Abstract
Cyclin-dependent kinase inhibition is considered a promising target for cancer treatment for its crucial role in cell cycle regulation. Pyrazolo pyrimidine derivatives were well established for their antitumor activity via CDK2 inhibition. In this research, new series of pyrazolopyrimidine derivatives (4–15) was designed and synthesised as novel CDK2 inhibitors. The anti-proliferative activities against MCF-7, HCT-116, and HepG-2 were used to evaluate their anticancer activity as novel CDK2 inhibitors. Most of the compounds showed superior cytotoxic activity against MCF-7 and HCT-116 compared to Sorafenib. Only compounds 8, 14, and 15 showed potent activity against HepG-2. The CDK2/cyclin A2 enzyme inhibitory activity was tested for all synthesised compounds. Compound 15 showed the most significant inhibitory activity with IC50 0.061 ± 0.003 µM. It exerted remarkable alteration in Pre G1 and S phase cell cycle progression and caused apoptosis in HCT cells. In addition, the normal cell line cytotoxicity for compound 15 was assigned revealing low cytotoxic results in normal cells rather than cancer cells. Molecular docking was achieved on the designed compounds and confirmed the two essential hydrogen binding with Leu83 in CDK2 active site. In silico ADMET studies and drug-likeness showed proper pharmacokinetic properties which helped in structure requirements prediction for the observed antitumor activity.
1. Introduction
Protein kinases represent a large group of structurally related enzymes that are essential and regulate cell cycle progression involved in cell divisionCitation1–3. Cyclin-dependent kinases (CDKs) are serine-threonine kinases responsible for cell cycle regulation and cell differentiationCitation2. Cyclin-dependent kinases (CDK) are mainly responsible for the phosphorylation process of proteinsCitation4–6. Cyclin is the regulatory protein bound by CDK leading to ATP binding region modificationCitation2. CDKs in absence of cyclin have less activity where the activation loop (known as T-loop) blocks the cleft, and the key amino acid residues are not optimally positioned for ATP bindingCitation2. CDK2 has a catalytic effect in cyclin-dependent protein kinase complexCitation7,Citation8. Protein phosphorylation has a critical role in cellular function regulation. This essential role during the cell cycle could be altered in tumour cellsCitation7–11. Alteration in kinases may lead to the development of many diseases, including cancer. Hence, the control of CDK dependent cell cycle is essential for tumour progression management. Where overexpression of CDK enzymes occurs in cancerCitation2. As uncontrolled CDK2 activation in human cancer is associated with overexpression of cyclins A and E in many human cancersCitation12. Thus, CDKs are considered critical targets for the development of novel anticancer drugsCitation9,Citation10.
Pyrazolopyrimidine moiety is a common nucleus heterocycle used in the design of different pharmaceutical compoundsCitation11,Citation12, with various medicinal applications including antimicrobial, antitumorCitation13, antidiabetic, anti-Alzheimer's disease, anti-inflammatoryCitation14,Citation15, and antioxidantCitation16–18.
Pyrazolopyrimidine is considered an appealing scaffold for the development of new antitumor agentsCitation19–24. This scaffold is considered an adenine bioisostere that retains the essential interactions of ATP at the kinase domainCitation25–28. The pyrazolo[3,4-d]pyrimidine bicycle showed great potential in CDK inhibition in drug discovery. It is a purine ring bioisostere with potent growth inhibitory activityCitation29,Citation30, via CDK inhibition as CDK1Citation31, CDK2Citation32, and 5-lipoxygenase enzymesCitation33. They also possess significant and specific pharmacological activity in CDK2/Cyclin A inhibitionCitation11. Among the known drugs of potent CDK2 inhibition are Dinaciclib, Ibrutinib, and RoscovitineCitation1,Citation9,Citation12,Citation25,Citation34,Citation35 ().
Figure 1. Structures of active drugs containing fused pyrimidine (I) Roscovitine, (II) Ibrutinib, and (III) Dinaciclib, Reported pyrazolo[3,4-d]pyrimidines derivatives (IV), (V), and (VI), and Reported pyrazolo[1,5-a]pyrimidines derivative (VII) as CDK2 inhibitors.
![Figure 1. Structures of active drugs containing fused pyrimidine (I) Roscovitine, (II) Ibrutinib, and (III) Dinaciclib, Reported pyrazolo[3,4-d]pyrimidines derivatives (IV), (V), and (VI), and Reported pyrazolo[1,5-a]pyrimidines derivative (VII) as CDK2 inhibitors.](/cms/asset/98e46170-6934-4fe6-abca-322230013f03/ienz_a_2086866_f0001_b.jpg)
The aim of this research was based on the previously reported work with a rationale for anticancer drug discovery research, to design and synthesise new pyrazolo[3,4-d]pyrimidine and its glycosyl amino derivatives as well. In addition to the thioxo-pyrazolo[4,3-e][1,2,4]triazolo [1,5-c]pyrimidine compounds and the thioglycoside derivatives, based on pyrazolo[3,4-d]pyrimidine scaffold to develop novel CDK2 inhibitorsCitation34,Citation35. The new derivatives were evaluated for their in vitro anti-proliferative activity against three cell lines: breast cancer (MCF-7), hepatocellular carcinoma (HepG-2), and colorectal carcinoma (HCT-116) cell lines. The potential inhibition of the most potent promising compounds on CDK2/Cyclin A was also tested. Also, molecular docking studies were applied to investigate the binding mode of the promising compounds by calculating their binding energies and visualising their orientations with respect to the active site of CDK-2 protein compared to the Roscovitine ligand. The computational results were in agreement with the reported observations.
1.1. Rational design
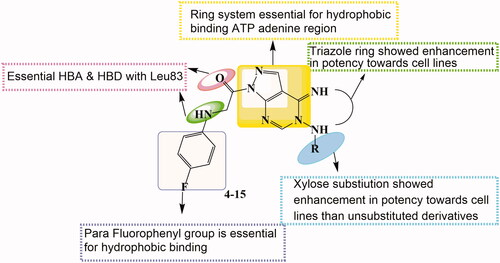
The design of the new pyrazolo[3,4-d]pyrimidine derivatives in compounds 4–6 and pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin derivatives in compounds 7–15 was based on lead compounds Roscovitine and compound VII optimisation by structural modifications according to their reported binding mode () whereCitation12.
Figure 2. Binding mode schematic illustration of Roscovitine ligand (I) (A) and Newly synthesised pyrazolopyrimidine derivative compound 13 (B) with key amino acids at CDK2 binding site.
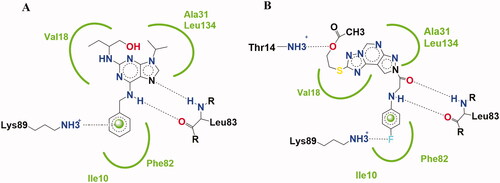
Figure 3. Features' similarities between Roscovitine ligand (I) and reported pyrazolo[1,5-a]pyrimidines derivative (VII) against the newly designed compounds as potent CDk2 inhibitors.
![Figure 3. Features' similarities between Roscovitine ligand (I) and reported pyrazolo[1,5-a]pyrimidines derivative (VII) against the newly designed compounds as potent CDk2 inhibitors.](/cms/asset/fd07e08e-1f7f-4b29-85fd-4929654f33bf/ienz_a_2086866_f0003_c.jpg)
Purine scaffold was bioisostericlly replaced by two different ring systems first: pyrazolo[3,4-d]pyrimidine in compounds 4–6 and second: pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin in compounds 7–15 through the hydrophobic binding with Leu134, Val18, Val64, Ala31, Ala144, Ile10, and Phe80. This is required to place the compound in the ATP adenine region.
The two essential hydrogen bonds in the phosphate binding region with Leu83,1 HBD by –NH of the 6-substituted amino moiety, 1 HBA through N7 in Roscovitine, were replaced by N arylglycyl moiety in all the presented compounds.
The N5 pyrazolopyrimidine substitution by amino group (in compound 4) or N-glycosyl (in compounds 5 and 6) replaced the N butanol at position 2 of Roscovitine.
The pyrazolopyrimidine hydrophobic binding with Ala31, Val18, and Leu134 was maintained as in lead compound and the triazole ring fusion was considered in compounds 7–15 with hydrophobic binding with Val18.
In compounds 7–13, an additional hydrogen bonding by the thio group was achieved as in compound 8 with Asp145. And a hydrogen bond for compound 13 via the acetyloxy moiety with Thr14 was observed.
The hydrophobic binding with Ile10 by the benzyl group in Roscovitine and para bromophenyl group in compound VII was maintained by the isosteric replacement by the para fluorophenyl group in all the new compounds. In addition to the hydrogen binding via the para fluoro group with Lys89 in compounds 8, 9, 11, and 13. As in reported compound VII where it was published that substitution of the phenyl ring with halogen atom is preferable to the un-substituted ring for its cytotoxic effect, with either bromo, fluoro, or chloro groupCitation25. While a halogen hydrogen bond formation by compound 11 with Glu8 and compounds 14 and 15 with Asp145 was observed too.
The thioglycoside group at position 2 in compounds 14 and 15 showed also additional Carbon- Hydrogen binding with Glu8.
2. Materials and methods
2.1. Chemistry
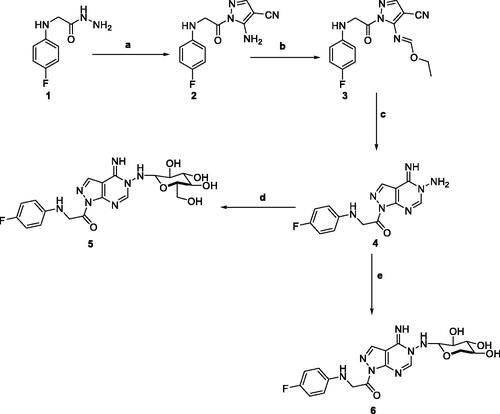
5-Amino-1-(4-fluorophenyl)glycyl)-1H-pyrazole-4-carbonitrile 2, was prepared by the reaction of 2-((4-fluorophenyl)amino)acetohydrazide 1 with ethoxymethylene malononitrile in ethanol under reflux. The isolated brown solid 2 was then treated with triethoxyorthoformate in acetic anhydride to afford ethyl-N-(4-cyano-1-((4-fluorophenyl)glycyl)-1H-pyrazol-5-yl)formimidate 3. The key starting material pyrazolopyrimidine 4 was obtained by treatment of the formimidate derivative 3 with hydrazine hydrate in ethanol. Subsequent reaction of pyrazolopyrimidine 4 with sugar aldoses namely; D-glucose and D-xylose, in the presence of a catalytic amount of acetic acid, afforded the amino-sugar products 5 and 6 respectively (Scheme 1). The structures of this set of novel amino sugars were confirmed by their spectral and elemental analysis data (see Experimental Section). 1H NMR spectra of the amino-sugar compounds 5 and 6 showed a signal attributed to H-1 at 5.13 and 5.10 ppm confirming the sp3 hybridisation of its corresponding carbon (C-1) and the cyclic form of the sugar moiety which agrees well with the reported results for such type of compounds following such mode of preparationCitation36.
Scheme 1. Reagents and conditions; (a) 2-(ethoxymethylene)-malononitrile, EtOH, reflux, 6 h. (b) triethyl orthoformate, acetic anhydride, reflux, 6 h. (c) hydrazine hydrate, EtOH, reflux, 6 h. (d) D-Glucose, EtOH, glacial acetic acid, reflux, 3 h. (e) D-Xylose, EtOH, glacial acetic acid, reflux, 3 h.
Adding a carbon disulphide to pyrazolopyrimidine 4 in the presence of potassium hydroxide in ethanol afforded the corresponding triazolomercapto derivative 7. The 1H NMR spectra of compound 7 revealed the disappearance of the NH2 and NH signals and instead another broad singlet at 11.66 ppm for the NH in the thione-thiol system appeared. The latter was utilised for the preparation of a series of acyclic nucleoside analogs via reaction with acyclic oxygenated halides. Thus, reaction with chloroacetonitrile, 2-chloro-1,1-dimethoxyethane, 2-chloropropane-1,2-diol, and 2-chloroethan-2-ol by stirring or by reflux in ethanol afforded the thioderivatives 8–10 and 12, respectively in good yields. In addition, the hydroxyl compounds 10 and 12 were acetylated with acetic anhydride to yield the corresponding O-acetylated acyclic analogs 11 and 13, respectively (Scheme 2). The 1H NMR spectra of the resulted acetylated derivatives showed the signals of the acetyl-methyl groups in addition to the disappearance of the hydroxyl signals which was also confirmed by their IR spectra. The latter showed the carbonyl bands of the ester groups and the disappearance of the characteristic bands of the hydroxyl groups (see Experimental Section).
Scheme 2. Reagents and condition; (a) CS2, KOH, EtOH, reflux, 6 h. (b) Chloroacetonitrile, K2CO3, DMF, 25 °C, 8 h. (c) Chloroacetaldhyde dimethyl acetal, K2CO3, DMF, 25 °C, 8 h. (d) 2-chloro-1,2-propandiol, KOH, EtOH, reflux, 3 h. (e) Acetic anhydride, pyridine, reflux, 2 h. (f) 2-chloroethanol, KOH, EtOH, reflux, 3 h.
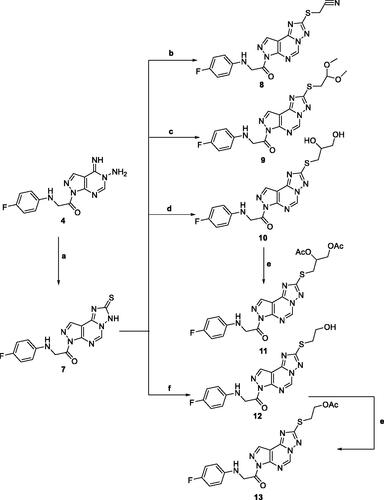
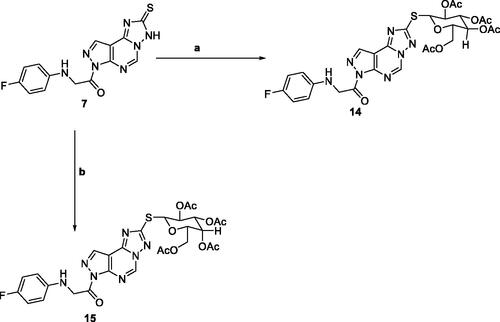
The reaction of triazolo pyrazolo pyridine derivatives 7 with two acetylated glycosyl halides, tetra-O-acetyl-α-D-gluco- and galactopyranosyl bromide, afforded the thioglycoside derivatives 14 and 15 as nucleoside analogs (Scheme 3). The 1H NMR of compound 14 revealed an increase in the integration of the aliphatic region at 2.23–5.76 ppm, while the 13C NMR spectrum showed the characteristic signals for the five carbons of C = O groups at δ 165 and 175 ppm.
3. Results and discussion
3.1. In vitro anti-proliferative activity
All the synthesised compounds were evaluated for their in vitro anti-proliferative activities using MTT assayCitation37,Citation38, against three cell lines named: breast cancer (MCF-7), hepatocellular carcinoma (HepG-2), and colorectal carcinoma (HCT-116). Results were compared to reference Sorafenib (). Most of the compounds 8, 9, 10, 11, 13, 14, and 15 showed superior cytotoxic activities against MCF-7 with IC50 range (7–95 nM). While compounds 6, 8, 9, 11, 13, 14, and 15 showed potent activity against HCT-116 with IC50 range (6–93 nM) compared to the reference (IC50:144 and 176 nM, respectively). Only compounds 8, 14, and 15 showed potent activity against HepG-2 with IC50 values of (8, 7, 7 nM, respectively) compared to the reference (IC50: 19 nM). Compounds 14 and 15 showed the most potent cytotoxic activities against the three cell lines and compound 13 showed moderate HepG-2 activity with IC50 74 nM and superior activity against both MCF-7 and HCT-116 with IC50 values of 55 and 9 nM, respectively ().
Figure 4. The IC50 of tested compounds against MCF-7, HepG-2, and HCT-116 cancer cell lines. Data expressed as mean ± SD from three independent repeats (n = 3). *Significant from sorafenib at p < 0.05.
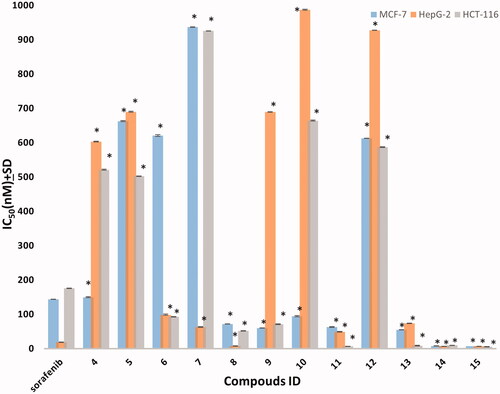
Table 1. The In vitro anti-proliferative activity of the synthesised compounds against human cell lines.
3.2. CDK2/cyclin A2 inhibitory activity
All the synthesised compounds 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, and 15 were tested for their in vitro CDK2/cyclin A2 assays using Promega Kinase-Glo Plus luminescence kinase assayCitation39. The assay depends on kinase reaction and ADP measurement, ADP is first transferred to ATP and then converted to light. The produced luminescent signal is directly proportional to the amount of ATP and inversely proportional to the kinase activity. The results of the enzymatic inhibitory activity of the tested compounds against CDK2/cyclin A2 were presented in . All the tested compounds showed a good inhibitory effect with IC50 values ranging from 0.061 ± 0.003 to 2.050 ± 0.05 µM compared to Sorafenib IC50: 0.184 ± 0.01 µM and Dinaciclib IC50: 0.029 ± 0.002. Results revealed that compounds 15, 11, 14, and 13 showed the most powerful inhibitory activity with IC50 values of 0.061 ± 0.003, 0.089 ± 0.005, 0.118 ± 0.007, and 0.13 ± 0.007 µM, respectively, compared to control drug Sorafenib and/or Dinaciclib. Where compound 15 showed the superior activity ().
Figure 5. Inhibitory activity of most potent compounds on CDK/Cyclin A2. *Significant from sorafenib at P < 0.05, # Significant from dinaciclib at P < 0.05.
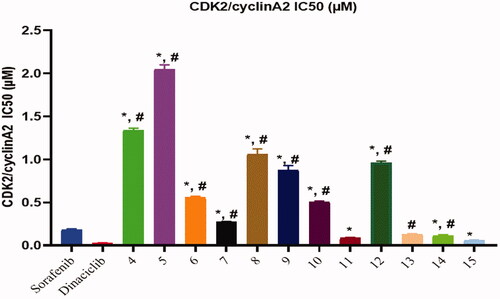
Table 2. CDK2/cyclin A2 inhibitory activity results of all synthesised compounds.
3.3. In vitro cytotoxicity against normal cells WI-38
The cytotoxic effect of the most active compound 15 against WI-38 normal lung cells was in vitro assessed () against Staurosporine as a reference standard. Compound 15 showed low cytotoxicity against normal cells compared to the reference standard with IC50 values of 69.60 ± 2.47 and 57.57 ± 1.42 µM for the most active compound 15 and reference standard, respectively. This data indicated that the synthesised compounds, more specifically that compound 15 exhibit low cytotoxicity against normal cells than cancer cell lines.
Table 3. The In vitro cytotoxicity of the most active compound 15 and Staurosporine against normal lung cells (WI-38).
3.4. Flow cytometry cell cycle analysis
Compound 15 showed the most potent enzymatic inhibitory activity against CDK2/cyclin A2 and potent anti-proliferative activity against the three selected cancer cell lines. Furthermore, compound 15 was selected for flow cytometry assay to evaluate cell cycle analysis against HCT-116 cells. The cell cycle analysis is used to investigate the mechanism of action of the new compoundsCitation40. presents the results showing Pre-G1 and cell growth arrest in the S phase. Results were compared to normal control cells, in Dip Pre-G1with an increase from 1.85 to 32.73% for compound 15 ().
Figure 6. Flow cytometric analysis for cell cycle distribution. (A) Control HCT, (B) Compound 15, and (C) graphical representation for cell cycle distribution analysis among differently treated cells.
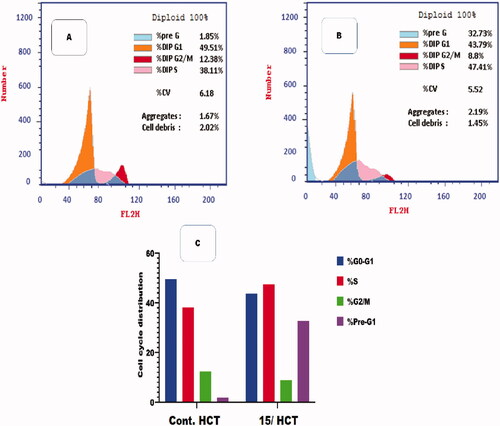
Table 4. Flow cytometric analysis for cell cycle distribution of compound 15 on HCT-116 cells.
3.5. Flow cytometric analysis of apoptosis
Flow cytometric analysis of apoptosis was performed on compound 15 to determine the potentiality of apoptosis induction against the HCT-116 cell lineCitation41.
and showed that compound 15 induced apoptosis by 32.73% (9.26 and 19.12 at early and late apoptosis, respectively), which was 17.69 times more than the standard control (1.85%).
Figure 7. Flow cytometric analysis of apoptosis among treated cells. (A) Control HCT, (B) Compound 15, and (C) Graphical illustration of apoptosis % among differently treated cells.
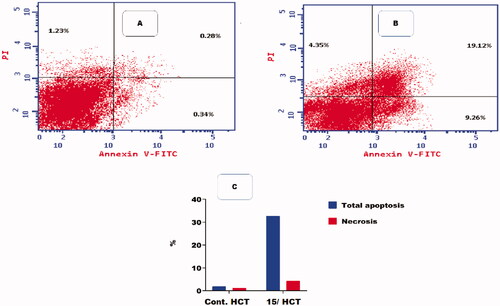
Table 5. Effect of compound 15 on apoptosis in HCT-116 cells.
3.6. Molecular docking
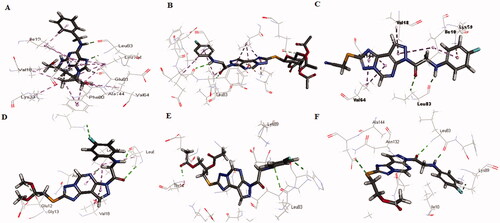
A molecular docking study using the C-Docker protocol in Discovery Studio 4.0 Software was performed on all the synthesised compounds. The compounds were prepared and then docked into the binding site of the CDK2 enzyme. An analysis study was applied to determine the binding modes of the designed compounds and to interpret the biological results. Hence, more explanation about the binding poses and interactions with the key amino acids in the binding site was noticed. X-ray crystallographic structure of CDK2 being complexed with Roscovitine (PDB ID: 2A4L) was downloaded from Protein Data BankCitation9,Citation12. It was known from the previous literature that there are two essential hydrogen bonds with Leu83 at the active site besides hydrophobic interactionsCitation9,Citation12. Validation was confirmed by re-docking of Roscovitine in the active site of CDK2 with RMSD value = 0.5 A°. Superimposition of co-crystallised ligand and ligand after docking indicates the perfect superimposition at the binding site (). The best pose out of ten for each compound was selected showing maximum similarity to the binding mode compared to the ligand. The presented docking study showed comparable binding modes to the lead compound for the docked molecules.
Figure 8. The superimposition 3D diagram of co-crystallised (green colour) and ligand after docking (purple colour) at the active site.
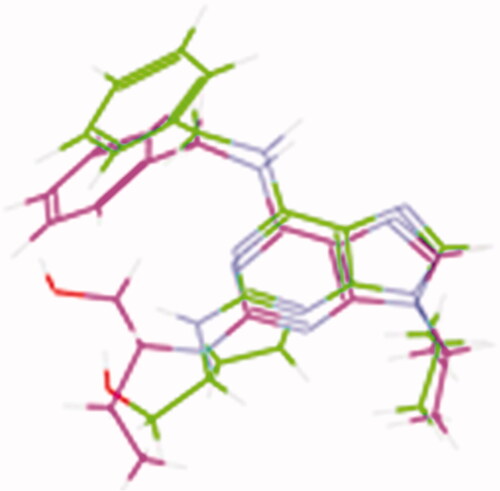
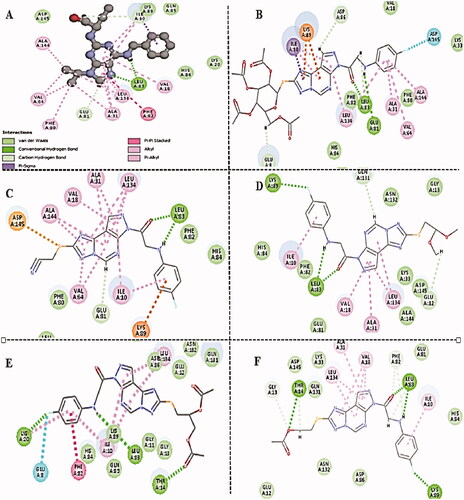
The interaction energy with the essential amino acids of the most biologically active synthesised compounds at the active site is demonstrated in . Results showed comparable binding interaction energy to the lead compound Roscovitine (E = −55.75 kcal/mol), ranging from 60.89 to 50.85 kcal/mol). Compound 15 revealed a binding score = −59.85 kcal/mol with a similar binding mode.
Figure 9. (A) The proposed binding mode of the most promising compounds after docking compared to lead compound: Roscovitine (A), compound 15 (B), compound 8 (C), compound 9 (D), compound 11 (E), and compound 13 (F) within the active site of CDK2 resulting from docking. (B) The proposed 3D binding mode diagram of the most promising compounds after docking compared to lead compound: Roscovitine (A), compound 15 (B), compound 8 (C), compound 9 (D), compound 11 (E), and compound 13 (F) within the active site of CDK2 resulting from docking.
All the docked molecules revealed a hydrogen bond with Leu83. Most of the compounds formed 1 HBD through –NH of glycyl group where binding distance range was observed to be within (2.02–2.10 Å) and 1 HBA through the oxygen atom of the carbonyl group of the glycyl moiety with a binding distance range of (2.20–2.74 Å) (). Compounds 4,5,6,10, and 12 showed different binding modes with either two hydrogen bonds with Leu83 as in compounds 4 and 6, or only one hydrogen bond as in compounds 5, 10, and 12. The sugar moiety in compounds 5 and 6 was responsible for the HBD interaction with Leu83 (Supplementary Material).
All the tested compounds showed binding via the parafluorophenyl group except compounds 4, 5, 7, 8, and 10. Either by a hydrogen bond with Lys89 as in compounds 9, 11, and 13 or by halogen hydrogen binding with Asp145 as in compounds 14 and 15 or extra binding with Glu81 as in compound 11. Compounds 6 and 12 undergo one halogen hydrogen bond with Glu81 and His84 via the parafluoro group, respectively. Compounds 7 and 11 showed only 1 1HBD with Leu83 (compound 11 showed better interaction energy after docking than compound 7). Also, compound 11 (the diacetoxy derivative) and compound 13 (the monoacetoxy derivative) showed an extra hydrogen bond with Thr14 via the acetyloxy group. Compounds 14 and 15 showed additional HBD with Glu81 via the –NH group of the glycyl moiety.
Also, the docked compounds showed comparable hydrophobic binding to lead compounds with Leu134, Val18, Val64, Ala31, Ala144, Ile10, and Phe80.
3.7. In silico predictive ADMET study
The design and development of novel drug molecules need early investigation of pharmacokinetic properties including absorption, distribution, metabolism, and excretion (ADME), at a stage where access to the physical samples is limitedCitation41. It was observed that the limited pharmacokinetic or toxicity profile of new drug-like molecules had a great influence on the low percentage of the drugs that could reach to clinical development stage to be ready to reach the marketCitation42.
Several ways for in silico predictive ADMET studies have been developed. Including web-based platforms, such as ADMETlab, Pro Tox-II, and SwissADME to predict ADMET propertiesCitation42.
Here in this research ADMET study was performed mainly using Discovery Studio 4.0 Software for all the synthesised compounds. Also, the ADMETlab web tool was used to confirm results on the highly potent molecules with promising molecular docking results only to assure the early investigation of their pharmacokinetic properties and compare to Roscovitine as a control drugCitation43.
The ADMET study using Discovery Studio 4.0 Software is mainly concerned with the chemical structure of the molecule, it includes several parameters calculation; Blood Brain Barrier Level, Absorption level, Atom-based Log P98 (A LogP 98), 2D polar surface area (ADMET 2D PSA), Cytochrome P450 2D6 (CYP2D6), Hepatotoxicity Probability, Aqueous solubility level, and Plasma protein binding logarithmic level (PPB Level).
Results shown in reveals that all the compounds had BBB level of 3 and 4. So, they are not able to pass the blood–brain barrier. Most of the compounds had absorption levels = 0 or 1, thus estimated to have good to moderate human intestinal absorption, whilst only compounds 4, 5, 14, and 15 showed low absorption. All the compounds showed ADME aqueous solubility levels between 2 and 3 which indicates good aqueous solubility except compounds 7, 8, and 9 which showed low aqueous solubility. The key property (PSA) was linked to drug bioavailability. Therefore, molecules that are passively absorbed and PSA < 140 are thought to have lower bioavailability. Most of the synthesised compounds were predicted to present good passive oral absorption except compounds 4, 5, 14, and 15 which showed good bioavailability results with a PSA range of 168.67–199.705. Compound 4 showed no hepatotoxicity. Also, all the compounds are considered non-inhibitors to Cytochrome P450 2D6 (CYP2D6) and are poorly bounded to plasma proteins except compounds 2 and 3. ().
Table 6. In silico ADMET predictions of the newly synthesised compounds using Discovery Studio.
Also, the ADMETlab web tool was used to predict the pharmacokinetics of the most promising synthesised compounds. The results were compared to the control drug (Roscovitine)Citation43.
ADMETlab was used to determine the Physicochemical Properties using different descriptors (logS, logD, and logP), Absorption: CaCO3 Permeability (CaCO2) (human colon carcinoma cell line Caco-2), Human Intestinal Absorption (HIA), P-gp inhibitor/substrate (Pgp), 20% bioavailability (as F20%), and 30% bioavailability (as F30%), Distribution: Blood Brain Barrier (BBB), Plasma Protein Binding (PPB) and Volume of Distribution (VD) and Excretion: as both Half-Life (T1/2) and Clearance (CL). Some properties were expressed numerically as CaCO2, VD, PPB, CL, and T1/2, while others were expressed categorically including the rest of the pharmacokinetic parametersCitation42.
Also, Toxicity profiles, such as H-HT (Human Hepatotoxicity) and CYP2D6 were predicted using ADMETlab.
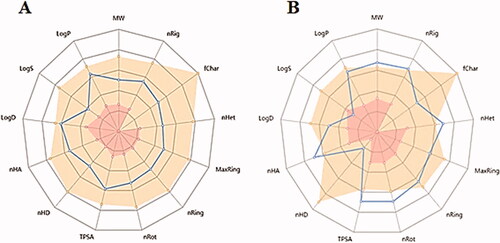
Lipinski’s rule of five is an essential tool for rational drug design. The low permeability or poor absorption for tested compounds was related to a disturbance in one of Lipinski’s rules of fiveCitation42. Molecular weight, LogP, and the number of hydrogen bond acceptors (NHBAs) should lie within the values of <500, 3, and 10, respectively. Hereby, all the compounds except compound 8 had NHBAs of 10 or more. Compounds 11, 14, and 15 showed low aqueous solubility and poor absorption. Where it was observed from log S results that all the tested drugs showed non-aqueous solubility values in the range of −5.401 to −4.731 compared to the control drug (optimal: −4–0.5 log mol/L). While, the logP results showed optimal lipophilicity values (Optimal: 0 < LogP < 3) ().
Table 7. Physicochemical properties predictions of the most potent promising compounds using ADMETlab.
Molecules having similar molecular mass showed similarities in Topological Polar Surface Area (TPSA, %Abs). Where molecules with a TPSA of 140 Å and more showed poor absorption (<10% fractional absorption) as in compounds 11, 14, and 15, while those with a TPSA of 60 Å showed good absorption (>90%)Citation42 ().
3.7.1. Pharmacokinetics properties
3.7.1.1. Absorption
To evaluate the absorption property, CaCO3 Permeability (Caco-2-Permeability), P-glycoprotein (P-gp), substrate or inhibitor (P-gpinh/P-gpsub), Human intestinal absorption (HIA), 20% bioavailability (F20), and 30% bioavailability (F30) were predicted and results were presented in .
Table 8. ADMETlab absorption, distribution, and excretion parameters of the most potent compounds with promising docking results.
It is found that the values of Caco-2-Permeability were in the range from −4.647 to −5.49 cm/s, suggesting to have optimal permeability except for compounds 14 and 15 which showed a −5.49 cm/s result. (optimal permeability more than −5.15 cm/s)
The predicted HIA data of the tested compounds are represented categorically as 1, where Category 1: HIA+ (HIA ≥ 30%); Category 0: HIA− (HIA < 30%). The output value indicated a high HIA probability for the tested compounds except for compounds 14, 15, and the control drug. Most of the compounds showed good bioavailability, Compounds 14 and 15 showed 20–30% bioavailability.
3.7.1.2. Distribution
To study the distribution of drug candidates some parameters were considered as Plasma Protein Binding (PPB), Blood Brain Barrier (BBB) permeability, and Volume of distribution (VD) (). The PPB was predicted to exceed 90% to bind significantly to blood proteins compared to the control drug. While BBB results showed that all the tested compounds have no BBB penetration effect except compounds 14 and 15.
The volume of distribution (VD) in L/Kg was predicted to be optimal when it lies in the range of: 0.04–20 L/kg. So, all compounds showed good VD with moderate clearance results (except compounds 11, 14, and 15 with low clearance results). The hydrophobic character of the compounds was contributed to good values of VD in tissues which confirmed absorption and distribution parameters. Also, all the tested compounds were non-inhibitors to CYP2D6 and showed no hepatotoxicity.
The most potent anti-proliferative compound 15, as compared to the control drug Roscovitine. Where related pharmacokinetic parameters showed the probability of the same physicochemical characteristics and related behaviour inside the human body with similar biological effects. showed related pharmacokinetic properties showing differences mainly in the number of nHD due to differences in their chemical structure.
3.8. Structure–activity relationship
The structure–activity relationship of the novel compounds was studied based on their biological evaluation in addition to the molecular studies results. Where in the pyrazolo[3,4-d]pyrimidine scaffold in compounds (4–6), it was observed that the N5 pyrazolopyrimidine D-Xylose substitution showed potent activity against HCT-116 cell lines and moderate activity against HepG-2 cell lines compared to D- Glucose derivative or the unsubstituted derivative that was inactive against the three cell lines ().
While after the triazole ring fusion in pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin scaffold (7–15), the in vitro anti-proliferative activity results revealed that compounds 14 and 15 showed the best cytotoxic results that might be related to the thioglycosidic substitution on triazole ring. The thioalkyl substituents as the monoacetoxy alkyl group in compound 13 and diacetoxy alkyl in 11 showed the most potent cytotoxic results against HCT-116 cell lines with good activity against MCF-7 and moderate activity against HepG-2cell lines compared to other derivatives. While the dihydroxy alkyl derivative 10 showed only potent activity against MCF-7 cell lines. The acetonitrile derivative 8 showed potent activity against HepG-2cell lines, compared to the dimethoxy alkyl substitution in compound 9 with no activity against HepG-2cell lines. The monohydroxy alkyl derivative 12 was inactive against the three cell lines. While the thioxo derivative 7 showed only moderate activity against HepG-2cell line ().
summarised the SAR investigation of the newly synthesised compounds after their in vitro and in silico evaluation.
4. Conclusion
New series of pyrazolo[3,4-d]pyrimidine and pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidine compounds (4–13) and the thioglycoside derivatives (14, 15) designed as novel CDK2 targeting compounds was synthesised. Most of the compounds showed superior cytotoxic activities against MCF-7 and HCT-116, and moderate activity against HepG-2 compared to Sorafenib. Compounds 14 and 15 showed the best cytotoxic activities against the three cell lines. Also, compounds 11, 13, 14, and 15 revealed the most significant inhibitory activity against CDK2/cyclin A2. Compound 15 showed cell arrest at the S phase and apoptosis in Pre-G1 phase on HCT cells.
A molecular docking study revealed that all the potent anti-proliferative tested compounds were of comparable binding mode to that of the ligand. In silico ADMET studies and drug-likeness showed good pharmacokinetic properties of the new potent compounds. This helped in the prediction of structure requirements necessary for the observed antitumor activity.
5. Experimental
5.1. Chemistry
All melting points were measured using a Reichert Thermovar apparatus and are uncorrected. The IR spectra were recorded on a Perkin-Elmer model 1720 FTIR spectrometer for KBr disc. Routine NMR measurements were made on a JEOL ECA-500 II spectrometer with operating frequencies of 400 MHz for 1H. Chemical shifts were reported in δ scale (ppm) relative to TMS as a reference standard and the coupling constants J values are given in Hz. 13C NMR were recorded at 125 M Hz. The progress of the reactions was monitored by TLC using aluminium silica gel plates 60 F245. Spectral measurements and Elemental analyses were performed at the Micro-analytical centre at the Faculty of Science, Mansoura University, Mansoura. Compound 1 was synthesised according to a reported procedureCitation44.
5.1.1. 5-Amino-1-((4-fluorophenyl)glycyl)-1H-pyrazole-4-carbonitrile (2)
A solution of the acid hydrazide 1 (1.8 g, 10 mmol) and 2(ethoxymethylene)malononitrile (1.2 g, 10 mmole) in absolute ethanol (20 ml) was refluxed for 6 h, then cooled to room temperature. The resulted solid was recrystallized from ethanol to afford the pyrazole compound 2 as brown solid, yield 75%, m.p. 131–132 °C; IR (KBr): ύ/cm−1: 3340, 3255 (NH, NH2), 2205 (CN), 1665 (C=O), 1620 (C=N); 1H-NMR (DMSO-d6) (ppm): 8.25 (s, 1H, pyrazolo-H3), 7.46 (brs, 1H, NH, D2O exchangeable), 6.91 (d, J = 7.4 Hz, 2H, Ar-H), 6.55 (d, J = 7.4 Hz, 2H, Ar-H), 6.10 (brs, 2H, NH2, D2O exchangeable), 4.12 (s, 2H, CH2); 13C NMR (DMSO-d6) δ: (166.6), (158.6), (157.9), (145.4), (142.3), (131.6), (130.5), (126.4), (106.2), (54.1); MS (m/z): 259; Anal. Calc. for (C12H10FN5O): C, 55.60; H, 3.89; N, 27.02%; Found C, C, 55.67; H, 3.95; N, 27.08%.
5.1.2. Ethyl -N-(4-cyano-1-((4-fluorophenyl)glycyl)-1H-pyrazol-5-yl)formimidate (3)
A mixture of the substituted pyrazole derivative 2 (2.6 g, 10 mmole) and triethyl orthoformate (1.4 g, 10 mmole) in acetic anhydride (20 ml) was heated under reflux for 6 h, then cooled to room temperature. The precipitated formed was filtered and recrystallized by ethanol to afford the ester derivative 3 as Brown solid, yield 67%, m.p. 185–186 °C; IR (KBr): ύ/cm−1: 3318 (NH), 2210 (CN), 1655 (C=O), 1620 (C=N); 1H-NMR (DMSO-d6) (ppm): 8.27 (s, 1H, pyrazolo-H3), 8.19 (s, 1H, N = CH), 7.42 (brs, 1H, NH, D2O exchangeable), 6.91 (d, J = 7.4 Hz, 2H, Ar-H), 6.52 (d, J = 7.4 Hz, 2H, Ar-H), 4.13 (s, 2H, CH2), 3.61 (q, J = 5.2 Hz, 2H, CH2), 1.2 (t, J = 5.2 Hz, 3H, CH3); 13C NMR (DMSO-d6) δ: (166.3), (163.9), (159.4), (158.3), (147.2), (143.4), (131.9), (130.2), (126.4), (106.9), (64.2), (54.8), (18.3);MS (m/z): 315; Anal. Calc. for (C15H14FN5O2): C, 57.14; H, 4.48; N, 22.21%; Found C, 57.20; H, 4.52; N, 22.25%.
5.1.3. 1-(5-Amino-4-imino-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-1-yl)-2-((4-fluorophenyl)amino)ethan-1-one (4)
To solution of the ethyl ester derivative 3 (3.1 g, 10 mmol) in absolute ethanol (20 ml), hydrazine hydrate (99%, 1 ml, 20 mmol) was added dropwise. The reaction mixture was heated at reflux for 6 h then allowed to cool down to room temperature. The formed solid was separated by filtration and crystallised from methanol to provide the desired product as brown crystals (59%), m.p. = 168–169 °C; IR (KBr) cm−1: 3350–3230 (NH2, NH), 1660 (C = O), 1630 (C = N); 1H-NMR (DMSO-d6) (ppm): 11.42 (brs, 1H, NH, D2O exchangeable), 8.77 (brs, 1H, NH, D2O exchangeable), 8.51 (s, 1H, pyrimidine-H2), 7.98 (s, 1H, pyrazolo-H3), 7.56 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.47 (d, J = 8 Hz, 2H, phenyl-H3,H5), 5.51 (brs, 2H, NH2, D2O exchangeable), 4.12 (s, 2H, CH2); 13C NMR (DMSO-d6) δ: 168.7, 161.4, 153.5, 148.3, 143.1, 133.0, 129.4, 127.1, 122.2, 118.4, 55.9; MS (m/z) 301; Anal. Calc. for: (C13H12FN7O): C, 51.83; H, 4.01; N, 32.54%; Found: C, 51.88; H, 4.05; N, 32.58%.
5.1.3.4. General procedure a
A mixture of aminopyrazolopyrimidine compound 4 (3 g, 10 mmol), the aldose sugar namely; D-glucose or D-xylose [10 mmol suspended in water (1 ml)] and glacial acetic acid (0.5 ml) in ethanol (25 ml) was allowed to be refluxed for 6 h. Half of the amount of the solvent was evaporated under reduced pressure and the resulting solution was cooled to room temperature and then left to stand in a refrigerator at 5–8 °C overnight. The resulting precipitated solid was filtered, dried, and recrystallized by methanol to afford the N-glycosyl compounds 5 or 6, respectively.
5.1.4. 1-(4-Imino-5-((ß-D-glucopyranosyl)amino)-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-1-yl)-2-((4-fluorophenyl)amino)ethan-1-one (5)
Brown solid, yield 66%, m.p. 183–184 °C;.IR (KBr): ύ/cm−1: 3455–3440 (OH), 3325 (NH), 1668 (C = O), 1615 (C = N); 1H-NMR (DMSO-d6) (ppm): 11.25 (brs, 1H, NH, D2O exchangeable), 8.74 (brs, 1H, NH, D2O exchangeable), 8.51 (s, 1H, pyrimidine-H2), 7.79 (s, 1H, pyrazolo-H3), 7.60 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.47 (d, J = 8 Hz, 2H, phenyl-H3,H5), 6.71 (brs, 1H, NH, D2O exchangeable), 5.10–5.13 (m, 1H), 4.97 (brs, 1H, OH, D2O exchangeable), 4.66 (brs, 1H, OH, D2O exchangeable), 4.63–4.62 (m, 2H), 4.43 (brs, 2H, 2OH, D2O exchangeable), 4.13 (s, 2H, CH2), 3.87–3.93 (m, 3H), 3.59–3.65 (m, 1H); 13C NMR (DMSO-d6) δ: 169.0, 161.4, 155.5, 146.8, 143.0, 134.3, 128.8, 127.1, 122.5, 118.3, 93.0, 83.6, 75.7, 72.2, 71.8, 62.1, 55.5; MS (m/z) 463; Anal. Calc. for: (C19H22FN7O6): C, 49.24; H, 4.79; N, 21.16%; Found: C, 49.31; H, 4.84; N, 21.20%.
5.1.5. 1-(4-Imino-5-((ß-D-xylopyranosyl)amino)-4,5-dihydro-1H-pyrazolo[3,4-d]pyrimidin-1-yl)-2-((4-fluorophenyl)amino)ethan-1-one (6)
Brown solid, yield 72%, m.p. 122–123 °C; IR (KBr): ύ/cm−1: 3460–3450 (OH), 3325 (NH), 1668 (C = O), 1622 (C = N); 1H-NMR (DMSO-d6) (ppm): 11.25 (brs, 1H, NH, D2O exchangeable), 8.72 (brs, 1H, NH, D2O exchangeable), 8.51 (s, 1H, pyrimidine-H2), 8.02 (s, 1H, pyrazolo-H3), 7.58 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.47 (d, J = 8 Hz, 2H, phenyl-H3,H5), 6.68 (brs, 1H, NH, D2O exchangeable), 5.08–5.10 (m, 1H), 4.83 (brs, 1H, OH, D2O exchangeable), 4.43 (brs, 2H, 2OH, D2O exchangeable), 4.12 (s, 2H, CH2), 3.90–3.96 (m, 2H), 3.71–3.76 (m, 3H); 13C NMR (DMSO-d6) δ: 168.7, 161.4, 155.4, 146.8, 143.0, 134.3, 128.8, 127.1, 122.5, 118.3, 93.0, 83.6, 75.7, 70.5, 62.1, 55.5; MS (m/z) 433; Anal. Calc. for: (C18H20FN7O5): C, 49.88; H, 4.65; N, 22.62%; Found: C, 49.95; H, 4.70; N, 22.67%.
5.1.6. 1-(2-Thioxo-2,3-dihydro-7H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-7-yl)-2-((4-fluorophenyl)amino)ethan-1-one (7)
To a well-stirred mixture of compound 4 (3 g, 10 mmol), potassium hydroxide (0.67 g, 12 mmol) in ethanol 25 (mL), carbon disulphide (0.4 ml, 12 mmol) was added dropwise over 30 min. The reaction mixture was heated in the water bath at 70 °C for 1 h and then refluxed for 10 h. The solvent was evaporated under a vacuum, then poured onto ice-cold water (50 ml) and the solution was acidified, the precipitate formed was filtered, washed with water, and dried. Crystallisation from ethanol gave the thione derivative 7 as yellow solid, yield 74%, m. p 178–179 °C; IR (KBr): ύ/cm−1: 3328–3310 (NH), 1670 (C = O), 1616 (C = N); 1H-NMR (DMSO-d6) (ppm): 11.66 (brs, 1H, NH, D2O exchangeable), 8.96 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 8.13 (s, 1H, pyrazolo-H3), 7.59 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.47 (d, J = 8 Hz, 2H, phenyl-H3,H5), 4.12 (s, 2H, CH2); 13C NMR (DMSO-d6) δ: 176.4, 170.5, 161.7, 154.2, 146.5, 143.0, 131.2, 129.1, 127.1, 122.2, 118.3, 56.2; MS (m/z) 343; Anal. Calc. for: (C14H10FN7OS): C, 48.98; H, 2.94; N, 28.56%; Found: C, 49.08; H, 3.01; N, 28.62%.
5.1.7. 2-((7-((4-Fluorophenyl)glycyl)-7H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-2-yl)thio)acetonitrile (8)
A mixture of 7 (3.4 g, 10 mmol), chloroacetonitrile (0.7 g, 11 mmol) and anhydrous potassium carbonate (1.4 g, 10 mmol) in dimethyl formamide (20 ml) was stirred for 8 h at room temperature. The reaction mixture was poured into ice-cold water (50 ml) with vigorous stirring for 30 min and the formed precipitate was filtered, dried and recrystallized from ethanol to give the nitrile derivative 8 as brown powder, yield 63%; m.p > 300 °C; IR (KBr): ύ/cm−1: 3315 (NH), 2218 (CN), 1655 (C = O), 1612 (C = N); 1H-NMR (DMSO-d6) (ppm): 8.74 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 8.0 (s, 1H, pyrazolo-H3), 7.58 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.47 (d, J = 8 Hz, 2H, phenyl-H3,H5), 4.54 (s, 2H), 4.12 (s, 2H, CH2); 13C NMR (DMSO-d6) δ: 169.8, 166.3, 161.4, 154.2, 146.8, 142.7, 131.2, 129.1, 127.1, 122.5, 118.06, 118.01, 54.5, 17.0; MS (m/z) 382; Anal. Calc. for: (C16H11FN8OS): C, 50.26; H, 2.90; N, 29.31%; Found: C, 50.33; H, 2.98; N, 29.38%.
5.1.7.1. General procedure b
A solution of pyrazolopyrimidine derivative 7 (3.4 g, 10 mmol), chloro-acetaldhyde dimethyl acetal (10 mmol), or 3-chloropropane-1,2-diol, and anhydrous potassium hydroxide (0.67 g, 10 mmol) in ethanol (25 ml) were heated under reflux with stirring for 10 h. The solvent was evaporated under reduced pressure and ice-cold water was added to the remaining residue with continuous stirring to afford a precipitate which was filtered, dried, and recrystallized from ethanol-water mixture 1:1 to afford the dimethoxy product 9 and 10, respectively.
5.1.8. 1-(2-((2,2-Dimethoxyethyl)thio)-7H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-7-yl)-2-((4-fluorophenyl)amino)ethan-1-one (9)
Pale yellow solid, yield 65%, m.p. 190–192 °C; IR (KBr): ύ/cm−1: 3295 (NH), 1668 (C = O), 1618 (C = N); 1H-NMR (DMSO-d6) (ppm): 8.74 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 8.0 (s, 1H, pyrazolo-H3), 7.60 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.47 (d, J = 8 Hz, 2H, phenyl-H3,H5), 4.87–4.90 (m, 1H, OCH), 4.12 (s, 2H), 3.65 (s, 6H), 3.09–3.12 (m, 2H, SCH2); 13C NMR (DMSO-d6) δ: 169.8, 166.3, 159.7, 157.2, 146.8, 142.7, 131.9, 129.4, 127.1, 122.9, 118.8, 96.8, 67.3, 55.9, 35.3; MS (m/z) 431; Anal. Calc. for: (C18H18FN7O3S): C, 50.11; H, 4.21; N, 22.73%; Found: C, 50.17; H, 4.26; N, 22.78%.
5.1.9. 1-{2-[(2,3-Dihydroxypropyl)thio]-7H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-7-yl}-2-((4-fluorophenyl)amino)ethan-1-one (10)
Brown solid, yield 72%, m.p > 300 °C; IR (KBr): ύ/cm−1: 3425 (OH), 3315 (NH), 1676 (C = O), 1622 (C = N); 1H-NMR (DMSO-d6) (ppm): 8.72 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 8.13 (s, 1H, pyrazolo-H3), 7.58 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.47 (d, J = 8 Hz, 2H, phenyl-H3,H5), 4.76 (brs, 1H, OH, D2O exchangeable), 4.57 (brs, 1H, OH, D2O exchangeable), 4.12 (s, 2H), 3.70–3.73 (m, 1H), 3.45–3.48 (m, 2H), 3.15–3.17 (m, 2H); 13C NMR (DMSO-d6) δ: 170.1, 166.3, 159.3, 155.2, 146.8, 142.7, 131.6, 129.4, 127.4, 122.2, 118.0, 71.2, 65.6, 55.5, 35.3; MS (m/z) 417; Anal. Calc. for: (C17H16FN7O3S): C, 48.92; H, 3.86; N, 23.49%; Found: C, 48.96; H, 3.89; N, 23.54%.
5.1.10. 3-((7-((4-Fluorophenyl)glycyl)-7H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-2-yl)thio)propane-1,2-diyl diacetate (11)
A mixture of compound 10 (4.1 g, 10 mmol) and acetic anhydride (5 ml) in pyridine (15 ml) was stirred at room temperature for 8 h then allowed to cool down to room temperature and poured on cold water (30 ml) with vigorous stirring for 2 h. The formed precipitate was filtered, washed with water then with potassium hydrogen carbonate and dried. Crystallisation form methanol afford the acetylated product as Brown solid, yield 73%, m.p. 260–262 °C; IR (KBr): ύ/cm−1: 3318 (NH), 1672 (C = O); 1H-NMR (DMSO-d6) (ppm): 8.80 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 8.07 (s, 1H, pyrazolo-H3), 7.60 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.48 (d, J = 8 Hz, 2H, phenyl-H3,H5), 4.32–4.37 (m, 1H), 4.12 (s, 2H), 3.42–3.48 (m, 2H), 3.12–3.22 (m, 2H), 2.20 (s, 6H); 13C NMR (DMSO-d6) δ: 172.2, 171.8, 169.7, 165.6, 159.0, 153.8, 146.8, 142.7, 131.2, 129.4, 127.4, 121.8, 115.2, 74.2, 67.7, 55.2, 37.1, 23.6, 17.3, 17.0; MS (m/z) 501; Anal. Calc. for: (C21H20FN7O5S): C, 50.30; H, 4.02; N, 19.55%; Found: C, 50.38; H, 4.09; N, 19.62%.
5.1.11. 1-(2-((2-Hydroxyethyl)thio)-7H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-7-yl)-2-((4-fluorophenyl)amino)ethan-1-one (12)
A solution of pyrazolopyrimidine derivative 7 (3.4 g, 10 mmol), 1-chloroethanol (0.8 g, 10 mmol) and anhydrous potassium hydroxide (0.67 g, 10 mmol) in ethanol (25 ml) was heated under reflux for 12 h. The solvent was evaporated under reduced pressure and ice-cold water was added to the remaining residue with continuous stirring to afford a precipitate which was filtered, dried and recrystallized from methanol-water mixture 1:1 to provide the hydroxyethoxy compound 12 as Brown solid, yield 65%, m.p. 199–200 °C; IR (KBr): ύ/cm−1: 3415 (OH), 3345 (NH), 1672 (C = O), 1618 (C = N); 1H-NMR (DMSO-d6) (ppm): 8.72 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 8.03 (s, 1H, pyrazolo-H3), 7.58 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.44 (d, J = 8 Hz, 2H, phenyl-H3,H5), 4.81 (brs, 1H, OH, D2O exchangeable), 4.12 (s, 2H), 3.68–3.71 (m, 2H), 3.48–3.51 (m, 2H); 13C NMR (DMSO-d6) δ: 170.1, 165.9, 159.4, 153.8, 146.5, 142.7, 131.2, 129.4, 127.1, 122.2, 115.3, 66.3, 55.2, 32.6; MS (m/z) 387; Anal. Calc. for: (C16H14FN7O2S): C, 49.61; H, 3.64; N, 25.31%; Found: C, 49.69; H, 3.71; N, 25.37%.
5.1.12. 2-((7-((4-Fluorophenyl)glycyl)-7H-pyrazolo[4,3-e][1,2,4]triazolo[1,5-c]pyrimidin-2-yl)thio)ethyl acetate (13)
To a mixture of compound 12 (3.8 g, 10 mmol) and acetic anhydride (5 ml) in pyridine (15 ml) was stirred at room temperature for 14 h then cooled to room temperature. Ice-cold water (35 ml) was added with vigorous stirring for 3 h. The formed precipitate was filtered, washed with water then with potassium hydrogen carbonate and dried. Crystallisation form methanol afford the acetylated products as brown powder, yield 67%, m.p. 172–173 °C; IR (KBr): ύ/cm−1: 3318 (NH), 1675 (C = O), 1620 (C = N); 1H-NMR (DMSO-d6) (ppm): 8.80 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 8.03 (s, 1H, pyrazolo-H3), 7.58 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.44 (d, J = 8 Hz, 2H, phenyl-H3,H5), 4.12 (s, 2H), 3.93–3.96 (m, 2H), 3.51–3.54 (m, 2H), 2.32 (s, 3H); 13C NMR (DMSO-d6) δ: 171.5, 169.4, 165.9, 159.7, 153.4, 146.1, 142.7, 131.2, 129.1, 127.1, 122.5, 115.3, 67.3, 55.5, 17.3; MS (m/z) 429; Anal. Calc. for: (C18H16FN7O3S): C, 50.35; H, 3.76; N, 22.83%; Found: C, 50.39; H, 3.79; N, 22.87%.
5.1.12.1. General procedure c
To a suspension of potassium hydroxide (0.67 g, 10 mmol) in water (1 ml), a solution of pyrazolopyrimidine derivative 7 (3.4 g, 10 mmol) in acetone (20 ml) was added with continuous stirring for about 15 min, then a solution of the glycosyl bromide namely; 2,3,4,6-tetra-O-acetyl-α-D-glucopyranosyl bromide or 2,3,4,6-tetra-O-acetyl-α-D-galactopyranosyl bromide (12 mmol) in acetone (10 ml) was added portion-wise and the resulting mixture was stirred at room temperature for 5 h. The solvent was evaporated under reduced pressure and the residue was washed with cold water (15 ml) to remove any remained inorganic substance. Extraction of the product with ethyl acetate (50 ml) followed by removal of the solvent afforded a solid material which was treated with petroleum ether/diethyl ether mixture 3:1 (30 ml) with stirring for 15 min for further purification. The formed solid product was filtered and dried to give compounds 14 or 15, respectively.
5.1.13. 1-(2-((Tetra-O-acetyl-D-glucopyranosyl)thio)-7H-pyrazolo[4,3e][1,2,4]triazolo[1,5-c]pyrimidin-7-yl)-2-((4fluorophenyl)amino)ethan-1-one (14)
Pale yellow solid, yield 79%, m.p. 168–169 °C; IR (KBr): ύ/cm−1: 3330 (NH), 1670 (C = O), 1740 (C = O), 1620 (C = N); 1H-NMR (DMSO-d6) (ppm): 8.66 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 7.98 (s, 1H, pyrazolo-H3), 7.58 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.42 (d, J = 8 Hz, 2H, phenyl-H3,H5), 5.71–5.76 (m, 1H), 5.01–5.07 (m, 2H), 4.29–4.34 (m, 3H), 4.12 (s, 2H), 3.68–3.73 (m, 1H), 2.23 (s, 12H); 13C NMR (DMSO-d6) δ: 173.6, 173.2, 171.8, 171.5, 169.4, 165.3, 159.4, 153.8, 146.1, 142.7, 131.6, 129.1, 127.1, 122.2, 115.6, 84.3, 80.8, 76.7, 71.5, 67.3, 63.1, 55.5, 18.7, 18.3, 17.7, 17.3; MS (m/z) 673; Anal. Calc. for: (C28H28FN7O10S): C, 49.92; H, 4.19; N, 14.56%; Found: C, 49.97; H, 4.25; N, 14.59%.
5.1.14. 1-(2-((Tetra-O-acetyl-D-galactopyranosyl)thio)-7H-pyrazolo[4,3e][1,2,4]triazolo[1,5-c]pyrimidin-7-yl)-2-((4-fluorophenyl)amino)ethan-1-one (15)
Pale yellow solid, yield 75%, m.p. 172–173 °C; IR (KBr): ύ/cm-1: 3325 (NH), 1745 (C = O), 1665 (C = O), 1620 (C = N); 1H-NMR (DMSO-d6) (ppm): 8.67 (brs, 1H, NH, D2O exchangeable), 8.50 (s, 1H, pyrimidine-H2), 7.98 (s, 1H, pyrazolo-H3), 7.58 (d, J = 8 Hz, 2H, phenyl-H2,H6), 7.44 (d, J = 8 Hz, 2H, phenyl-H3,H5), 5.82–5.74 (m, 1H), 5.01–5.07 (m, 2H), 4.29–4.38 (m, 3H), 4.12 (s, 2H), 3.68–3.73 (m, 1H), 2.23 (s, 12H); 13C NMR (DMSO-d6) δ: 173.6, 173.2, 171.8, 171.5, 169.4, 165.6, 159.4, 153.8, 146.1, 142.7, 131.6, 129.1, 127.1, 122.2, 115.6, 84.3, 80.8, 76.7, 71.5, 67.3, 63.1, 55.5, 18.7, 18.3, 17.7, 17.3; MS (m/z) 673; Anal. Calc. for: (C28H28FN7O10S): C, 49.92; H, 4.19; N, 14.56%; Found: C, 49.99; H, 4.27; N, 14.61%.
5.2. Biological assays
5.2.1. In vitro anti-proliferative activity
The In vitro cytotoxicity of all the newly synthesised compounds against three cancer cell lines named: Human breast adenocarcinoma (MCF-7) cell line, hepatocellular carcinoma (HepG-2), and colorectal carcinoma (HCT-116) were performed. Cells were purchased from the American Type Culture Collection (ATCC, Rockville, MD, USA) (Supplementary Material). Results were compared to the reference Sorafenib. Also, the in vitro cytotoxic effect of compound 15 against WI-38 normal lung cells was compared to Staurosporine as a reference standard. The MTT assay was performed according to published procedureCitation37,Citation38. Briefly, 5000–10,000 cells per well were plated in a 96-well plate and allowed for growth for 24 h, then treated with the media of increased concentrations of tested compounds in 100 µl amount, complete growth medium was further mixed with 100 µl for each compound per well for 48 h before the assay. Then, the media were withdrawn, and 100 µl of MTT was added to each well, which was incubated for 4 h. The formed formazan crystals were solubilised by the addition of 100 µl of dimethyl sulfoxide (DMSO) solution. The cell viability was evaluated by measuring the optical density (OD) per well of the developed purple colour using spectrophotometric determination at 570 nm using an ELISA microplate reader (Epoc-2 C micro-plate reader, Bio Tek, VT, USA). The optical density of the produced colour was measured at 570 nm. The IC50 values (the required concentration to inhibit cell viability by 50%) were calculated. The data was expressed as a percentage of control cells (100 percent of cell viability).
5.2.2. CDK2/cyclin A2 assay
The In vitro assay of CDK2/cyclin A2 protein kinase was carried out on all the synthesised compounds 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, and 15. The assay was proceeded in Egypt applying Kinase-Glo Plus luminescence kinase Assay kit (Promega)Citation39. The Protocol steps were applied by enzyme and substrate dilution. Where, 1 µl of inhibitor, 5% DMSO, 2 µl of the enzyme, and 2 µl of substrate/ATP mix were added after ATP and inhibitors were diluted in Kinase Buffer. Incubation for 10 min at room temperature was allowed, then 5 µl of Reagent ADP‐Glo™ was added and incubated for another 40 min. After that, an amount equivalent to 10 µl of Kinase Detection Reagent was added to be incubated at room temperature for another 30 min. Luminescence was (where Integration time range 0.5–1 s). The luminescent signal is in direct relation to the present quantity of ATP and in inverse relation to the activity of the kinase enzyme.
5.2.3. Flow cytometry cell cycle analysis
The cell cycle analysis protocol was performed on compound 14 on HCT-116 cells. The test is based on the content of DNA measurement via staining using propidium iodide. Cells were first washed in PBS before being kept at 4 °C for 3 min through the dropwise using vortex addition of cold 70% ethanol, this is to avoid cell clumping and to ensure fixation as well. Then 50 µl from a stock of 100 µg/ml ribonuclease was added to selectively stain only DNA. Finally, an amount of 200 µl from a stock solution of 50 µg/ml of propidium iodide was addedCitation40.
5.2.4. Flow cytometric analysis of apoptosis
Annexin V—FITC—apoptosis detection kit (PN IM3546) was used to detect apoptosis in treated cells. Flow cytometric analysis was further performed according to manufacturer protocol. Where HCT-116 cells were allowed to grow in a 25 cm3 flask until 70–80% confluence. Then HCT-116 cells were treated with compound 15 for 48 h followed by a wash in PBS and suspended in 1× binding buffer. One µL of annexin V-FITC solution together with 5 µL of dissolved PI were added to 100 µL of cell suspensions, then incubated for 15 min in the dark. An aliquot of 400 µL of ice-cold 1× binding buffer was gently added and mixed. The flow cytometric analysis method for the determination of the percentage of apoptotic cells was performed on a COULTER® EPICS® XL™ Flow Cytometer (USA)Citation41.
5.3. Molecular modeling studies
A molecular docking study using CDOCKER protocol in Discovery Studio 4.0 Software was carried out. The targeted compounds were docked into the CDK2 active site. The X-ray crystallographic structure of CDk2 complexed with Roscovitine (PDB ID: 2A4L) was downloaded from PDBCitation9,Citation12. The binding mode of the designed compounds was studied to explain their biological results and to detect the essential hydrogen bonding with Leu83. Where the best pose out of ten for each compound was selected compared to the ligand binding mode. In silico ADMET studies using Discovery Studio 4.0 Software and ADMETlab web tool were carried out to predict the pharmacokinetic properties of the targeted compoundsCitation43, which helped in structure requirements prediction for the observed antitumor activity.
Supplemental Material
Download PDF (2.1 MB)Acknowledgements
The authors extend their appreciation to the Research center at Al Maarefa University for funding this work. Acknowledgement for Future University in Egypt (FUE) Computer Aided Drug Design labs.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Supporting data is supplied with the manuscript.
References
- Baillache DJ, Unciti-Broceta A. Recent developments in anticancer kinase inhibitors based on the pyrazolo [3,4-d] pyrimidine scaffold. RSC Med Chem 2020;11:1112–35. https://pubs.rsc.org/en/content/articlelanding/2020/md/d0md00227e
- Abdelgawad MA, Elkanzi NA, Nayl AA, et al. Targeting tumor cells with pyrazolo [3,4-d] pyrimidine scaffold: a literature review on synthetic approaches, structure activity relationship, structural and target-based mechanisms. Arab J Chem 2022;15:103781.
- Cohen P. Protein kinases—the major drug targets of the twenty-first century? Nat Rev Drug Discov 2002;1:309–15.
- Abd El-Sattar NE, Badawy EH, AbdEl-Hady WH, et al. Design and synthesis of new CDK2 inhibitors containing thiazolone and thiazolthione scafold with apoptotic activity. Chem Pharm Bull 2021;69:106–17.
- Chen J, Pang L, Wang W, et al. Decoding molecular mechanism of inhibitor bindings to CDK2 using molecular dynamics simulations and binding free energy calculations. J Biomol Struct Dyn 2020;38:985–96.
- Liang SS, Liu XG, Cui YX, et al. Molecular mechanism concerning conformational changes of CDK2 mediated by binding of inhibitors using molecular dynamics simulations and principal component analysis. SAR QSAR Environ Res 2021;32:573–94.
- Kontopidis G, McInnes C, Pandalaneni SR, et al. Differential binding of inhibitors to active and inactive CDK2 provides insights for drug design. Chem Biol 2006;13:201–11.
- Li Y, Zhang J, Gao W, et al. Insights on structural characteristics and ligand binding mechanisms of CDK2. Int J Mol Sci 2015;16:9314–40.
- Husseiny EM. Synthesis, cytotoxicity of some pyrazoles and pyrazolo[1,5-a]pyrimidines bearing benzothiazole moiety and investigation of their mechanism of action. Bioorg Chem 2020;102:104053.
- Wee S, Dhanak D, Li H, Armstrong, et al. Targeting epigenetic regulators for cancer therapy. Ann N Y Acad Sci 2014;1309:30–6.
- Cherukupalli S, Karpoormath R, Chandrasekaran B, et al. An insight on synthetic and medicinal aspects of pyrazolo [1,5-a] pyrimidine scaffold. Eur J Med Chem 2017;126:298–352.
- Ali GM, Ibrahim DA, Elmetwali AM, et al. Design, synthesis and biological evaluation of certain CDK2 inhibitors based on pyrazole and pyrazolo [1,5-a] pyrimidine scaffold with apoptotic activity. Bioorg Chem 2019;86:1–4.
- Farag AM, Fahim AM. Synthesis, biological evaluation and DFT calculation of novel pyrazole and pyrimidine derivatives. J Mol Struct 2019;1179:304–14.
- Gökhan-Kelekçi N, Yabanoğlu S, Küpeli E, et al. A new therapeutic approach in Alzheimer disease: some novel pyrazole derivatives as dual MAO-B inhibitors and antiinflammatory analgesics. Bioorg Med Chem 2007;15:5775–86.
- Nithyabalaji R, Krishnan H, Sribalan R. Synthesis, molecular structure and multiple biological activities of N-(3-methoxyphenyl)-3-(pyridin-4-yl)-1H-pyrazole-5-carboxamide. J Mol Struct 2019;1186:1–10.
- Hernández-Vázquez E, Salgado-Barrera S, Ramírez-Espinosa JJ, et al. Synthesis and molecular docking of N′-arylidene-5-(4-chlorophenyl)-1-(3,4-dichlorophenyl)-4-methyl-1H-pyrazole-3-carbohydrazides as novel hypoglycemic and antioxidant dual agents. Bioorg Med Chem 2016;24:2298–306.
- Nassar IF, El Farargy AF, Abdelrazek FM, et al. Design, synthesis and anticancer evaluation of novel pyrazole, pyrazolo [3,4-d] pyrimidine and their glycoside derivatives. Nucleosides Nucleotides Nucleic Acids 2017;36:275–91.
- Nassar IF, Atta-Allah SR, Elgazwy AS. A convenient synthesis and molecular modeling study of novel pyrazolo[3,4-d]pyrimidine and pyrazole derivatives as anti-tumor agents. J Enzyme Inhib Med Chem 2015;30:396–405.
- Rashad AE, Mahmoud AE, Ali MM. Synthesis and anticancer effects of some novel pyrazolo [3,4-d] pyrimidine derivatives by generating reactive oxygen species in human breast adenocarcinoma cells. Eur J Med Chem 2011;46:1019–26.
- Hassan GS, Kadry HH, Abou-Seri SM, et al. Synthesis and in vitro cytotoxic activity of novel pyrazolo [3,4-d] pyrimidines and related pyrazole hydrazones toward breast adenocarcinoma MCF-7 cell line. Bioorg Med Chem 2011;19:6808–17.
- El-Naggar M, Hassan AS, Awad HM, et al. Design, synthesis and antitumor evaluation of novel pyrazolopyrimidines and pyrazoloquinazolines. Molecules 2018;23:1249.
- Radi M, Dreassi E, Brullo C, et al. Design, synthesis, biological activity, and ADME properties of pyrazolo [3,4-d] pyrimidines active in hypoxic human leukemia cells: a lead optimization study. J Med Chem 2011;54:2610–26.
- Curran KJ, Verheijen JC, Kaplan J, et al. Pyrazolopyrimidines as highly potent and selective, ATP-competitive inhibitors of the mammalian target of rapamycin (mTOR): optimization of the 1-substituent. Bioorg Med Chem Lett 2010;20:1440–4.
- Li Y, Gao W, Li F, et al. An in silico exploration of the interaction mechanism of pyrazolo [1,5-a] pyrimidine type CDK2 inhibitors. Mol Biosyst 2013;9:2266–81.
- Almehmadi SJ, Alsaedi AM, Harras MF, et al. Synthesis of a new series of pyrazolo[1,5-a]pyrimidines as CDK2 inhibitors and anti-leukemia. Bioorg Chem 2021;117:105431.
- Ismail NS, Ali EM, Ibrahim DA, et al. Pyrazolo [3,4-d] pyrimidine based scaffold derivatives targeting kinases as anticancer agents. Future J Pharm Sci 2016;2:20–30.
- Chauhan M, Kumar R. Medicinal attributes of pyrazolo[3,4-d]pyrimidines: a review. Bioorg Med Chem 2013;21:5657–68.
- Schenone S, Radi M, Musumeci F, et al. Biologically driven synthesis of pyrazolo[3,4-d]pyrimidines as protein kinase inhibitors: an old scaffold as a new tool for medicinal chemistry and chemical biology studies. Chem Rev 2014;114:7189–238.
- Bocci G, Fioravanti A, La Motta C, et al. Antiproliferative and proapoptotic activity of CLM3, a novel multiple tyrosine kinase inhibitor, alone and in combination with SN-38 on endothelial and cancer cells. Biochem Pharmacol 2011;81:1309–16.
- Abdelgawad MA, Bakr RB, Alkhoja OA, et al. Design, synthesis and antitumor activity of novel pyrazolo [3,4-d] pyrimidine derivatives as EGFR-TK inhibitors. Bioorg Chem 2016;66:88–96.
- Le Brazidec JY, Pasis A, Tam B, et al. Synthesis, SAR and biological evaluation of 1,6-disubstituted-1H-pyrazolo[3,4-d]pyrimidines as dual inhibitors of Aurora kinases and CDK1. Bioorg Med Chem Lett 2012;22:2070–4.
- Cherukupalli S, Chandrasekaran B, Aleti RR, et al. Synthesis of 4, 6-disubstituted pyrazolo [3,4-d] pyrimidine analogues: cyclin-dependent kinase 2 (CDK2) inhibition, molecular docking and anticancer evaluation. J Mol Struct 2019;1176:538–51.
- Rahmouni A, Souiei S, Belkacem MA, et al. Synthesis and biological evaluation of novel pyrazolopyrimidines derivatives as anticancer and anti-5-lipoxygenase agents. Bioorg Chem 2016;66:160–8.
- Cherukupalli S, Chandrasekaran B, Kryštof V, et al. Synthesis, anticancer evaluation, and molecular docking studies of some novel 4,6-disubstituted pyrazolo[3,4-d]pyrimidines as cyclin-dependent kinase 2 (CDK2) inhibitors. Bioorg Chem 2018;79:46–59.
- Elgazwy AS, Ismail NS, Elzahabi HS. A convenient synthesis and molecular modeling study of novel purine and pyrimidine derivatives as CDK2/cyclin A3 inhibitors. Bioorg Med Chem 2010;18:7639–50.
- Abdel‐Aal MT, El‐Sayed WA, El‐Kosy SM, et al. Synthesis and antiviral evaluation of novel 5‐(N‐aryl‐aminomethyl‐1,3,4‐oxadiazol‐2‐yl) hydrazines and their sugars, 1,2,4‐triazoles, tetrazoles and pyrazolyl derivatives. Arch Pharm Chem Life Sci 2008;341:307–13.
- Girgis AS, Stawinski J, Ismail NS, et al. Synthesis and QSAR study of novel cytotoxic spiro[3H-indole-3,2′(1′H)-pyrrolo[3,4-c]pyrrole]-2,3′,5′ (1H,2′aH,4′H)-triones. Eur J Med Chem 2012;47:312–22.
- Fawzy IM, Youssef KM, Lasheen DS, et al. Design, synthesis and 3D QSAR based pharmacophore study of novel imatinib analogs as antitumor-apoptotic agents. Future Med Chem 2018;10:1421–33.
- Hennek J, Alves J, Yao E, et al. Bioluminescent kinase strips: a novel approach to targeted and flexible kinase inhibitor profiling. Anal Biochem 2016;495:9–20.
- Pozarowski P, Darzynkiewicz Z. Analysis of cell cycle by flow cytometry. In: Checkpoint controls and cancer. Humana Press; 2004. p. 301–11. doi:10.1385/1-59259-811-0:301.
- Ismail A, Doghish AS, Elsadek BE, et al. Hydroxycitric acid potentiates the cytotoxic effect of tamoxifen in MCF-7 breast cancer cells through inhibition of ATP citrate lyase. Steroids 2020;160:108656.
- Bitew M, Desalegn T, Demissie TB, et al. Pharmacokinetics and drug-likeness of antidiabetic flavonoids: molecular docking and DFT study. PLoS One 2021;16:e0260853.
- Dong J, Wang NN, Yao ZJ, et al. ADMETlab: a platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J Cheminformatics 2018;10:1.
- Zou W, Ma X, Hua W, et al. BRIP1 inhibits the tumorigenic properties of cervical cancer by regulating RhoA GTPase activity. Oncol Lett 2016;11:551–8.