering anti-cancer molecules with improved anti-cancer effect, selectivity, and reduced toxicity. For example (), a series of β-carboline derivatives with nitrogen mustard moieties synthesised by Sun et al. showed potent inhibitory activities in human breast carcinoma cells (MCF-7 and MDA-MB-231); especially, compound A containing benzoic acid mustard possessed significant anti-proliferative activity against MCF-7 cellsCitation18. Compound B, which was obtained by introducing a benzoic acid mustard fragment displayed the highest anti-proliferative properties against cervical cancer HeLa cellsCitation19. In addition, Han et al. synthesised a series of novel conjugates of brefeldin A and nitrogen mustards and found that compound C was the most active derivative against Bel-7402 cellsCitation20. Compound D, which was synthesised by conjugating the D-ring-derived androstene oxime with benzoic acid mustard, exhibited the most outstanding effect on inhibiting the growth of ovarian cancer IGROV1 cellsCitation21. These findings, coupled with the anti-cancer profiles of DSG and benzoic acid mustard, promoted us to further explore the anti-tumour potential of DSG–benzoic acid mustard hybrids.
Figure 2. Chemical structures of several examples of natural product and benzoic acid mustard hybrids.
To the best of our knowledge, there are few reports on the synthesis and biological activity of DSG–benzoic acid mustard derivatives with amide-amide linkages. Therefore, in this study, with the aim of finding new kind of DSG derivatives with improved anti-tumour activity, selectivity, and reduced toxicity, and further explore their structure–activity relationships, fifteen novel DSG–benzoic acid mustard hybrids bearing diversified linkers were designed and synthesised using the molecular hybridisation strategy. The cytotoxic activities of hybrids were evaluated against three cancer cell lines (HepG-2, MCF-7, and HeLa) and normal GES-1 cells. Furthermore, in-depth anti-proliferative mechanisms of the most potent compound, 14f, including cell cycle progression, expression of cell cycle-related genes and proteins, induction of apoptosis, changes in mitochondrial membrane potential (MMP) and expression of apoptosis-related genes and proteins were explored as well. Taken together, these new results might give researchers some inspiration for structural modification of other natural products, to facilitate discovery of novel anti-tumour agents.
2. Experimental
2.1. Materials and methods
All reagents were obtained from chemical and biological companies. Anhydrous solvents were dried through routine protocols. DSG derivatives 5, 6, and 11 were prepared using our published proceduresCitation22. The purity of the compounds was measured by HPLC by using a Waters Symmetry C18 (4.6 × 250 mm, 5 µm) column and its peak UV detection was 254 nm. HPLC conditions: methanol/water with 90:10; flow rate 1.0 ml/min. The purity of target compounds was >95% by the analysis of HPLC. Nuclear magnetic resonance (NMR) spectra were recorded using a AVANCE NEO 600 (1H, 600 MHz; 13 C, 150 MHz, Bruker) spectrometer. High-resolution mass spectra (HRMS) (ESI) data were measured on TripleTOF® 4600 (AB SCIEX, USA) and Accurate-Mass Q-TOF 6530 (Agilent, USA). The reactions were detected using SGF254 silica gel thin layer chromatography (TLC) (Yantai Huayang New Material Technology Co. Ltd., China). Preparative TLC were performed on layer plates of SGF254 silica gel, 1 mm (Yantai Huayang New Material Technology Co. Ltd.).
2.1.1. (22r,25R)-3β-hydroxy-5-en-furostan-26-ol (7)
A mixture of intermediate 6 (200 mg, 0.436 mmol) and KOH (48.8 mg, 0.872 mmol) in MeOH (8 ml) was stirred at room temperature for 6 h. After confirming the progress of the reaction using TLC, the mixture was poured into 40 ml of H2O and filtered. The obtained precipitate was washed with water and dried to yield intermediate 7 (172.4 mg, 95.0%) as a white powder. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.33 (d, J = 5.0 Hz, 1H, H-6), 4.30 (m, 1H, H-16), 3.53–3.47 (m, 2H, H-26, H-3), 3.43 (dd, J = 10.6, 6.0 Hz, 1H, H-26), 3.32 (td, J = 8.3, 3.8 Hz, 1H, H-22), 1.02 (s, 3H, 19-CH3), 0.99 (d, J = 6.7 Hz, 3H, 21-CH3), 0.91 (d, J = 6.7 Hz, 3H, 27-CH3), 0.80 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 140.94 (C-5), 121.56 (C-6), 90.50 (C-22), 83.36 (C-16), 71.83 (C-3), 68.17 (C-26), 65.23 (C-17), 57.11 (C-14), 50.23 (C-9), 42.41 (C-4), 40.84 (C-12), 39.59 (C-13), 38.04 (C-20), 37.39 (C-1), 36.76 (C-10), 35.86 (C-25), 32.37 (C-7), 32.13 (C-15), 31.73 (C-2), 31.73 (C-8), 30.55 (C-24), 30.25 (C-23), 20.83 (C-11), 19.55 (C-19), 19.06 (C-21), 16.77 (C-27), 16.58 (C-18). HRMS (ESI): m/z calculated for C27H45O3 [M + H]+: 417.3363, found: 417.3354.
2.1.2. Synthesis of compounds 8 and 9
Compound 7 (60 mg, 0.145 mmol) was dissolved in dry CH2Cl2 (6 ml), following which benzoic acid mustard (90.8 mg, 0.348 mmol), EDCI (110.8 mg, 0.58 mmol), and catalytic amount of DMAP were added to it. After 24 h of stirring at room temperature, the obtained solvent was concentrated under vacuum to obtain the crude product, which was purified using preparative TLC (PE:EtOAc = 2:1, v/v), to obtain compounds 8 and 9.
2.1.2.1. (22 R,25R)-3β-hydroxy-5-en-furostan-26–4-(bis(2-chloroethyl)amino)benzoate (8)
White powder, yield 35.6%. HPLC purity 97.44%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.94 (d, J = 8.9 Hz, 2H, Ph-H), 6.66 (d, J = 8.9 Hz, 2H, Ph-H), 5.34 (d, J = 4.8 Hz, 1H, H-6), 4.30 (m, 1H, H-16), 4.17 (dd, J = 10.6, 5.6 Hz, 1H, H-26), 4.08 (dd, J = 10.6, 6.6 Hz, 1H, H-26), 3.80 (t, J = 7.0 Hz, 4H, NCH2CH2Cl, ×2), 3.65 (t, J = 7.0 Hz, 4H, NCH2CH2Cl, ×2), 3.52 (m, 1H, H-3), 3.32 (m, 1H, H-22), 1.02 (s, 3H, 19-CH3), 1.01 (d, J = 6.6 Hz, 3H, 21-CH3), 0.99 (d, J = 6.7 Hz, 3H, 27-CH3), 0.80 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 166.69 (–COO), 149.67 (Ph-C), 140.95 (C-5), 131.91 (Ph-C, ×2), 121.59 (C-6), 119.48 (Ph-C), 110.96 (Ph-C, ×2), 90.36 (C-22), 83.35 (C-16), 71.87 (C-3), 69.31 (C-26), 65.28 (C-17), 57.11 (C-14), 53.46 (NCH2CH2Cl, ×2), 50.24 (C-9), 42.42 (C-4), 40.84 (C-12), 40.23 (NCH2CH2Cl, ×2), 39.59 (C-13), 38.07 (C-20), 37.40 (C-1), 36.77 (C-10), 33.23 (C-24), 32.39 (C-7), 32.16 (C-15), 31.76 (C-8), 31.04 (C-23), 30.73 (C-25), 29.41 (C-2), 20.84 (C-11), 19.57 (C-19), 19.11 (C-21), 17.15 (C-27), 16.58 (C-18). HRMS (ESI): m/z calculated for C38H56Cl2NO4 [M + H]+: 660.3581, found: 660.3578.
2.1.2.2. (22 R,25R)-3β-((4-(bis(2-chloroethyl)amino)benzoyl)oxy)-5-en-furostan-26–4-(bis(2-chloroethyl)amino) benzoate (9)
White powder, yield 27.2%. HPLC purity 99.56%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.94 (d, J = 8.9 Hz, 2H, Ph-H), 7.93 (d, J = 8.8 Hz, 2H, Ph-H), 6.67 (d, J = 8.8 Hz, 2H, Ph-H), 6.66 (d, J = 8.9 Hz, 2H, Ph-H), 5.40 (d, J = 4.8 Hz, 1H, H-6), 4.80 (m, 1H, H-3), 4.31 (m, 1H, H-16), 4.17 (dd, J = 10.9, 5.7 Hz, 1H, H-26), 4.08 (dd, J = 10.9, 6.8 Hz, 1H, H-26), 3.80 (t, J = 7.0 Hz, 8H, NCH2CH2Cl, ×4), 3.65 (t, J = 7.0 Hz, 8H, NCH2CH2Cl, ×4), 3.33 (m, 1H, H-22), 1.08 (s, 3H, 19-CH3), 1.02 (d, J = 6.8 Hz, 3H, 21-CH3), 1.00 (d, J = 6.7 Hz, 3H, 27-CH3), 0.81 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 166.68 (-COO), 166.02 (-COO), 149.67 (Ph-C), 149.62 (Ph-C), 140.04 (C-5), 131.90 (Ph-C, ×4), 122.48 (C-6), 119.75 (Ph-C), 119.48 (Ph-C), 110.96 (Ph-C, ×2), 110.93 (Ph-C, ×2), 90.36 (C-22), 83.35 (C-16), 73.98 (C-3), 69.31 (C-26), 65.27 (C-17), 57.06 (C-14), 53.46 (NCH2CH2Cl, ×4), 50.16 (C-9), 40.84 (C-12), 40.25 (NCH2CH2Cl, ×4), 39.56 (C-13), 38.45 (C-4), 38.09 (C-20), 37.22 (C-1), 36.93 (C-10), 33.23 (C-24), 32.39 (C-7), 32.18 (C-15), 31.75 (C-8), 31.05 (C-23), 30.74 (C-25), 27.35 (C-2), 20.81 (C-11), 19.57 (C-19), 19.10 (C-21), 17.15 (C-27), 16.59 (C-18). HRMS (ESI): m/z calculated for C49H67Cl4N2O5Na [M + Na]+: 925.3618, found: 925.3587.
2.1.3. Synthesis of (22 R,25R)-3β-acetoxy-5-en-furostan-26–4-(bis(2-chloroethyl)amino)benzoate (10)
Compound 6 (40 mg, 0.087 mmol) was used to synthesise compound 10, according to the method described to prepare compounds 8 and 9 from 7.
White powder, yield 50.8%. HPLC purity 97.32%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.94 (d, J = 9.0 Hz, 2H, Ph-H), 6.66 (d, J = 9.0 Hz, 2H, Ph-H), 5.37 (d, J = 4.9 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.30 (m, 1H, H-16), 4.17 (dd, J = 10.8, 5.7 Hz, 1H, H-26), 4.08 (dd, J = 10.8, 6.7 Hz, 1H, H-26), 3.80 (t, J = 7.1 Hz, 4H, NCH2CH2Cl, ×2), 3.65 (t, J = 7.1 Hz, 4H, NCH2CH2Cl, ×2), 3.32 (m, 1H, H-22), 2.03 (s, 3H, Ac-CH3), 1.03 (s, 3H, 19-CH3), 1.01 (d, J = 6.7 Hz, 3H, 21-CH3), 0.99 (d, J = 6.7 Hz, 3H, 27-CH3), 0.80 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 170.69 (Ac-COO), 166.69 (-COO), 149.67 (Ph-C), 139.84 (C-5), 131.92 (Ph-C, ×2), 122.53 (C-6), 119.48 (Ph-C), 110.96 (Ph-C, ×2), 90.36 (C-22), 83.35 (C-16), 74.03 (C-3), 69.31 (C-26), 65.27 (C-17), 57.04 (C-14), 53.46 (NCH2CH2Cl, ×2), 50.14 (C-9), 40.83 (C-12), 40.24 (NCH2CH2Cl, ×2), 39.54 (C-13), 38.24 (C-4), 38.09 (C-20), 37.14 (C-1), 36.86 (C-10), 33.24 (C-24), 32.38 (C-7), 32.14 (C-15), 31.72 (C-8), 31.05 (C-23), 30.74 (C-25), 27.89 (C-2), 21.58 (Ac-CH3), 20.79 (C-11), 19.49 (C-19), 19.10 (C-21), 17.15 (C-27), 16.58 (C-18). HRMS (ESI): m/z calculated for C40H58Cl2NO5 [M + H]+: 724.3506, found: 724.3474.
2.1.4. General procedure for synthesising 12a–12f
To a stirred solution of intermediate 11 (80 mg, 0.169 mmol) and N-Boc-protected amines (0.338 mmol) in CH2Cl2 (5 ml), 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethylaminium tetrafluoroborate (TBTU; 108.8 mg, 0.338 mmol) and N, N-diisopropylethylamine (DIPEA; 43.7 mg, 0.338 mmol) were added, following which the solution was stirred for 8 h at room temperature. After the reaction was completed, the solvent was evaporated at reduced pressure to get a residue, which was purified using preparative TLC (PE:EtOAc = 1:1, v/v), to obtain compounds 12a–12f.
2.1.4.1. (22 R,25R)-(boc-(2-aminoethyl))-3β-acetoxy-5-en-furostan 26-amide (12a)
White oil, yield 84.6%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 6.46 (br s, 1H, NH), 5.36 (d, J = 4.6 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.31 (m, 1H, H-16), 3.34 (m, 2H, NHCH2), 3.30 (m, 1H, H-22), 3.25 (m, 2H, NHCH2), 2.34 (m, 1H, H-25), 2.03 (s, 3H, Ac-CH3), 1.43 (s, 9H, Boc-CH3, ×3), 1.12 (d, J = 6.6 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.98 (d, J = 6.7 Hz, 3H, 27-CH3), 0.79 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 177.49 (-CONH), 170.69 (Ac-COO), 156.86 (Boc-COO), 139.87 (C-5), 122.44 (C-6), 90.82 (C-22), 83.54 (C-16), 79.70 (Boc-quart.-C), 74.02 (C-3), 64.85 (C-17), 57.03 (C-14), 50.12 (C-9), 40.91 (NHCH2), 40.82 (C-12), 40.34 (NHCH2), 39.52 (C-13), 38.36 (C-4), 38.23 (C-20), 37.13 (C-1), 36.86 (C-10), 32.59 (C-24), 32.47 (C-7), 32.12 (C-15), 31.70 (C-8), 31.00 (C-23), 28.54 (Boc-CH3, ×3), 27.88 (C-2), 21.57 (Ac-CH3), 20.77 (C-11), 19.48 (C-19), 18.86 (C-21), 18.06 (C-27), 16.62 (C-18). HRMS (ESI): m/z calculated for C36H58N2O6Na [M + Na]+: 637.4193, found: 637.4162.
2.1.4.2. (22 R,25R)-(boc-(2-aminopropyl))-3β-acetoxy-5-en-furostan 26-amide (12 b)
Yellowish oil, yield 80.9%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.36 (d, J = 4.7 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.30 (m, 1H, H-16), 3.30 (m, 4H, NHCH2, ×2), 3.13 (m, 1H, H-22), 2.36 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.43 (s, 9H, Boc-CH3, ×3), 1.13 (d, J = 6.6 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.97 (d, J = 6.8 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 177.31 (-CONH), 170.67 (Ac-COO), 156.59 (Boc-COO), 139.87 (C-5), 122.43 (C-6), 90.85 (C-22), 83.52 (C-16), 79.33 (Boc-quart.-C), 74.00 (C-3), 64.81 (C-17), 57.01 (C-14), 50.11 (C-9), 40.99 (NHCH2), 40.90 (NHCH2), 40.80 (C-12), 39.50 (C-13), 38.37 (C-4), 38.22 (C-20), 37.12 (C-1), 36.84 (C-10), 35.97 (C-25), 32.63 (C-24), 32.47 (C-7), 32.11 (C-15), 31.71 (C-8), 31.12 (C-23), 28.56 (Boc-CH3, ×3), 28.52 (NHCH2CH2), 27.87 (C-2), 21.56 (Ac-CH3), 20.76 (C-11), 19.48 (C-19), 18.87 (C-21), 18.23 (C-27), 16.55 (C-18). HRMS (ESI): m/z calculated for C37H60N2O6Na [M + Na]+: 651.4349, found: 651.4307.
2.1.4.3. (22 R,25R)-(3-(boc-amino)azetidinyl))-3β-acetoxy-5-en-furostan 26-amide (12c)
White oil, yield 83.5%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.37 (d, J = 4.4 Hz, 1H, H-6), 4.60 (m, 1H, H-3), 4.46 (m, 1H, NHCH), 4.37 (m, 1H, NCHH), 4.28 (m, 2H, H-16, NCHH), 3.94 (m, 1H, NCHH), 3.78 (m, 1H, NCHH), 3.28 (m, 1H, H-22), 2.34 (m, 1H, H-25), 2.03 (s, 3H, Ac-CH3), 1.44 (s, 9H, Boc-CH3, ×3), 1.07 (d, J = 6.6 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.98 (d, J = 6.5 Hz, 3H, 27-CH3), 0.79 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 176.61 (-CON), 170.68 (Ac-COO), 155.00 (Boc-COO), 139.85 (C-5), 122.49 (C-6), 90.27 (C-22), 83.37 (C-16), 80.42 (Boc-quart.-C), 74.03 (C-3), 65.21 (C-17), 57.02 (C-14), 55.07 (NCH2, ×2), 50.13 (C-9), 40.81 (C-12), 40.39 (NHCH), 39.52 (C-13), 38.92 (C-4), 38.25 (C-20), 37.13 (C-1), 36.85 (C-10), 35.63 (C-25), 32.42 (C-7), 32.13 (C-15), 31.72 (C-8), 31.48 (C-24), 31.34 (C-23), 28.46 (Boc-CH3, ×3), 27.88 (C-2), 21.57 (Ac-CH3), 20.78 (C-11), 19.48 (C-19), 19.07 (C-21), 17.60 (C-27), 16.62 (C-18). HRMS (ESI): m/z calculated for C37H59N2O6 [M + H]+: 627.4368, found: 627.4350.
2.1.4.4. (22 R,25R)-((S)-3-(boc-amino)pyrrolidyl)-3β-acetoxy-5-en-furostan 26-amide (12d)
White oil, yield 79.8%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.37 (d, J = 4.5 Hz, 1H, H-6), 4.60 (m, 1H, H-3), 4.58 (m, 1H, NHCH), 4.29 (m, 1H, H-16), 3.75 (m, 1H, NCHH), 3.60 (m, 1H, NCHH), 3.50 (m, 1H, NCHH), 3.33 (m, 1H, NCHH), 3.29 (m, 1H, H-22), 2.58 (m, 1H, H-25), 2.03 (s, 3H, Ac-CH3), 1.44 (s, 9H, Boc-CH3, ×3), 1.09 (d, J = 6.5 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.97 (d, J = 6.3 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 175.63 (-CON), 170.68 (Ac-COO), 155.34 (Boc-COO), 139.82 (C-5), 122.53 (C-6), 90.38 (C-22), 83.34 (C-16), 80.18 (Boc-quart.-C), 74.03 (C-3), 65.22 (C-17), 57.02 (C-14), 52.54 (NCH2), 50.96 (NHCH), 50.13 (C-9), 43.74 (NCH2), 40.80 (C-12), 39.52 (C-13), 38.26 (C-4), 38.23 (C-20), 38.19 (NCH2CH2), 37.74 (C-25), 37.13 (C-1), 36.85 (C-10), 32.39 (C-7), 32.13 (C-15), 31.71 (C-8), 31.52 (C-24), 31.48 (C-23), 28.49 (Boc-CH3, ×3), 27.88 (C-2), 21.57 (Ac-CH3), 20.78 (C-11), 19.48 (C-19), 19.08 (C-21), 17.79 (C-27), 16.57 (C-18). HRMS (ESI): m/z calculated for C38H60N2O6Na [M + Na]+: 663.4349, found: 663.4314.
2.1.4.5. (22 R,25R)-(boc-1-piperazinyl)-3β-acetoxy-5-en-furostan 26-amide (12e)
White powder, yield 86.0%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.36 (d, J = 4.0 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.27 (m, 1H, H-16), 3.46–3.63 (m, 4H, NCH2, ×2), 3.57–3.43 (m, 4H, NCH2, ×2), 3.29 (m, 1H, H-22), 2.75 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.46 (s, 9H, Boc-CH3, ×3), 1.09 (d, J = 6.8 Hz, 3H, 21-CH3), 1.02 (s, 3H, 19-CH3), 0.97 (d, J = 6.6 Hz, 3H, 27-CH3), 0.77 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 175.23 (-CON), 170.65 (Ac-COO), 154.75 (Boc-COO), 139.83 (C-5), 122.46 (C-6), 90.15 (C-22), 83.34 (C-16), 80.34 (Boc-quart.-C), 74.00 (C-3), 65.17 (C-17), 56.99 (C-14), 50.11 (C-9), 45.44 (NCH2, ×2), 41.66 (NCH2, ×2), 40.78 (C-12), 39.49 (C-13), 38.23 (C-4), 38.21 (C-20), 37.11 (C-1), 36.83 (C-10), 35.39 (C-25), 32.38 (C-7), 32.11 (C-15), 31.70 (C-8), 31.53 (C-24), 31.18 (C-23), 28.51 (Boc-CH3, ×3), 27.87 (C-2), 21.56 (Ac-CH3), 20.76 (C-11), 19.46 (C-19), 19.01 (C-21), 17.96 (C-27), 16.57 (C-18). HRMS (ESI): m/z calculated for C38H60N2O6Na [M + Na]+: 663.4349, found: 663.4312.
2.1.4.6. (22 R,25R)-(boc-1-homopiperazinyl)-3β-acetoxy-5-en-furostan 26-amide (12f)
White oil, yield 84.9%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.36 (d, J = 4.5 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.28 (m, 1H, H-16), 3.61 (m, 2H, NCH2), 3.53 (m, 2H, NCH2), 3.46 (m, 2H, NHCH2), 3.37 (m, 2H, NHCH2), 3.29 (t, J = 7.5 Hz, 1H, H-22), 2.70 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.45 (s, 9H, Boc-CH3, ×3), 1.10 (d, J = 6.8 Hz, 3H, 21-CH3), 1.02 (s, 3H, 19-CH3), 0.97 (d, J = 6.6 Hz, 3H, 27-CH3), 0.77 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 176.49 (-CON), 170.65 (Ac-COO), 155.21 (Boc-COO), 139.83 (C-5), 122.48 (C-6), 90.31 (C-22), 83.34 (C-16), 79.90 (Boc-quart.-C), 74.01 (C-3), 65.20 (C-17), 56.99 (C-14), 53.95 (NCH2), 50.12 (C-9), 49.16 (NCH2), 48.86 (NCH2), 48.68 (NCH2), 40.77 (C-12), 39.50 (C-13), 38.24 (C-4), 38.22 (C-20), 37.12 (C-1), 36.84 (C-10), 35.83 (C-25), 32.35 (C-7), 32.12 (C-15), 31.98 (C-8), 31.70 (C-24), 31.43 (C-23), 29.41 (NCH2CH2), 28.55 (Boc-CH3, ×3), 27.87 (C-2), 21.56 (Ac-CH3), 20.76 (C-11), 19.47 (C-19), 19.04 (C-21), 18.29 (C-27), 16.55 (C-18). HRMS (ESI): m/z calculated for C39H63N2O6 [M + H]+: 655.4681, found: 655.4672.
2.1.5. General procedure for synthesising 13a–13f
To a solution of compounds 12a–12f (0.12 mmol) in dry CH2Cl2 (6 ml), CF3COOH (0.5 ml) was added. The resulting mixture was stirred for 4 h at room temperature. The reaction solution was adjusted to a pH in the range of 7–9 using an aqueous saturated solution of NaHCO3 (40 ml), extracted with CH2Cl2 (5 ml × 3), following which the organic layer was separated, combined, and washed thrice with water (25 ml) and an aqueous saturated solution of NaCl (25 ml), dried over anhydrous Na2SO4, filtered, and evaporated under reduced pressure, to yield compounds 13a–13f.
2.1.5.1. (22 R,25R)-(2-aminoethyl)-3β-acetoxy-5-en-furostan 26-amide (13a)
White powder, yield 72.4%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.59 (s, 1H, NH), 5.36 (br s, 1H, H-6), 4.59 (m, 1H, H-3), 4.29 (m, 1H, H-16), 3.60 (br s, 1H, NHCHH), 3.42 (br s, 1H, NHCHH), 3.30 (m, 1H, H-22), 3.12 (br s, 2H, NH2CH2), 2.35 (m, 1H, H-25), 2.03 (s, 3H, Ac-CH3), 1.10 (d, J = 5.8 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.97 (d, J = 6.0 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 178.56 (-CONH), 170.66 (Ac-COO), 139.84 (C-5), 122.45 (C-6), 90.22 (C-22), 83.37 (C-16), 73.99 (C-3), 65.10 (C-17), 57.04 (C-14), 50.13 (C-9), 40.82 (C-12), 40.07 (NHCH2), 39.53 (C-13), 38.25 (C-4), 38.04 (C-20), 37.45 (NH2CH2), 37.14 (C-1), 36.84 (C-10), 36.10 (C-25), 32.40 (C-7), 32.13 (C-15), 31.70 (C-8), 31.56 (C-23), 31.19 (C-24), 27.89 (C-2), 21.57 (Ac-CH3), 20.80 (C-11), 19.51 (C-19), 19.04 (C-21), 17.93 (C-27), 16.67 (C-18). HRMS (ESI): m/z calculated for C31H51N2O4 [M + H]+: 515.3843, found: 515.3809.
2.1.5.2. (22 R,25R)-(2-aminopropyl)-3β-acetoxy-5-en-furostan 26-amide (13 b)
White powder, yield 73.0%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.36 (br s, 1H, H-6), 4.59 (m, 1H, H-3), 4.30 (m, 1H, H-16), 3.40 (m, 2H, NHCH2), 3.31 (m, 1H, H-22), 2.38 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.10 (d, J = 5.8 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.97 (d, J = 5.8 Hz, 3H, 27-CH3), 0.77 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 178.13 (-CONH), 170.67 (Ac-COO), 139.88 (C-5), 122.37 (C-6), 90.82 (C-22), 83.54 (C-16), 73.98 (C-3), 64.77 (C-17), 57.01 (C-14), 50.10 (C-9), 40.82 (C-12), 40.61 (NHCH2), 39.50 (NH2CH2), 39.47 (C-13), 38.32 (C-4), 38.22 (C-20), 37.11 (C-1), 36.83 (C-10), 36.24 (C-25), 32.51 (C-7), 32.44 (NHCH2CH2), 32.11 (C-15), 31.70 (C-8), 31.05 (C-24), 30.98 (C-23), 27.87 (C-2), 21.56 (Ac-CH3), 20.75 (C-11), 19.49 (C-19), 18.87 (C-21), 18.18 (C-27), 16.61 (C-18). HRMS (ESI): m/z calculated for C32H53N2O4 [M + H]+: 529.4000, found: 529.3973.
2.1.5.3. (22 R,25R)-(3-aminoazetidinyl)-3β-acetoxy-5-en-furostan 26-amide (13c)
White powder, yield 70.2%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.37 (d, J = 4.5 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.37 (m, 1H, NCHH), 4.29 (m, 1H, H-16), 4.24 (m, 1H, NCHH), 3.90 (m, 2H, NCHH, NH2CH), 3.74 (m, 1H, NCHH), 3.28 (m, 1H, H-22), 2.35 (m, 1H, H-25), 2.03 (s, 3H, Ac-CH3), 1.07 (d, J = 6.0 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.98 (d, J = 6.2 Hz, 3H, 27-CH3), 0.79 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 176.94 (-CON), 170.68 (Ac-COO), 139.86 (C-5), 122.49 (C-6), 90.27 (C-22), 83.35 (C-16), 74.02 (C-3), 65.24 (C-17), 57.36 (NCH2, ×2), 57.01 (C-14), 50.13 (C-9), 42.20 (NH2CH2), 40.81 (C-12), 39.52 (C-13), 38.24 (C-4), 38.18 (C-20), 37.14 (C-1), 36.86 (C-10), 35.67 (C-25), 32.41 (C-7), 32.13 (C-15), 31.73 (C-8), 31.42 (C-24), 31.17 (C-23), 27.89 (C-2), 21.58 (Ac-CH3), 20.78 (C-11), 19.49 (C-19), 19.09 (C-21), 17.59 (C-27), 16.60 (C-18). HRMS (ESI): m/z calculated for C32H51N2O4 [M + H]+: 527.3843, found: 527.3842.
2.1.5.4. (22 R,25R)-((S)-3-aminopyrrolidyl)-3β-acetoxy-5-en-furostan 26-amide (13d)
White powder, yield 73.7%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.37 (d, J = 4.7 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.27 (m, 1H, H-16), 3.76 (m, 1H, NCHH), 3.68 (m, 2H, NH2CH, NCHH), 3.50 (m, 1H, NCHH), 3.32 (m, 1H, NCHH), 3.29 (m, 1H, H-22), 2.57 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.12 (d, J = 7.1 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.98 (d, J = 6.8 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 175.73 (-CON), 170.68 (Ac-COO), 139.86 (C-5), 122.49 (C-6), 90.36 (C-22), 83.33 (C-16), 74.02 (C-3), 65.21 (C-17), 57.01 (C-14), 51.75 (NCH2), 50.12 (C-9), 49.79 (NH2CH), 44.80 (NCH2), 40.80 (C-12), 39.52 (C-13), 38.23 (C-4), 38.23 (C-20), 38.16 (NCH2CH2), 37.78 (C-25), 37.13 (C-1), 36.85 (C-10), 32.40 (C-7), 32.13 (C-15), 31.71 (C-8), 31.46 (C-24), 31.40 (C-23), 27.88 (C-2), 21.57 (Ac-CH3), 20.78 (C-11), 19.48 (C-19), 19.09 (C-21), 17.73 (C-27), 16.57 (C-18). HRMS (ESI): m/z calculated for C33H53N2O4 [M + H]+: 541.4000, found: 541.3998.
2.1.5.5. (22 R,25R)-(piperazinyl)-3β-acetoxy-5-en-furostan 26-amide (13e)
White powder, yield 76.1%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.35 (d, J = 4.7 Hz, 1H, H-6), 4.58 (m, 1H, H-3), 4.27 (m, 1H, H-16), 3.50–3.64 (m, 4H, NCH2, ×2), 3.29 (m, 1H, H-22), 2.87–2.84 (m, 4H, NHCH2, ×2), 2.73 (m, 1H, H-25), 2.01 (s, 3H, Ac-CH3), 1.08 (d, J = 6.9 Hz, 3H, 21-CH3), 1.02 (s, 3H, 19-CH3), 0.97 (d, J = 6.7 Hz, 3H, 27-CH3), 0.77 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 175.06 (-CON), 170.69 (Ac-COO), 139.82 (C-5), 122.46 (C-6), 90.19 (C-22), 83.31 (C-16), 74.02 (C-3), 65.15 (C-17), 56.98 (C-14), 50.10 (C-9), 45.96 (NCH2, ×2), 42.56 (NHCH2, ×2), 40.77 (C-12), 39.48 (C-13), 38.20 (C-4), 38.16 (C-20), 37.09 (C-1), 36.82 (C-10), 35.23 (C-25), 32.36 (C-7), 32.09 (C-15), 31.68 (C-8), 31.47 (C-24), 31.22 (C-23), 27.85 (C-2), 21.55 (Ac-CH3), 20.74 (C-11), 19.45 (C-19), 19.00 (C-21), 17.95 (C-27), 16.55 (C-18). HRMS (ESI): m/z calculated for C33H53N2O4 [M + H]+: 541.4000, found: 541.3962.
2.1.5.6. (22 R,25R)-(1-homopiperazinyl)-3β-acetoxy-5-en-furostan 26-amide (13f)
White powder, yield 74.4%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 5.36 (d, J = 4.0 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.28 (m, 1H, H-16), 3.65 (m, 2H, NCH2), 3.58 (m, 2H, NCH2), 3.29 (t, J = 8.0 Hz, 1H, H-22), 2.97 (m, 2H, NHCH2), 2.93 (m, 1H, NHCHH), 2.87 (m, 1H, NHCHH), 2.73 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.11 (d, J = 6.8 Hz, 3H, 21-CH3), 1.02 (s, 3H, 19-CH3), 0.97 (d, J = 6.6 Hz, 3H, 27-CH3), 0.77 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 176.37 (-CON), 170.65 (Ac-COO), 139.84 (C-5), 122.47 (C-6), 90.34 (C-22), 83.33 (C-16), 74.01 (C-3), 65.20 (C-17), 57.00 (C-14), 50.60 (NCH2), 50.12 (C-9), 49.14 (NHCH2), 48.13 (NCH2), 46.81 (NHCH2), 40.77 (C-12), 39.50 (C-13), 38.22 (C-4), 38.2 (C-20), 37.11 (C-1), 36.84 (C-10), 35.81 (C-25), 32.35 (C-7), 32.11 (C-15), 31.85 (C-8), 31.70 (C-24), 31.46 (C-23), 29.29 (NCH2CH2), 27.87 (C-2), 21.55 (Ac-CH3), 20.76 (C-11), 19.46 (C-19), 19.05 (C-21), 18.41 (C-27), 16.55 (C-18). HRMS (ESI): m/z calculated for C34H55N2O4 [M + H]+: 555.4156, found: 555.4181.
2.1.6. General procedure for synthesising 14a–14f
To a solution of 13a–13f (0.07 mmol) in dry CH2Cl2 (5 ml), benzoic acid mustard (21 mg, 0.08 mmol), 1-ethyl-3–(3-dimethylaminopropyl) carbodiimide hydrochloride (EDCI; 20 mg, 0.105 mmol), and 4-dimethylaminopyridine (DMAP; catalytic amount) were added. After being stirred for 24 h at room temperature, the mixture was concentrated in vacuo. The product was purified using preparative TLC, with a petroleum ether/ethyl acetate (1:2, v/v) system, to achieve the compounds 14a–14f.
2.1.6.1. (22 R,25R)-(2-aminoethyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-acetoxy-5-en-furostan-26-amide (14a)
White powder, yield 88.0%. HPLC purity 95.16%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.75 (d, J = 8.8 Hz, 2H, Ph-H), 7.52 (s, 1H, NH), 6.95 (s, 1H, NH), 6.65 (d, J = 8.8 Hz, 2H, Ph-H), 5.35 (d, J = 3.8 Hz, 1H, H-6), 4.58 (m, 1H, H-3), 4.27 (m, 1H, H-16), 3.76 (t, J = 6.8 Hz, 4H, NCH2CH2Cl, ×2), 3.63 (t, J = 6.8 Hz, 4H, NCH2CH2Cl, ×2), 3.52 (m, 2H, NHCH2), 3.49 (m, 2H, NHCH2), 3.24 (t, J = 7.9 Hz, 1H, H-22), 2.40 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.10 (d, J = 6.6 Hz, 3H, 21-CH3), 1.02 (s, 3H, 19-CH3), 0.88 (d, J = 6.6 Hz, 3H, 27-CH3), 0.74 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 178.96 (-CONH), 170.65 (Ac-COO), 167.60 (-CONH), 148.62 (Ph-C), 139.84 (C-5), 129.26 (Ph-C, ×2), 122.69 (C-6), 122.35 (Ph-C), 111.17 (Ph-C, ×2), 90.89 (C-22), 83.52 (C-16), 73.97 (C-3), 64.71 (C-17), 56.97 (C-14), 53.45 (NCH2CH2Cl, ×2), 50.06 (C-9), 40.76 (C-12), 40.64 (NHCH2), 40.30 (NCH2CH2Cl, ×2), 39.42 (C-13), 39.34 (NHCH2), 38.31 (C-4), 38.19 (C-20), 37.08 (C-1), 36.80 (C-10), 36.17 (C-25), 32.48 (C-7), 32.06 (C-15), 31.69 (C-8), 31.66 (C-23), 30.87 (C-24), 27.84 (C-2), 21.53 (Ac-CH3), 20.71 (C-11), 19.45 (C-19), 18.73 (C-21), 18.04 (C-27), 16.54 (C-18). HRMS (ESI): m/z calculated for C42H62Cl2N3O5 [M + H]+: 758.4061, found: 758.4044.
2.1.6.2. (22 R,25R)-(3-aminopropyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-acetoxy-5-en- furostan-26-amide (14 b)
White oil, yield 85.6%. HPLC purity 98.17%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.84 (d, J = 8.7 Hz, 2H, Ph-H), 6.69 (d, J = 8.7 Hz, 2H, Ph-H), 5.36 (d, J = 4.8 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.32 (m, 1H, H-16), 3.78 (t, J = 7.0 Hz, 4H, NCH2CH2Cl, ×2), 3.64 (t, J = 7.0 Hz, 4H, NCH2CH2Cl, ×2), 3.45 (m, 2H, NHCH2), 3.34 (m, 2H, NHCH2), 3.32 (m, 1H, H-22), 2.45 (m, 1H, H-25), 2.03 (s, 3H, Ac-CH3), 1.15 (d, J = 6.9 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.97 (d, J = 6.8 Hz, 3H, 27-CH3), 0.77 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 178.04 (-CONH), 170.68 (Ac-COO), 167.21 (-CONH), 148.61 (Ph-C), 139.90 (C-5), 129.35 (Ph-C, ×2), 123.06 (Ph-C), 122.37 (C-6), 111.29 (Ph-C, ×2), 91.00 (C-22), 83.61 (C-16), 73.99 (C-3), 64.71 (C-17), 57.01 (C-14), 53.51 (NCH2CH2Cl, ×2), 50.10 (C-9), 40.87 (NHCH2), 40.82 (C-12), 40.31 (NCH2CH2Cl, ×2), 39.47 (C-13), 38.45 (C-4), 38.22 (C-20), 37.12 (C-1), 36.84 (C-10), 35.80 (NHCH2), 35.79 (C-25), 32.84 (NHCH2CH2), 32.52 (C-7), 32.10 (C-15), 31.71 (C-8), 31.02 (C-24), 30.15 (C-23), 27.86 (C-2), 21.56 (Ac-CH3), 20.75 (C-11), 19.49 (C-19), 18.83 (C-21), 18.28 (C-27), 16.57 (C-18). HRMS (ESI): m/z calculated for C43H64Cl2N3O5 [M + H]+: 772.4218, found: 772.4202.
2.1.6.3. (22 R,25R)-(3-aminoazetidinyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-acetoxy-5- en-furostan-26-amide (14c)
White oil, yield 81.4%. HPLC purity 95.12%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.77 (d, J = 8.5 Hz, 2H, Ph-H), 6.67 (d, J = 8.5 Hz, 2H, Ph-H), 5.35 (d, J = 4.0 Hz, 1H, H-6), 4.83 (m, 1H, NHCH), 4.58 (m, 1H, H-3), 4.54 (m, 1H, NCHH), 4.37 (m, 1H, NCHH), 4.28 (m, 1H, H-16), 4.05 (m, 1H, NCHH), 3.95 (m, 1H, NCHH), 3.79 (t, J = 6.9 Hz, 4H, NCH2CH2Cl, ×2), 3.65 (t, J = 6.9 Hz, 4H, NCH2CH2Cl, ×2), 3.28 (m, 1H, H-22), 2.37 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.09 (d, J = 6.9 Hz, 3H, 21-CH3), 1.04 (s, 3H, 19-CH3), 0.98 (d, J = 6.9 Hz, 3H, 27-CH3), 0.76 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 176.91 (-CON), 170.68 (Ac-COO), 167.02 (-CONH), 149.06 (Ph-C), 139.82 (C-5), 129.42 (Ph-C, ×2), 122.50 (C-6), 122.13 (Ph-C), 111.23 (Ph-C, ×2), 90.24 (C-22), 83.41 (C-16), 74.04 (C-3), 65.17 (C-17), 57.02 (C-14), 54.40 (NCH2, ×2), 53.42 (NCH2CH2Cl, ×2), 50.13 (C-9), 40.80 (NHCH), 40.76 (C-12), 40.26 (NCH2CH2Cl, ×2), 39.52 (C-13), 38.23 (C-4), 38.18 (C-20), 37.12 (C-1), 36.85 (C-10), 35.55 (C-25), 32.40 (C-7), 32.12 (C-15), 31.72 (C-8), 31.46 (C-24), 31.11 (C-23), 27.88 (C-2), 21.56 (Ac-CH3), 20.78 (C-11), 19.47 (C-19), 19.05 (C-21), 17.53 (C-27), 16.61 (C-18). HRMS (ESI): m/z calculated for C43H62Cl2N3O5 [M + H]+: 770.4061, found: 770.4100.
2.1.6.4. (22 R,25R)-((S)-3-aminopyrrolidyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-acetoxy-5-en-furostan-26-amide (14d)
White powder, yield 82.4%. HPLC purity 95.21%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.69 (d, J = 7.8 Hz, 2H, Ph-H), 6.65 (d, J = 7.8 Hz, 2H, Ph-H), 6.30 (s, 1H, NH), 5.34 (d, J = 4.3 Hz, 1H, H-6), 4.68 (m, 1H, NH2CH), 4.58 (m, 1H, H-3), 4.28 (m, 1H, H-16), 3.94 (m, 1H, NCHH), 3.77 (t, J = 6.9 Hz, 4H, NCH2CH2Cl, ×2), 3.70 (m, 1H, NCHH), 3.63 (t, J = 6.9 Hz, 4H, NCH2CH2Cl, ×2), 3.57 (m, 1H, NCHH), 3.47 (m, 1H, NCHH), 3.28 (m, 1H, H-22), 2.58 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.10 (d, J = 7.0 Hz, 3H, 21-CH3), 1.02 (s, 3H, 19-CH3), 0.97 (d, J = 6.8 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 175.83 (-CON), 170.66 (Ac-COO), 167.07 (-CONH), 148.93 (Ph-C), 139.79 (C-5), 129.22 (Ph-C, ×2), 124.32 (Ph-C), 122.47 (C-6), 111.23 (Ph-C, ×2), 90.34 (C-22), 83.34 (C-16), 74.00 (C-3), 65.14 (C-17), 56.97 (C-14), 53.40 (NCH2CH2Cl, ×2), 52.63 (NCH2), 50.22 (NH2CH), 50.09 (C-9), 44.10 (NCH2), 40.76 (C-12), 40.26 (NCH2CH2Cl, ×2), 39.49 (C-13), 38.24 (C-4), 38.20 (C-20), 38.10 (NCH2CH2), 37.62 (C-25), 37.09 (C-1), 36.81 (C-10), 32.34 (C-7), 32.09 (C-15), 31.66 (C-8), 31.57 (C-24), 31.40 (C-23), 27.85 (C-2), 21.55 (Ac-CH3), 20.74 (C-11), 19.45 (C-19), 19.03 (C-21), 17.75 (C-27), 16.58 (C-18). HRMS (ESI): m/z calculated for C44H64Cl2N3O5 [M + H]+: 784.4218, found: 784.4247.
2.1.6.5. (22 R,25R)-(1-piperazinyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-acetoxy-5-en-furostan-26-amide (14e)
White oil, yield 90.6%. HPLC purity 99.55%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.37 (d, J = 8.3 Hz, 2H, Ph-H), 6.68 (d, J = 8.3 Hz, 2H, Ph-H), 5.36 (d, J = 4.3 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.27 (m, 1H, H-16), 3.77 (t, J = 7.0 Hz, 4H, NCH2CH2Cl, ×2), 3.66 (m, 4H, NCH2, ×2), 3.64 (t, J = 7.0 Hz, 4H, NCH2CH2Cl, ×2), 3.57 (m, 4H, NCH2, ×2), 3.30 (m, 1H, H-22), 2.78 (m, 1H, H-25), 2.02 (s, 3H, Ac-CH3), 1.11 (d, J = 6.7 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.98 (d, J = 6.9 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 175.32 (-CON), 170.87 (-CON), 170.65 (Ac-COO), 147.74 (Ph-C), 139.86 (C-5), 129.96 (Ph-C, ×2), 123.65 (Ph-C), 122.45 (C-6), 111.38 (Ph-C, ×2), 90.14 (C-22), 83.36 (C-16), 73.99 (C-3), 65.14 (C-17), 56.99 (C-14), 53.47 (NCH2CH2Cl, ×2), 50.11 (C-9), 45.61 (NCH2, ×2), 42.01 (NCH2, ×2), 40.79 (C-12), 40.28 (NCH2CH2Cl, ×2), 39.48 (C-13), 38.25 (C-4), 38.22 (C-20), 37.11 (C-1), 36.83 (C-10), 35.37 (C-25), 32.4 (C-7), 32.12 (C-15), 31.71 (C-8), 31.54 (C-24), 31.14 (C-23), 27.86 (C-2), 21.56 (Ac-CH3), 20.76 (C-11), 19.48 (C-19), 19.00 (C-21), 17.93 (C-27), 16.59 (C-18). HRMS (ESI): m/z calculated for C44H64Cl2N3O5 [M + H]+: 784.4218, found: 784.4257.
2.1.6.6. (22 R,25R)-(4–(1-homopiperazinyl)-4-(bis(2-chloroethyl)amino)benzamido)- 3β-acetoxy-5-en-furostan-26-amide (14f)
White oil, yield 89.1%. HPLC purity 95.91%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.33 (br s, 2H, Ph-H), 6.69 (br s, 2H, Ph-H), 5.37 (d, J = 3.8 Hz, 1H, H-6), 4.59 (m, 1H, H-3), 4.28 (m, 1H, H-16), 3.76 (t, J = 6.8 Hz, 4H, NCH2CH2Cl, ×2), 3.65 (t, J = 6.8 Hz, 4H, NCH2CH2Cl, ×2), 3.63–3.52 (m, 8H, NCH2, ×4), 3.29 (t, J = 8.0 Hz, 1H, H-22), 2.74 (m, 1H, H-25), 2.03 (s, 3H, Ac-CH3), 1.11 (d, J = 6.8 Hz, 3H, 21-CH3), 1.03 (s, 3H, 19-CH3), 0.97 (d, J = 6.6 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 176.48 (-CON), 172.03 (-CON), 170.68 (Ac-COO), 147.18 (Ph-C), 139.87 (C-5), 129.66 (Ph-C, ×2), 122.48 (C-6), 121.17 (Ph-C), 111.74 (Ph-C, ×2), 90.29 (C-22), 83.37 (C-16), 74.02 (C-3), 65.18 (C-17), 57.00 (C-14), 53.73 (NCH2CH2Cl, ×2), 53.68 (NCH2), 50.12 (C-9), 49.32 (NCH2), 48.47 (NCH2), 48.15 (NCH2), 40.79 (C-12), 40.31 (NCH2CH2Cl, ×2), 39.51 (C-13), 38.26 (C-4), 38.23 (C-20), 37.13 (C-1), 36.85 (C-10), 35.89 (C-25), 32.37 (C-7), 32.14 (C-15), 31.90 (C-8), 31.72 (C-24), 31.45 (C-23), 29.20 (NCH2CH2), 27.88 (C-2), 21.57 (Ac-CH3), 20.77 (C-11), 19.49 (C-19), 19.07 (C-21), 18.76 (C-27), 16.59 (C-18). HRMS (ESI): m/z calculated for C45H66Cl2N3O5 [M + H]+: 798.4374, found: 798.4410.
2.1.7. General procedure for synthesising 15a–15f
A solution of compounds 14a–14f in aqueous NaOH (4 N) in methanol:tetrahydrofuran (1:1.5, v/v) was stirred for 6 h at room temperature. After concentration in vacuo, 30 ml of water was added to it and it was extracted with CH2Cl2 (6 ml × 3). The CH2Cl2 extract was washed with aqueous saturated NaCl (25 ml) and dried over anhydrous Na2SO4, following which the solids were removed by means of filtration. The reaction mixture was evaporated under reduced pressure and purified using preparative TLC with a CH2Cl2/MeOH (10:1, v/v) system, to obtain the compounds 15a–15f.
2.1.7.1. (22 R,25R)-(2-aminoethyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-hydroxy-5-en-furostan-26-amide (15a)
White powder, yield 92.3%. HPLC purity 95.05%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.74 (d, J = 9.0 Hz, 2H, Ph-H), 7.45 (s, 1H, NH), 6.86 (s, 1H, NH), 6.66 (d, J = 9.0 Hz, 2H, Ph-H), 5.33 (d, J = 4.7 Hz, 1H, H-6), 4.28 (m, 1H, H-16), 3.77 (t, J = 7.1 Hz, 4H, NCH2CH2Cl, ×2), 3.64 (t, J = 7.1 Hz, 4H, NCH2CH2Cl, ×2), 3.53 (m, 2H, NHCH2), 3.52 (m, 1H, H-3), 3.49 (m, 2H, NHCH2), 3.25 (t, J = 7.9 Hz, 1H, H-22), 2.40 (m, 1H, H-25), 1.10 (d, J = 7.0 Hz, 3H, 21-CH3), 1.01 (s, 3H, 19-CH3), 0.89 (d, J = 6.6 Hz, 3H, 27-CH3), 0.75 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 178.99 (-CONH), 167.64 (-CONH), 148.63 (Ph-C), 141.00 (C-5), 129.26 (Ph-C, ×2), 122.71 (Ph-C), 121.41 (C-6), 111.19 (Ph-C, ×2), 90.92 (C-22), 83.57 (C-16), 71.82 (C-3), 64.71 (C-17), 57.07 (C-14), 53.46 (NCH2CH2Cl, ×2), 50.18 (C-9), 42.38 (C-4), 40.79 (C-12), 40.64 (NHCH2), 40.32 (NCH2CH2Cl, ×2), 39.49 (C-13), 39.31 (NHCH2), 38.32 (C-20), 37.37 (C-1), 36.71 (C-10), 36.05 (C-25), 32.52 (C-7), 32.10 (C-15), 31.72 (C-8), 31.06 (C-23), 30.83 (C-24), 27.33 (C-2), 20.79 (C-11), 19.57 (C-19), 18.75 (C-21), 18.03 (C-27), 16.57 (C-18). HRMS (ESI): m/z calculated for C40H60Cl2N3O4 [M + H]+: 716.3955, found: 716.3978.
2.1.7.2. (22 R,25R)-(3-aminopropyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-hydroxy-5-en- furostan-26-amide (15b)
White powder, yield 89.9%. HPLC purity 97.56%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.84 (d, J = 6.7 Hz, 2H, Ph-H), 6.70 (d, J = 6.7 Hz, 2H, Ph-H), 5.34 (br s, 1H, H-6), 4.32 (br s, 1H, H-16), 3.78 (t, J = 6.7 Hz, 4H, NCH2CH2Cl, ×2), 3.64 (t, J = 6.7 Hz, 4H, NCH2CH2Cl, ×2), 3.51 (m, 1H, H-3), 3.45 (m, 2H, NHCH2), 3.35–3.33 (m, 3H, NHCH2, H-22), 2.45 (m, 1H, H-25), 1.16 (d, J = 6.0 Hz, 3H, 21-CH3), 1.02 (s, 3H, 19-CH3), 0.97 (d, J = 5.5 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 178.10 (-CONH), 167.29 (-CONH), 148.64 (Ph-C), 141.01 (C-5), 129.38 (Ph-C, ×2), 122.98 (Ph-C), 121.44 (C-6), 111.31 (Ph-C, ×2), 91.00 (C-22), 83.64 (C-16), 71.86 (C-3), 64.72 (C-17), 57.10 (C-14), 53.51 (NCH2CH2Cl, ×2), 50.20 (C-9), 42.40 (C-4), 40.87 (NHCH2), 40.84 (C-12), 40.32 (NCH2CH2Cl, ×2), 39.54 (C-13), 38.44 (C-20), 37.39 (C-1), 36.76 (C-10), 35.89 (NHCH2), 35.86 (C-25), 32.81 (NHCH2CH2), 32.54 (C-7), 32.13 (C-15), 31.75 (C-8), 31.01 (C-24), 30.16 (C-23), 27.35 (C-2), 20.81 (C-11), 19.59 (C-19), 18.84 (C-21), 18.26 (C-27), 16.59 (C-18). HRMS (ESI): m/z calculated for C41H62Cl2N3O4 [M + H]+: 730.4112, found: 730.4132.
2.1.7.3. (22 R,25R)-(3-aminoazetidinyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-hydroxy-5-en-furostan-26-amide (15c)
White powder, yield 91.0%. HPLC purity 95.57%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.81 (br s, 2H, Ph-H), 6.68 (br s, 2H, Ph-H), 5.33 (d, J = 4.5 Hz, 1H, H-6), 4.88 (m, 1H, NHCH), 4.56 (m, 1H, NCHH), 4.46 (m, 1H, NCHH), 4.28 (m, 1H, H-16), 4.14 (m, 1H, NCHH), 4.08 (m, 1H, NCHH), 3.79 (t, J = 6.7 Hz, 4H, NCH2CH2Cl, ×2), 3.64 (t, J = 6.7 Hz, 4H, NCH2CH2Cl, ×2), 3.51 (m, 1H, H-3), 3.28 (m, 1H, H-22), 2.38 (m, 1H, H-25), 1.10 (d, J = 5.2 Hz, 3H, 21-CH3), 1.01 (s, 3H, 19-CH3), 0.98 (d, J = 5.0 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 177.01 (-CON), 167.07 (-CONH), 149.08 (Ph-C), 140.91 (C-5), 129.51 (Ph-C, ×2), 122.06 (Ph-C), 121.56 (C-6), 111.31 (Ph-C, ×2), 90.19 (C-22), 83.41 (C-16), 71.86 (C-3), 65.15 (C-17), 57.09 (C-14), 53.92 (NCH2, ×2), 53.43 (NCH2CH2Cl, ×2), 50.22 (C-9), 42.40 (C-4), 40.80 (C-12), 40.31 (NCH2CH2Cl, ×2), 39.82 (NHCH), 39.57 (C-13), 38.22 (C-20), 37.38 (C-1), 36.74 (C-10), 35.56 (C-25), 32.39 (C-7), 32.12 (C-15), 31.75 (C-8), 31.73 (C-24), 31.38 (C-23), 27.35 (C-2), 20.84 (C-11), 19.57 (C-19), 19.05 (C-21), 17.57 (C-27), 16.65 (C-18). HRMS (ESI): m/z calculated for C41H60Cl2N3O4 [M + H]+: 728.3955, found: 728.3965.
2.1.7.4. (22 R,25R)-((S)-3-aminopyrrolidyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-hydroxy-5-en-furostan-26-amide (15d)
White powder, yield 90.7%. HPLC purity 97.31%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.69 (d, J = 8.8 Hz, 2H, Ph-H), 6.66 (d, J = 8.8 Hz, 2H, Ph-H), 6.19 (s, 1H, NH), 5.33 (d, J = 4.7 Hz, 1H, H-6), 4.66 (m, 1H, NHCH), 4.28 (m, 1H, H-16), 3.83 (m, 1H, NCHH), 3.78 (t, J = 6.7 Hz, 4H, NCH2CH2Cl, ×2), 3.69 (m, 1H, NCHH), 3.64 (t, J = 6.7 Hz, 4H, NCH2CH2Cl, ×2), 3.57 (m, 1H, NCHH), 3.51 (m, 1H, H-3), 3.46 (m, 1H, NCHH), 3.29 (m, 1H, H-22), 2.60 (m, 1H, H-25), 1.07 (d, J = 6.9 Hz, 3H, 21-CH3), 1.01 (s, 3H, 19-CH3), 0.97 (d, J = 6.7 Hz, 3H, 27-CH3), 0.78 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 175.85 (-CON), 167.06 (-CONH), 148.96 (Ph-C), 140.95 (C-5), 129.19 (Ph-C, ×2), 122.45 (Ph-C), 121.54 (C-6), 111.26 (Ph-C, ×2), 90.35 (C-22), 83.37 (C-16), 71.84 (C-3), 65.19 (C-17), 57.08 (C-14), 53.42 (NCH2CH2Cl, ×2), 52.62 (NCH2), 51.03 (NHCH), 50.21 (C-9), 44.01 (NCH2), 42.40 (C-4), 40.79 (C-12), 40.27 (NCH2CH2Cl, ×2), 39.57 (C-13), 38.24 (C-20), 38.15 (NCH2CH2), 37.39 (C-1), 36.75 (C-10), 35.88 (C-25), 32.38 (C-7), 32.13 (C-15), 31.73 (C-8), 31.57 (C-24), 31.43 (C-23), 27.34 (C-2), 20.82 (C-11), 19.57 (C-19), 19.07 (C-21), 17.79 (C-27), 16.61 (C-18). HRMS (ESI): m/z calculated for C42H62Cl2N3O4 [M + H]+: 742.4112, found: 742.4132.
2.1.7.5. (22 R,25R)-(1-piperazinyl-4-(bis(2-chloroethyl)amino)benzamido)-3β-hydroxy-5-en-furostan-26-amide (15e)
White powder, yield 93.8%. HPLC purity 98.33%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.36 (d, J = 8.8 Hz, 2H, Ph-H), 6.67 (d, J = 8.8 Hz, 2H, Ph-H), 5.33 (d, J = 4.0 Hz, 1H, H-6), 4.28 (m, 1H, H-16), 3.77 (t, J = 7.0 Hz, 4H, NCH2CH2Cl, ×2), 3.71–3.51 (m, 12H, NCH2CH2Cl, ×2, NCH2, ×4), 3.50 (m, 1H, H-3), 3.29 (m, 1H, H-22), 2.77 (m, 1H, H-25), 1.11 (d, J = 6.7 Hz, 3H, 21-CH3), 1.01 (s, 3H, 19-CH3), 0.97 (d, J = 6.7 Hz, 3H, 27-CH3), 0.77 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 175.32 (-CON), 170.88 (-CON), 147.75 (Ph-C), 141.00 (C-5), 129.94 (Ph-C, ×2), 123.59 (Ph-C), 121.47 (C-6), 111.33 (Ph-C, ×2), 90.13 (C-22), 83.36 (C-16), 71.79 (C-3), 65.14 (C-17), 57.06 (C-14), 53.43 (NCH2CH2Cl, ×2), 50.19 (C-9), 45.59 (NCH2, ×2), 42.38 (C-4), 42.01 (NCH2, ×2), 40.79 (C-12), 40.28 (NCH2CH2Cl, ×2), 39.53 (C-13), 38.23 (C-20), 37.37 (C-1), 36.74 (C-10), 35.36 (C-25), 32.41 (C-7), 32.13 (C-15), 31.74 (C-8), 31.52 (C-24), 31.13 (C-23), 27.33 (C-2), 20.80 (C-11), 19.56 (C-19), 19.00 (C-21), 17.92 (C-27), 16.59 (C-18). HRMS (ESI): m/z calculated for C42H62Cl2N3O4 [M + H]+: 742.4112, found: 742.4124.
2.1.7.6. (22 R,25R)-(4–(1-homopiperazinyl)-4-(bis(2-chloroethyl)amino)benzamido)-3β-hydroxy-5-en-furostan-26-amide (15f)
White powder, yield 94.5%. HPLC purity 98.57%. 1H NMR (600 MHz, Chloroform-d, TMS) δH: 7.32 (d, J = 8.7 Hz, 2H, Ph-H), 6.64 (d, J = 8.7 Hz, 2H, Ph-H), 5.33 (d, J = 4.7 Hz, 1H, H-6), 4.28 (m, 1H, H-16), 3.75 (t, J = 7.0 Hz, 4H, NCH2CH2Cl, ×2), 3.69–3.50 (m, 12H, NCH2CH2Cl, ×2, NCH2, ×4), 3.49 (m, 1H, H-3), 3.29 (m, 1H, H-22), 2.74 (m, 1H, H-25), 1.11 (d, J = 6.8 Hz, 3H, 21-CH3), 1.01 (s, 3H, 19-CH3), 0.97 (d, J = 6.5 Hz, 3H, 27-CH3), 0.77 (s, 3H, 18-CH3). 13 C NMR (150 MHz, Chloroform-d, TMS) δC: 176.46 (-CON), 171.97 (-CON), 147.31 (Ph-C), 140.99 (C-5), 130.00 (Ph-C, ×2), 124.85 (Ph-C), 121.49 (C-6), 111.30 (Ph-C, ×2), 90.27 (C-22), 83.35 (C-16), 71.81 (C-3), 65.19 (C-17), 57.06 (C-14), 53.56 (NCH2), 53.46 (NCH2CH2Cl, ×2), 50.20 (C-9), 50.03 (NCH2), 48.43 (NCH2), 47.98 (NCH2), 42.39 (C-4), 40.78 (C-12), 40.32 (NCH2CH2Cl, ×2), 39.54 (C-13), 38.22 (C-20), 37.38 (C-1), 36.75 (C-10), 35.86 (C-25), 32.36 (C-7), 32.13 (C-15), 31.87 (C-8), 31.74 (C-24), 31.42 (C-23), 29.44 (NCH2CH2), 27.33 (C-2), 20.81 (C-11), 19.56 (C-19), 19.05 (C-21), 18.72 (C-27), 16.57 (C-18). HRMS (ESI): m/z calculated for C43H64Cl2N3O4 [M + H]+: 756.4268, found: 756.4281.
2.2. Cells and cell culture conditions
Human hepatoma carcinoma HepG2, breast carcinoma MCF-7, cervical cancer HeLa, and gastric epithelial GES-1 cells were acquired from America Type Culture Collection (USA). HepG2 and GES-1 cells were cultured in Dulbecco’s Modified Eagle’s Medium supplemented with 10% foetal bovine serum (FBS). MCF-7 cells were incubated in Minimum Essential Medium supplemented with 10% FBS. HeLa cells were cultured in Roswell Park Memorial Institute medium supplemented with 10% FBS. All cells were maintained in a humidified incubator with 5% CO2, at 37 °C.
2.3. MTT assay
The inhibitory activities of the target hybrids (8–10, 14a–14f, and 15a–15f) were tested using the MTT method, as described previouslyCitation23. HepG2, MCF-7, HeLa, and GES-1 cells were seeded into 96-well plates, at a density of 5 × 103/well. After 24 h, the cells were treated with serially diluted concentrations of the test compounds. After treatment for 48 h, 10 µL of MTT (5 mg/mL) was added into each well and incubation was continued for another 4 h. The medium was then removed and 100 µL of dimethyl sulfoxide (DMSO) was added into the wells, to dissolve the formazan crystals. Finally, the absorbance was read on a microplate reader (iMark™, Bio-Rad, USA), at the wavelength of 492 nm.
2.4. Flow cytometric analysis of cell cycle distribution
HepG2 cells were seeded into 6-well plates and allowed to grow for a period of 12 h. After incubation with different concentrations of 14f (0, 5, 10, and 20 µM) for 24 h, the cells were trypsinized, washed with phosphate-buffered saline (PBS), and fixed in 1.5 ml of 75% ethanol for the night, at 4 °C, following which RNase and propidium iodide (PI) (Multi Sciences Biotech Co. Ltd.) were added to the cells. The cell cycle distribution and data processing were analysed using a flow cytometer (FACSCalibur™, BD Biosciences, USA).
2.5. Assessment of changes in cell morphology
First, HepG2 cells were plated into 6-well plates, at a density of 1 × 106 cells/well. Thereafter, 14f (0, 5, 10, and 20 µM) was added to the cells and they were incubated with it for 48 h; 0.1% DMSO was used as a vehicle control. The cellular morphology was then observed and photographed using a light microscope (Olympus, 1 × 51, Japan). Next, HepG2 cells were incubated with 14f (0, 5, 10, and 20 µM) for 48 h, after which the cells were collected and stained with a mixture of Hoechst 33342 and PI in buffer solution, for 20 min at room temperature. Finally, the cell samples were studied and photographed under a fluorescence microscope (Obeserve.A1, Zeiss, Germany).
2.6. Assessment of apoptosis using flow cytometry
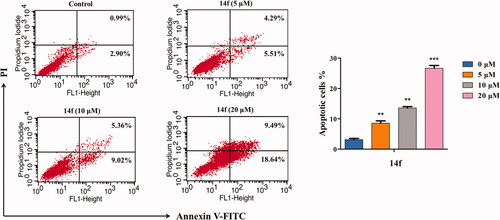
Briefly, HepG2 cells were incubated in 6-well plates for 24 h, and then treated with 14f (0, 5, 10, and 20 µM) for 48 h. Following that, the cells were washed once with pre-cold PBS, re-suspended in 300 µL of buffer solution, and stained with 10 µL of Annexin V-FITC and 5 µL of PI staining solution, under the conditions of 37 °C and absence of any light source, for 10 min. Finally, the samples were immediately measured using a flow cytometer. The percentages of apoptotic cells were determined on the CellQuest™ software (BD Biosciences).
2.7. Evaluation of MMP (δψm)
HepG2 cells were plated into 6-well plates for 12 h. Following treatment with varying concentrations of 14f for 48 h, the cells were incubated with JC-1 dyeing working solution for 0.5 h at room temperature, then washed with staining buffer 3 times and re-suspended in 300 µL of staining buffer. The cells were finally assessed and analysed using a flow cytometer (FACSCalibur™).
2.8. Quantitative real-time polymerase chain reaction (qRT-PCR) assay
Total RNA was extracted from HepG2 cells using a Trizol™ reagent (Invitrogen, USA) and qRT-PCR was conducted on it, as documented beforeCitation24. The purity and concentration of total RNA were checked using a spectrophotometer, in terms of A260 nm/A280 nm. A PrimeScript™ RT reagent Kit (Takara Bio, Japan) was applied to synthesise cDNA, according to the manufacturer’s instructions. A Detection System (Q5, ABI, USA) was used to carry out the qRT-PCR, using a TB Green™ Kit (Takara Bio) and the cycling conditions recommended for all genes in the kit instructions. Assessment of gene expression was carried out using the 2−ΔΔCt method. The primer sequences used are presented in .
Table 1. Primer sequences for qRT-PCR.
2.9. Determination of caspase activities
The activities of caspase 9 and caspase 3 in the cell lysates were evaluated as described previouslyCitation25. The harvested HepG2 cells treated with varying concentrations of 14f for 48 h were lysed using chilled 100 µL lysis buffer, to extract the total protein. The protein concentration was assessed using a Bradford Kit. The protein samples were incubated with assay buffer and colorimetric substrate (Ac-DEVD-pNA or Ac-LEHD-pNA) at room temperature for 2 h, according to the manufacturer’s protocol. The released pNAs were measured at the wavelength of 405 nm using a Multiskan® Spectrum system. The experiments were performed in triplicate.
2.10. Western blot
To analyse the effect of compound 14f on protein levels, western blot assay was employed, as elaborated previouslyCitation26. The proteins were acquired by collecting HepG2 cells treated with varying concentrations of 14f for 48 h and adding them to lysis buffer, in an ice bath for 30 min. After quantitation of protein concentration using BCA assay, equal amounts of total protein were separated using 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride membranes. The membranes were incubated with 5% bovine serum albumin solution for 1.5 h at 37 °C, followed by incubation with primary antibody overnight, and finally secondary antibody for 2 h at 37 °C. The blots were detected using enhanced chemiluminescence reagent with a fully automatic gel imaging system (ChemiDoc™ MP, Bio-Rad). The relative expression was calculated by normalising the expression of the control group to 1 for comparison.
2.11. Molecular docking study
Molecular docking studies were performed using GLIDE (2016, Schrödinger Suite)Citation27. The crystal structures of Bcl-2 (PDB: 2O2F)Citation28 were retrieved from the RCSB Protein Data Bank, and further prepared using the Protein Preparation Wizard tool implemented in the Schrödinger Suite, by adding all hydrogen atoms as well as missing side chains of residues and deleting all bound water. The ligands were built within Maestro BUILD (2016, Schrödinger Suite) and prepared using the LIGPREP module (2016, Schrödinger Suite)Citation27. The Glide Grid was built using an inner box of dimensions 15 × 15 × 15 Å3 around the centroid of the ligand, assuming that the ligands to be docked were of a size similar to that of the co-crystallized ligand. This docking methodology has been validated by extracting the crystallographic bound ligand and re-docking it with the Glide module using extra precision. Different docking poses of ligands were generated and analysed for interpretation of the final results.
3. Results and discussion
3.1. Chemistry
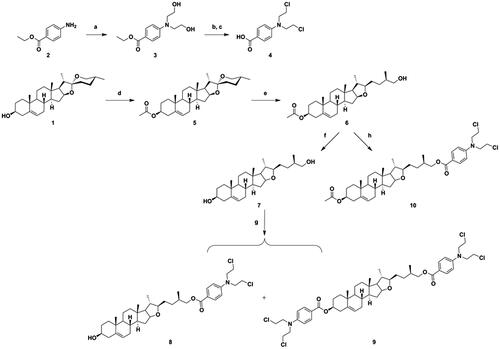
The structures and reaction conditions for the preparation of the new hybrids of DSG–benzoic acid mustard are depicted in Schemes 1 and 2. The benzoic acid mustard 4 was prepared according to a previously described methodCitation20. DSG derivative 6, which was obtained using a procedure reported previously by our research groupCitation22, was saponified to obtain intermediate 7. Intermediate 6 or 7 was coupled with benzoic acid mustard in CH2Cl2 using EDCI and DMAP as catalysts, to obtain the target DSG–benzoic acid mustard hybrids 8, 9, and 10 (Scheme 1).
Scheme 1. Synthesis of diosgenin–benzoic acid mustard trihybrids 8−10. Reagents and conditions: (a) ethylene oxide, H2O, CH3COOH, rt, 24 h; (b) POCl3, 50 °C, 0.5 h; (c) 10% HCl, 12 h; (d) Ac2O, dry pyridine, dry CH2Cl2, rt, 6 h; (e) NaBH3CN, AcOH, CH2Cl2, rt, 8 h; (f) KOH, CH3OH, rt, 6 h; (g) (h) Benzoic acid mustard, EDCI, DMAP, CH2Cl2, rt, 24 h.
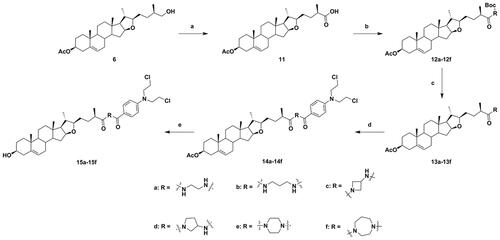
Next, the C-26 hydroxyl of intermediate 6 was oxidised using Jones reagent, to obtain intermediate 11. Further, the C-26 carboxyl of intermediate 11 was reacted with various N-Boc-protected amines using TBTU and DIPEA as coupling catalysts, to provide conjugates 12a–12f (Scheme 2), which were converted to 13a–13f by using CF3COOH for deprotection. The DSG–benzoic acid mustard hybrids 14a–14f were prepared using the same methods as those for 10. Finally, saponification of hybrids 14a–14f resulted in the corresponding target DSG–benzoic acid mustard hybrids 15a–15f. The structures of all new hybrids were fully corroborated using various spectroscopic methods, including HRMS and NMR spectroscopy (1H NMR and 13 C NMR).
Scheme 2. Synthesis of diosgenin–benzoic acid mustard hybrids 14a−14f and 15a−15f. Reagents and conditions: (a) Jones reagent, THF/acetone (1/1), rt, 3 h; (b) N-Boc-protected amines, TBTU, DIPEA, CH2Cl2, rt, 8 h; (c) CF3COOH, CH2Cl2, rt, 4 h; (d) Benzoic acid mustard, DMAP, EDCI, CH2Cl2, rt, 24 h; (e) NaOH, CH3OH/THF (1/1.5), rt, 6 h.
3.2. Biological testing
3.2.1. In vitro anti-proliferative activity
The anti-proliferative activities of the synthesised target hybrids towards three human cancer cells HepG2 (hepatoma), MCF-7 (breast cancer), HeLa (cervical cancer) and normal GES-1 (stomach) cells were evaluated using MTT assay. Mitomycin C was selected as the positive control drug, and the results of this experiment are listed in . Benzoic acid mustard (4) was firstly introduced on the scaffold of DSG derivatives 6 and 7 via an ester bond. Unfortunately, the resulting hybrids 8–10 showed decreased inhibitory activities against the three cancer cells.
Table 2. Cytotoxic activities of target hybrids 8– 10, 14a–14f and 15a–15f in different cell lines.
Compared to DSG and 4, all the target hybrids 14a–14f and 15a–15f (except for 15d) displayed moderate to potent inhibitory activities in HepG2 cells, with IC50 values in the range of 2.26–25.29 µM. The results showed that introducing amide-amide bonds into the two pharmacophores of DSG and 4, to afford hybrids 14a–14f and 15a–15f, could improve their anti-proliferative potency. However, changing the linkers does not seem to regularly affect activities. Among them, compound 14f (IC50 = 2.26 µM), which comprised of DSG derivative 11 substituted by the moiety of 4 through a homopiperazinyl linker, showed obvious selective anti-proliferative activity against HepG2 cells. Its potency was approximately 15.0-fold higher than that of DSG (IC50 = 33.87 µM), and 14.4-fold higher than that of Mitomycin C (IC50 = 32.63 µM). The cytotoxic activity of 14f was greatly improved in comparison with those of the counterparts of DSG–amino acid–benzoic acid mustard trihybrids 12a–12g (IC50 > 10.43 µM)Citation25.
In MCF-7 cell line, hybrids 14a–14f (IC50 > 11.07 µM) generally exhibited higher inhibitory activity than hybrids 15a–15f (IC50 > 37.20 µM). These data showed that the group of acetyl substituted at the C-3 OH was beneficial for compounds with inhibitory potency against MCF-7 cell line. Among them, only hybrid 14f possessing a homopiperazinyl linker exhibited moderate anti-proliferative activity, with an IC50 value of 11.07 µM, and was about 1.7-fold more active than DSG (IC50 = 23.91 µM). The results also confirmed that the incorporation of homopiperazinyl between DSG derivative 11 and 4 results in favourable enhancement of anti-proliferative activity against MCF-7 cells, as compared to those of nitrogen-containing groups.
In HeLa cell line, conjugates 14a (IC50 = 17.24 µM) and 15e (IC50 = 28.46 µM) showed relatively stronger cytotoxicity than other conjugates and DSG (IC50 = 55.05 µM).
The selectivity index (SI), one of the important pharmaceutical parameters, was calculated to determine the toxicities of these hybrids against human normal gastric epithelial GES-1 cell line, as compared to those against hepatoma HepG2 cell line (). Among them, the most potent anti-proliferative hybrid 14f showed considerable safety (SI > 44.25).
Taken together, among these DSG–benzoic acid mustard hybrids, hybrid 14f, with the linkage of homopiperazinyl, showed higher anti-proliferative activity against HepG2 and MCF-7 cancer cells, which suggested that the introduction of homopiperazinyl between DSG derivative 11 and benzoic acid mustard is beneficial for anti-proliferative activity. Our findings are in good agreement with a report by Wolfram et al., which also showed that the linker of homopiperazinyl is essential for mitocanic triterpenoidic rhodamine B adducts that show high cytotoxicity against a panel of human tumour cell linesCitation29. Hybrid 14f showed the lowest IC50 value of 2.26 µM and low anti-proliferative activity against normal GES-1 cells, with an IC50 value > 100 µM. Hence, 14f was selected for further investigation of the possible cellular mechanisms in HepG2 cell line.
3.2.2. Effect of 14f on the cell cycle
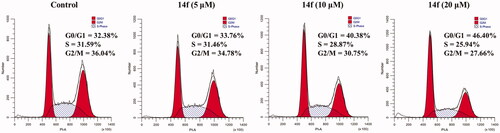
Uninterrupted cell cycle progression is vital to the biochemical processes surrounding cell division and replication. Thus, blockade of the cell cycle is considered as an effective strategy in cancer therapyCitation30. Previous studies have demonstrated that DSG arrests cell cycle and induces apoptosis in different cancer cellsCitation5,Citation7,Citation31,Citation32. To determine whether the anti-proliferative effects of hybrid 14f are caused by cell cycle arrest at a certain phase, the effects of different concentrations of 14f on cell cycle progression were examined in HepG2 cells. Upon treatment with different concentrations (0, 5, 10, and 20 µM) of 14f, 32.38, 33.76, 40.38, and 46.40% of the cells, respectively, were found to be in the G0/G1 phase (). The results demonstrated that 14f arrested the cell cycle at the G0/G1 phase in HepG2 cells.
Figure 3. Analysis of the effects of 14f on the cell cycle in HepG2 cells. HepG2 cells were treated with indicated concentrations (5, 10, and 20 μM) of 14f for 24 h, harvested, stained with PI, and assessed using a flow cytometer.
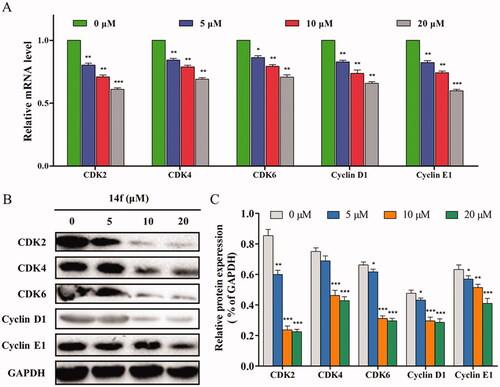
Subsequently, the specific mechanism by which 14f regulates the G0/G1 phase was further investigated at the transcription and translation levels. The effect of 14f on the expression levels of several key cell cycle-related genes in HepG2 cells was determined using qRT-PCR. It was found that 14f decreased the genes levels of CDK2, CDK4, CDK6, cyclin D1 and cyclin E1 (), all of which appear in the G0/G1 phase. In addition, upon performing western blot to detect the G0/G1 phase-related proteins, it was found that 14f could concentration-dependently downregulate the expression levels of CDK2, CDK4, CDK6, cyclin D1, and cyclin E1 (). These findings suggested that the inhibition of HepG2 cells by 14f was related to the induction of G0/G1 phase arrest.
Figure 4. Effect of 14f on the levels of cell cycle-related genes and proteins in HepG2 cells. GAPDH served as the loading control. (A) Relative mRNA level. (B) The expression of proteins was detected using Western blot. Values have been represented as mean ± SD (n = 3). *p<0.05; **p<0.01; ***p<0.001 versus control group.
3.2.3. 14f promoted apoptosis in HepG2 cells
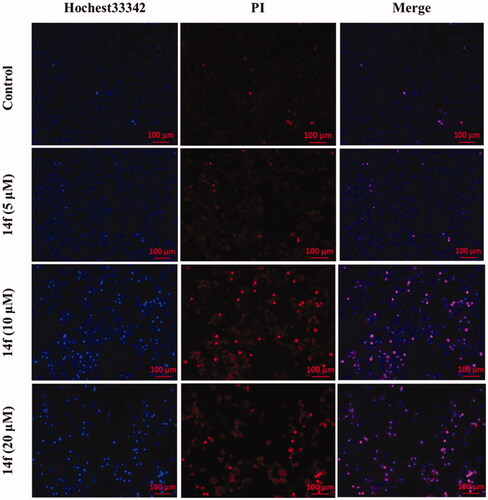
Inducing apoptosis is an important strategy for cancer treatment. To assess whether the 14f could promote the apoptosis of HepG2 cells, we detected the apoptosis of HepG2 cells upon treatment with various concentrations of 14f. First, we observed the morphological changes in HepG2 cells under an inversion microscope. As seen in Figure S1, microscopic observation showed that HepG2 cells presented a significant round shape with shrinkage of the cell membrane upon treatment with 14f, accompanied by a prominent decrease in the number of cells. These phenomena were further examined using the Hoechst 33342/PI staining assay, the results for which showed that upon treatment with 14f, there was an increase in the apoptosis of HepG2 cells, indicated by deep blue and red fluorescence, accompanied by the observation of apoptosis bodies (). Collectively, the above observation demonstrated that 14f treatment induced apoptosis in HepG2 cells.
Figure 5. Fluorescence microscopy images of HepG2 cells stained using Hoechst 33342/PI. HepG2 cells were treated with different concentrations (5, 10, and 20 μM) of 14f for 48 h.
To further obtain an accurate evaluation of the 14f-mediated apoptosis of HepG2 cells, cells treated with different concentrations of 14f were analysed using flow cytometry, post Annexin V-FITC/PI double staining. As seen in , with increasing concentrations of 14f, the number of apoptotic cells increased from 3.89% to 9.80%, 14.38%, and 28.13%, while the number of surviving cells decreased correspondingly, in a concentration-dependent manner. These data demonstrated that 14f promoted apoptosis in HepG2 cells as well.
3.2.4. 14f decreased the MMP (δψm) of HepG2 cells
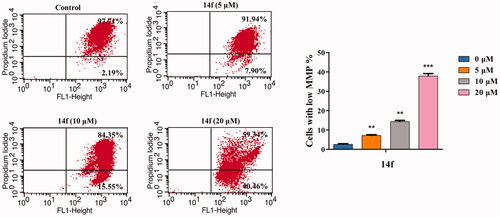
Mitochondrion plays an important role in regulating cellular functions. Loss of MMP is a sign of the apoptotic process in cells. MMP collapse represents the initiation and activation of apoptosis by the intrinsic pathwayCitation33. In the present study, JC-1 staining was conducted to investigate whether 14f-induced HepG2 cell apoptosis was associated with the loss of MMP. As seen in , there was a 14f dose-dependent increase in the percentage of mitochondrial depolarisation (7.90% at 5 µM, 15.55% at 10 µM, and 40.46% at 20 µM). The result indicated that 14f induced apoptosis in HepG2 cells by destroying the mitochondrial integrity and decreasing MMP.
Figure 7. 14f induced the collapse of mitochondrial membrane potential (ΔΨm) in HepG2 cells. After treatment with 14f for 48 h, the cells stained with JC-1 were determined by means of flow cytometric analysis. Values have been represented as mean ± SD (n = 3). **p<0.01; ***p<0.001 versus control group.
3.2.5. 14f induced apoptosis in HepG2 cells through the mitochondrial pathway
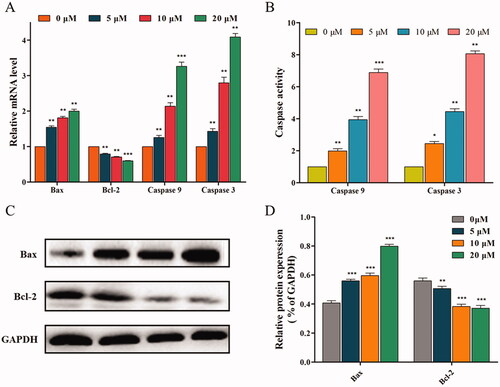
Based on the results, we hypothesised that 14f might be a MMP disruptor, and further examined the effect of 14f on mitochondrial pathway-related factors (such as anti-apoptotic protein Bcl-2 and pro-apoptotic protein Bax) and apoptotic effectors (such as caspase 9 and caspase 3), which play important roles in cell apoptosisCitation22. Firstly, the expression of mitochondrial pathway-related genes was evaluated using qRT-PCR. As seen in , 14f treatment led to high gene expression of Bax, caspase 9, and caspase 3, while the gene expression of Bcl-2 was suppressed. Next, based on the qRT-PCR results, ELISA was further used to confirm whether the apoptosis-related enzymes, caspase 9 and caspase 3, are involved in the regulation of apoptosis. The results indicated that 14f upregulated the enzyme activity of caspase 9 and caspase 3 (). Finally, western blot was conducted to investigate the protein expression levels of Bax and Bcl-2. As shown in , 14f upregulated the levels of Bax with associated downregulation of Bcl-2 levels (). Moreover, the quantitative analysis results were consistent with the trend of mRNA level expression (). These results suggested that 14f triggered the mitochondrial pathway of apoptosis in HepG2 cells.
Figure 8. 14f activated the mitochondrial pathway, to regulate the apoptosis of HepG2 cells. (A) Relative mRNA level. (B) The activities of caspase 9 and caspase 3 were detected using ELISA. (C) The relative protein expression levels were detected using western blot analyses. (D) Quantitative analysis of the protein expression data mentioned in (C). Values have been represented as mean ± SD (n = 3). **p<0.01; ***p<0.001 versus control group.
3.2.6. Molecular docking studies
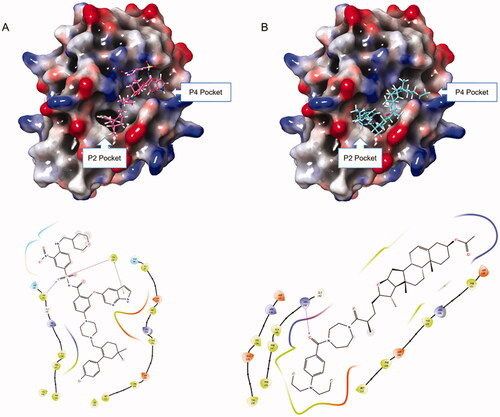
Based on the downregulation of Bcl-2 by hybrid 14f, we next explored the potential binding mode of 14f with Bcl-2. High-affinity binding of pro-apoptotic proteins to Bcl-2 is largely mediated by protein-protein interactions in the P2 and P4 hydrophobic pockets. Thus, the Bcl-2-inhibiting inhibitory potency of small molecules is determined by the interactions of these inhibitors with P2 and P4 pocketsCitation34,Citation35. Venetoclax, also known as ABT-199, was the first Bcl-2-selective inhibitorCitation35. Our docking research showed that the tetrahydro-1, 1′-biphenyl fragment and 1H-pyrrolo[[2,3-b]] pyridine fragment of ABT-199 could occupy the P2 and P4 pockets, respectively (). Coincidentally, similar behaviours could be observed in the bonding mode of 14f to Bcl-2 protein. 14f could lay on the deep hydrophobic binding pocket in the Bcl-2 groove, with the benzoic acid mustard moiety accommodating the P2 pocket and the acetyl moiety entering the P4 pocket. The experimental results showed that 14f might have the ability to interact with Bcl-2, thereby inducing HepG2 cells apoptosis.
Altogether, the above results indicated that 14f arrested the cell cycle at the G0/G1 phase and induced apoptosis in HepG2 cells through the mitochondrial apoptosis pathway. To comprehend the exact regulatory mechanism of 14f against hepatoma, there is a need to carry out well-designed in vivo studies in the future.
4. Conclusion
In summary, fifteen novel DSG-benzoic acid mustard hybrids (8–10, 14a–14f, and 15a–15f) were designed and synthesised in the present study. This was followed by testing the cytotoxicity of these target hybrids against human cancer HepG2, MCF-7, and HeLa cell lines and human normal GES-1 cells. Most of them did not display significant cytotoxic activity. Several of these hybrids exhibited moderate to potent inhibitory activities against HepG2 cells. Among them, hybrid 14f (IC50 = 2.26 µM) exhibited the most potent anti-proliferative activity in HepG2 cells, and displayed an efficacy that was 14.4-fold higher than that of DSG (IC50 = 32.63 µM). Moreover, 14f exhibited good anti-proliferative selectivity between normal and tumour cells. SAR studies showed that the introduction of homopiperazinyl between DSG derivative 11 and benzoic acid mustard was beneficial for enhancing the anti-proliferative activity. Further investigations on the anti-tumour mechanism in HepG2 cells indicated that 14f arrested the cell cycle at the G0/G1 phase, by regulating the cell cycle-related proteins (CDK4, CDK6, cyclin D1, CDK2, and cyclin E1), and induced apoptosis by mediating the apoptosis-related proteins (Bax, Bcl-2, caspase 9, and caspase 3). Furthermore, molecular docking revealed that hybrid 14f could bind at the deep hydrophobic binding pocket in the Bcl-2 groove, indicating that the potential target of 14f might be Bcl-2. Therefore, conjugating DSG with benzoic acid mustard might be an efficient strategy for the modification of this class of natural products.
Supplemental Material
Download PDF (1.8 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Newman DJ, Cragg GM. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 2020;83:1299–803.
- Zolottsev VA, Latysheva АS, Pokrovsky VS, et al. Promising applications of steroid сonjugates for cancer research and treatment. Eur J Med Chem 2021;210:113089.
- Nahar L, Sarker SD. A review on steroid dimers: 2011–2019. Steroids 2020;164:108736.
- Ma LW, Wang HJ, Wang J, et al. Novel steroidal 5α,8α-endoperoxide derivatives with semicarbazone/thiosemicarbazone side-chain as apoptotic inducers through an intrinsic apoptosis pathway: design, synthesis and biological studies. Molecules 2020;25:1209.
- Liao WL, Lin JY, Shieh JC, et al. Induction of G2/M phase arrest by diosgenin via activation of Chk1 kinase and Cdc25C regulatory pathways to promote apoptosis in human breast cancer cells. Int J Mol Sc 2019;21:172.
- Li SY, Shang J, Mao XM, et al. Diosgenin exerts anti-tumor effects through inactivation of cAMP/PKA/CREB signaling pathway in colorectal cancer. Eur J Pharmacol 2021;908:174370.
- Mao XM, Zhou P, Li SY, et al. Diosgenin suppresses cholangiocarcinoma cells via inducing cell cycle arrest and mitochondria-mediated apoptosis. Onco Targets Ther 2019;12:9093–104.
- Long C, Chen J, Zhou H, et al. Diosgenin exerts its tumor suppressive function via inhibition of Cdc20 in osteosarcoma cells. Cell Cycle 2019;18:346–58.
- Liu YL, Zhou ZJ, Yan JZ, et al. Diosgenin exerts antitumor activity via downregulation of skp2 in breast cancer cells. BioMed Res Int 2020;2020:8072639–10.
- Singh RK, Kumar S, Prasad DN, et al. Therapeutic journey of nitrogen mustard as alkylating anticancer agents: historic to future perspectives. Eur J Med Chem 2018;151:401–33.
- Yan X, Song JL, Yu MX, et al. Synthesis of flavonoids nitrogen mustard derivatives and study on their antitumor activity in vitro. Bioorg Chem 2020;96:103613.
- Martín-Cámara O, Arribas M, Wells G, et al. Multitarget hybrid fasudil derivatives as a new approach to the potential treatment of amyotrophic lateral sclerosis. J Med Chem 2022;65:1867–1882.
- Pandey AR, Rai D, Singh SP, et al. Synthesis and evaluation of galloyl conjugates of flavanones as BMP-2 upregulators with promising bone anabolic and fracture healing properties. J Med Chem 2021;64:12487–505.
- Harrison JR, Brand S, Smith V, et al. A molecular hybridization approach for the design of potent, highly selective, and brain-penetrant N-myristoyltransferase inhibitors. J Med Chem 2018;61:8374–89.
- Zhang L, Xu LJ, Chen HH, et al. Structure-based molecular hybridization design of Keap1-Nrf2 inhibitors as novel protective agents of acute lung injury. Eur J Med Chem 2021;222:113599.
- Hou QQ, Lin X, Lu X, et al. Discovery of novel steroidal-chalcone hybrids with potent and selective activity against triple-negative breast cancer. Bioorg Med Chem 2020;28:115763.
- Kerru N, Singh P, Koorbanally N, et al. Recent advances (2015–2016) in anticancer hybrids. Eur J Med Chem 2017;142:179–212.
- Sun JN, Wang JS, Wang XY, et al. Design and synthesis of β-carboline derivatives with nitrogen mustard moieties against breast cancer. Bioorg Med Chem 2021;45:116341.
- Xu YZ, Gu XY, Peng SJ, et al. Design, synthesis and biological evaluation of novel sesquiterpene mustards as potential anticancer agents. Eur J Med Chem 2015;94:284–97.
- Han T, Tian KT, Pan HQ, et al. Novel hybrids of Brefeldin A and nitrogen mustards with improved antiproliferative selectivity: design, synthesis and antitumor biological evaluation. Eur J Med Chem 2018;150:53–63.
- Acharya PC, Bansal R. Synthesis of androstene oxime-nitrogen mustard bioconjugates as potent antineoplastic agents. Steroids 2017;123:73–83.
- Zhang JL, Wang XM, Yang JF, et al. Novel diosgenin derivatives containing 1,3,4-oxadiazole/thiadiazole moieties as potential antitumor agents: design, synthesis and cytotoxic evaluation. Eur J Med Chem 2020;186:111897.
- Wang XD, Sun YY, Zhao C, et al. 12-Chloracetyl-PPD, a novel dammarane derivative, shows anti-cancer activity via delay the progression of cell cycle G2/M phase and reactive oxygen species-mediate cell apoptosis. Eur J Pharmacol 2017;798:49–56.
- Ma LW, Zhang JL, Guo LN, et al. Design and synthesis of diosgenin derivatives as apoptosis inducers through mitochondria-related pathways. Eur J Med Chem 2021;217:113361.
- Wang WB, Li C, Chen Z, et al. Novel diosgenin-amino acid-benzoic acid mustard trihybrids exert antitumor effects via cell cycle arrest and apoptosis. J Steroid Biochem Mol Biol 2022;216:106038.
- Ma LW, Chen Z, Li J, et al. DP from Euphorbia fischeriana S. mediated apoptosis in leukemia cells via the PI3k/Akt signaling pathways. J Ethnopharmacol 2021;279:113889.
- Schrodinger, LLC. Glide. New York, NY: Schrodinger, LLC; 2016
- Bruncko M, Oost TK, Belli BA, et al. Studies leading to potent, dual inhibitors of Bcl-2 and Bcl-xL. J Med Chem 2007;50:641–62.
- Wolfram RK, Fischer L, Kluge R, et al. Homopiperazine-rhodamine B adducts of triterpenoic acids are strong mitocans. Eur J Med Chem 2018;155:869–79.
- Gao X, Li J, Wang MY, et al. Novel enmein-type diterpenoid hybrids coupled with nitrogen mustards: Synthesis of promising candidates for anticancer therapeutics. Eur J Med Chem 2018;146:588–98.
- Sikka S, Shanmugam MK, Siveen KS, et al. Diosgenin attenuates tumor growth and metastasis in transgenic prostate cancer mouse model by negatively regulating both NF-κB/STAT3 signaling cascades. Eur J Pharmacol 2021;906:174274.
- Guo WH, Chen YJ, Gao JS, et al. Diosgenin exhibits tumor suppressive function via down-regulation of EZH2 in pancreatic cancer cells. Cell Cycle 2019;18:1745–58.
- Wang FF, Shi C, Yang Y, et al. Medicinal mushroom Phellinus igniarius induced cell apoptosis in gastric cancer SGC-7901 through a mitochondria-dependent pathway. Biomed Pharmacother 2018;102:18–25.
- Sattler M, Liang H, Nettesheim D, et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 1997;275:983–6.
- Ashkenazi A, Fairbrother WJ, Leverson JD, et al. From basic apoptosis discoveries to advanced selective BCL-2 family inhibitors. Nat Rev Drug Discov 2017;16:273–84.