Abstract
A new series of 1H-pyrrole (6a–c, 8a–c), pyrrolo[3,2-d]pyrimidines (9a–c) and pyrrolo[3,2-e][1, 4]diazepines (11a–c) were designed and synthesised. These compounds were designed to have the essential pharmacophoric features of EGFR Inhibitors, they have shown anticancer activities against HCT116, MCF-7 and Hep3B cancer cells with IC50 values ranging from 0.009 to 2.195 µM. IC50 value of doxorubicin is 0.008 µM, compounds 9a and 9c showed IC50 values of 0.011 and 0.009 µM respectively against HCT-116 cells. Compound 8b exerted broad-spectrum activity against all tested cell lines with an IC50 value less than 0.05 µM. Compound 8b was evaluated against a panel of kinases. This compound potently inhibited CDK2/Cyclin A1, DYRK3 and GSK3 alpha kinases with 10–23% compared to imatinib (1–10%). It has also arrested the cell cycle of MCF-7 cells at the S phase. Its antiproliferative activity was further augmented by molecular docking into the active sites of EGFR and CDK2 cyclin A1.
1. Introduction
Cancer is clearly associated with an increased incidence and rates of mortality, in addition to its devastating social and economic effectsCitation1,Citation2. Cancer is a generic term for a large group of diseases that can affect any part of the human bodyCitation2,Citation3. The complexity of cancer pathologies manifests in oncogenic mutations, severe and occasionally fatal drug side effects, multi-drug resistance and activation of compensatory pathwaysCitation4–6. Thus, there is an urgent need to develop more efficient anticancer candidates, relying on various biological and molecular aspects of neoplastic transformation. Accordingly, many efforts were done to reach promising anticancer candidatesCitation1,Citation7–10.
Protein kinases constitute one of the largest and most functionally diverse gene families that control a diverse set of cellular processes. Thus, they perform a major role in the proliferation, metastasis and survival of human tumour cellsCitation11. Tyrosine kinase is the most important protein kinases sub-family. This family comprises epidermal growth factor receptor (EGFR)Citation12, vascular endothelial growth factor receptor (VEGFR)Citation13, fibroblast growth factor receptor (FGFR)Citation14 and cyclin-dependent kinases (CDK)Citation15. Three of the protein kinases have been crystallised in active conformations (cAPK, PhK, and CK1), one in a partially active conformation (CDK2 in complex with cyclin A), and five in inactive conformations (CDK2, MAPK, IRK, twitchin kinase, and CaMKI). The structures of the kinases in the active conformation all showed equivalent positions for essential catalytic site residues Lys-72, Asp-166, and Asp-184 in cAPK. Their positions and the correct orientation of the Mg2+/ATP and the protein substrate appear crucial for catalysis and are dependent upon the tertiary structure of both lobes and the correct relative orientation of the lobes. In the partially active CDK2–cyclin A complex, the charge grouping is compensated by glutamate, Glu-162, (two residues removed from the phosphorylatable threonine Thr-160) and by interactions of arginyl residues with the main chain carbonyl groups from cyclin A. In PhK, there is only one basic group, the arginine adjacent to the catalytic base, and charge compensation can be satisfactorily accomplished by a carboxylate groupCitation16.
The epidermal growth factor receptor (EGFR) is a transmembrane protein receptor endowed with tyrosine kinase activity, occupying an important position in cancer progression and tumour cell biology. EGFR plays a role in cellular phenotyping and provides tumour cells with significant growthCitation17. Elevated levels of EGFR and its associated ligands (EGF and transforming growth factor (TGF)) have been found to be a common feature of a variety of cancer types. In many cases, abnormal EGFR activation appears to be a key element in carcinogenesis and a major driving force for cancer growthCitation18. Increased EGFR expression is thus expected to be a powerful prognostic factor in a variety of tumour forms and blocking its cellular functions looks to have significant therapeutic effectsCitation19. As a result, EGFR is being regarded as a viable target for the development of novel anticancer drugsCitation20–23.
The cyclin-dependent kinases (CDKs) are a group of enzymes involved in cell cycle progression and cellular proliferationCitation24. They work by phosphorylating critical serine and threonine residues in host proteins, which then can activate themCitation25,Citation26. It is commonly believed that inhibiting CDKs could help the limitation of the uncontrolled cellular proliferation seen in some malignanciesCitation27. The majority of CDK inhibitors bind to the ATP pocket as ATP-competitive inhibitors with essential structural hydrogen-bonding motifsCitation28. Cyclin-dependent kinase 2 (CDK2) belongs to the serine/threonine protein kinase family, and the CDK2 activity is found to be typically high in different types of human cancers. Studies have reported that CDK2 overexpression indicates poor prognosis in patients with HCC, and inhibition of CDK2 activity could reverse the malignant phenotype of cancer cells. Many studies revealed a key role of CDK2 in EGF-induced cell transformation and the associated signal transduction pathwaysCitation29. The literature survey revealed that EGFR and CDK2 were the key targets for many antitumor agents e.g. cinobufaginCitation30. In addition, biological and computational evidence supported that anticancer agents such as benzamide-substituted chalcones exerted their anti-proliferative effects via dual EGFR/CDK2 inhibitory activitiesCitation31.
It is worth mentioning that, dual-specificity tyrosine phosphorylation-regulated kinase 3 (DYRK3) is a regulator of phase transition during mitosisCitation32. Furthermore, it is able to promote mTORC1 activity, which is associated with resistance to EGFR-mediated endocrine therapy, and other forms of targeted therapy, also it regulates fundamental cellular functions including transcription, translation, proliferation, growth and survivalCitation33. Moreover, Glycogen synthase kinase 3 (GSK3) is involved in modulating numerous signalling pathways affecting metabolism, tumorigenesis, proliferation, apoptosis, autophagy, development, and differentiation involved in metabolic regulationCitation34. GSK3 inhibitors were reported to suppress breast tumour growth in pre-clinical modelsCitation35,Citation36.
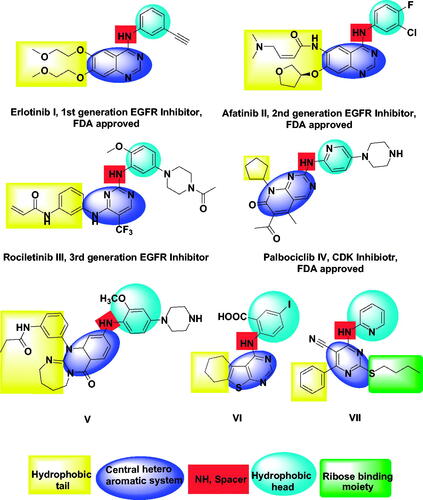
Until now, there are many FDA-approved drugs for the treatment of cancer targeting EGFR as erlotinib I and afatinib II. The first generation of these inhibitors (as erlotinib I) gave good benefits in the treatment of non-small-cell lung cancerCitation37. The second generation of EGFR inhibitors (as afatinib II) was approved to overcome EGFR mutation-related resistanceCitation38. The third generation of EGFR inhibitors (as rociletinib III) had overcome induced toxicity caused by 2nd generationCitation39–41. Palbociclib IV is the first FDA-approved cyclin-dependent kinase inhibitor that acts by binding to the ATP pocket with an IC50 in the range of 9–15 nMCitation27 (.
Figure 1. FDA approved and reported kinases inhibitors with their essential pharmacophoric features.
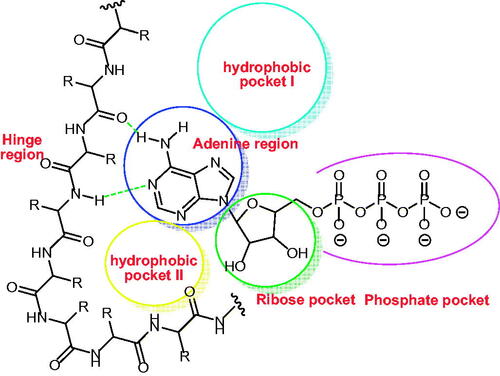
EGFR inhibitors have some pharmacophoric features which are essential for maximal affinity against the ATP binding site of EGFR. These features include i) a flat hetero aromatic ring system that occupies the adenine binding pocketCitation42, ii) terminal hydrophobic head which can occupy the hydrophobic region I, iii) imino group (NH, spacer) which can occupy the space between the adenine binding region and the hydrophobic region ICitation43, iv) hydrophobic tail which occupies the hydrophobic region IICitation44,Citation45, ribose binding moiety which can occupy the ribose binding pocket. Till now, there are limitations in research that target the ribose binding pocketCitation46 (.
Figure 2. ATP binding site of EGFR cavity composed of five main partsCitation47–49.
Privileged structures of diazepine have been widely used as effective templates for anticancer agents especially targeting EGFR enzymeCitation50. Xu et al., described in 2013 a compound with a 1,3‐diazepine moiety (compound V) presenting a potent inhibition activity against the EGFR790M mutant (IC50 = 3.36 nM) and the double EGFRL858R/T790M mutant (IC50 = 1.69 nM)Citation51. Also, in 2019, a series of thieno[2,3-d]pyrimidine derivatives were designed and synthesised as EGFR and HER2 tyrosine kinase inhibitors. Compound VI exhibited promising cytotoxic activityCitation47. In 2020, a new series of pyrimidine-5-carbonitrile derivatives were designed and synthesised as EGFR inhibitors. Compound VII showed high inhibitory activities against EGFRWT and EGFRT790MCitation48 (.
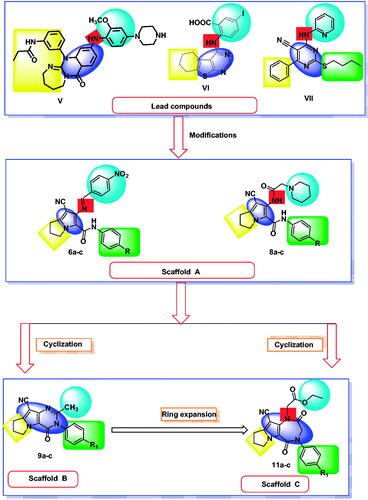
Based on the mentioned facts and as an extension of the previous efforts in the design and synthesis of new anticancer agentsCitation48,Citation52–59 especially to target EGFRCitation47,Citation48,Citation60 we used the 1H-pyrrole, pyrrolo[3,2-d]pyrimidine, and pyrrolo[3,2-e][1, 4]diazepine moieties as building blocks for the design and synthesis of new EGFR and CDK inhibitors (.
1.1. Rationale of molecular design
In the present work, the previously active compounds (V, VI and VII) were used as lead compounds to design new EGFR inhibitors. Dramatic modifications were achieved to reach more promising active candidates against EGFR and CDK. The modifications were performed on four features. The flat hetero aromatic system was modified to be 1H-pyrrole (compounds 6a–c and 8a–c) (Scaffold A), pyrrolo[3,2-d]pyrimidine (9a–c) (Scaffold B), and pyrrolo[3,2-e][1, 4]diazepine (compounds 11a–c) (Scaffold C). ii) For the terminal hydrophobic head, we used different hydrophobic moieties as substituted phenyl groups (compounds 6a–c) or aliphatic moieties (compounds 8a–c, 9a–c, and 11a–c). Regarding the hydrophobic tail, we used the pyrrolidine moiety in all the designed compounds. The NH spacer was kept as it is in compounds 8a–c, whereas it was changed into -N = moiety in compounds 6a–c, and deleted in compounds 9a–c relaying on the two nitrogen atoms of pyrimidine moiety to act as hydrogen bond centres. Moreover, in compounds 11a–c, it was modified to be inside the ring structure as a heteroatom. To occupy ribose binding moieties, we used different aromatic structures. Different structure modifications were then achieved to obtain a SAR study as a second aim in our work.
To confirm our rationale, the synthesised compounds were investigated for their antiproliferative activities against a panel of human cancer cell lines (Hep3B, HCT116, and MCF-7). In addition, the most active anti-proliferative agent was further subjected to a Kinase profiling test to assess its activity against EGFR, CDK, and other kinases. Furthermore, to reach a good insight into the activity of the most active candidate at the molecular level, cell cycle analysis was carried out. Finally, in silico, docking, ADMET, and toxicity studies were performed to predict the possible binding interaction of the synthesised compounds against the prospective targets (EGFR and CDK) as well as to calculate the proposed kinetic and toxicity profile.
2. Results and discussion
2.1. Chemistry
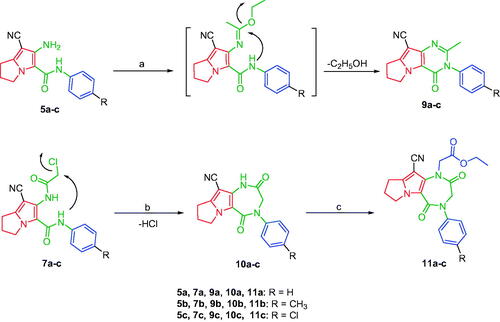
Compounds 8Citation61 and 10a–cCitation62 were synthesised according to reported methods, which were then used to afford the intermediates 11a–c according to the reported procedureCitation63. Condensation of compounds 5a–c (Scheme 1) with 4-nitrobenzaldehyde in absolute ethanol afforded three Schiff base derivatives (6a–c, Scheme 2). The IR spectra of compounds 6a–c showed sharp absorption bands at 2210–2214 cm−1, indicating the cyano group. Furthermore, the 1H NMR spectra of compounds 6a–c showed two doublet signals at a range of 8.10–8.42 ppm due to the aromatic protons of the 4-nitrobenzylidene moiety. Moreover, a singlet signal appeared at a range of 9.28–9.31 corresponding to the N = CH proton. While their 13 C NMR spectra revealed two signals at a range of 156.3–158 ppm indicating the N=CH and the carbonyl carbon in each of the three derivatives. The free amino group of compounds 5a–c was further acylated with 2-chloroacetyl chloride in benzene to give compounds 7a–c. Compounds 8a–c were prepared by N-alkylation of piperidine with the terminal chlorinated side chain of 7a–c in absolute ethanol, using sodium bicarbonate to neutralise the hydrogen chloride liberated from this reaction (Scheme 2). The 1H NMR spectra of compounds 8a–c confirmed the aliphatic protons of the piperidin-1-yl ring; multiplet and triplet signals at a range of 1.50–2.74 ppm. Their 13 C NMR spectra further revealed three additional signals in the range of 23.4–55.0 ppm due to the aliphatic carbons of the piperidin-1-yl ring.
Scheme 1. Construction of compounds 5a–c. Reagents and conditions: (a) (CH3)2SO4, benzene, CH2(CN)2, reflux, 6 h; (b) ClCH2COCl, glacial acetic acid, CH2COONa, 30–40 °C, 2 h; (c) acetone, K2CO3, reflux, 24 h.
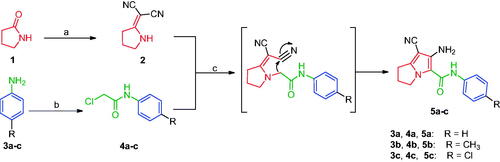
Scheme 2. Synthesis of compounds 6a–c, 7a–c and 8a–c. Reagents and conditions: (a) 4-Nitrobenzaldehyde, absolute ethanol, glacial acetic acid; (b) ClCH2COCl, benzene; r.t.; 48 h; (c) piperidine, NaHCO3, absolute ethanol, reflux, 8 h.
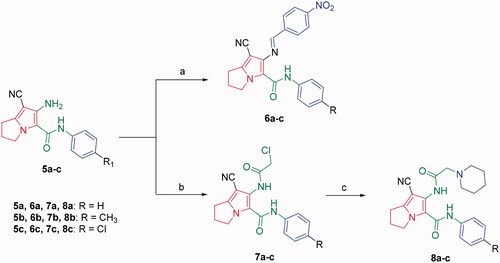
Moreover, several studies reported the use of triethyl orthoformate to produce cyclized derivativesCitation64. The tricyclic pyrrolo[3,2-d]pyrimidine derivatives 9a–c were achieved by refluxing compounds 5a–c with excess triethyl orthoformate (Scheme 3). The NHs stretching bands disappeared in the IR spectra of compounds 9a–c. Further, the 1H NMR spectra of compounds 9a–c confirmed disappearance of the exchangeable protons’ signals, corresponding to compounds 5a–c NHs protons. Additionally, they showed singlet signals at a range of 2.25–2.27 ppm corresponding to the aliphatic methyl protons. Furthermore, the 13 C NMR spectra revealed the presence of two additional signals; the first one at the range of 24.3–24.4 ppm due to the aliphatic methyl group at C-2 and a second signal at the range of 153.7–154.1 ppm assigned for the C-2. Whereas, the pyrrolo[3,2-e][1, 4]diazepine derivatives 10a–c were obtained by intramolecular cyclisation of compounds 7a–c using potassium carbonate in dimethylformamide (Scheme 3). Finally, N-alkylation of compounds 10a–c with ethyl chloroacetate afforded the ethyl ester derivatives 11a–c using potassium carbonate in acetone (Scheme 3). The disappearance of the amidic protons (CONH) of the starting materials 10a–c was observed in the 1H NMR spectra of compounds 11a–c, in addition to the presence of triplet signals at 1.28–1.29 ppm and quartette signals around 4.24 ppm due to the ethyl group. The 13 C NMR spectra of compounds 11a–c revealed three signals in the range of 159.6–167.6 ppm that were assigned for the carbon atoms of the three carbonyl groups.
2.2. Biological evaluation
2.2.1. Anticancer activity
Evaluation of the anticancer activity was performed against liver (Hep3B), colon (HCT116), and breast (MCF-7) cancer cell lines in the Centre of Genetic Engineering, Al-Azhar University, Cairo, Egypt, using Sulforhodamine-B (SRB) assayCitation65. Doxorubicin (DOX) was used as a reference drug. The survival curve was obtained by plotting concentrations of the compound under investigation against the survival fraction of the tumour cells. Then, results were expressed in half maximal inhibitory concentration (IC50).
All the tested derivatives showed potent antiproliferative activities (IC50 = 0.009 − 2.195 µM) against the three cancer cell lines (). A closer look at the results revealed that compound 8b exhibited the highest cytotoxic activity against Hep3B and MCF-7 cell lines with IC50 values of 0.049 and 0.043 µM, respectively. On the other hand, compound 8b displayed the highest cytotoxic activity against the HCT116 cell line with IC50 values of 0.011 µM.
Table 1. IC50 values of the new compounds (6a–c, 8a–c, 9a–c, and 11a–c) against Hep3B, HCT116 and MCF-7 cell lines.
2.2.2.1. Structure-Activity Relationship (SAR)
Inspecting the results of the anti-proliferative activity of the designed candidates, we concluded a valuable SAR. With reference to their cytotoxic activity, it was noticed that counterparts incorporating 2,3-dihydro-1H-pyrrolizine moiety (scaffold A) were slightly more advantageous than the 3H-pyrimido[4,5-b]pyrrolizine derivatives (scaffold B). However, hexahydro-[1, 4]diazepino[5,6-b]pyrrolizin containing derivatives (scaffold C) displayed less potent inhibitory activity against the tested cell lines.
Concerning the di-aryl compounds 6a–c, compound 6a (unsubstituted-N-phenylcarbamoyl derivatives) displayed higher activity than 6b and 6c (4-methyl and 4-chloro-N-phenylcarbamoyl derivatives, respectively) against Hep3B (IC50 = 0.219 µM) and HCT116 (IC50 = 0.213 µM) cancer cell lines. While the methyl analogue 6b was the most active member against MCF-7 cell line with IC50 value of 0.336 µM. Replacement of the aromatic 6–4-nitrobenzylidene)amino side chain in compounds 6a–c by the 2-(piperidin-1-yl)acetamido moiety (compounds 8a–c) resulted in a slightly enhanced cytotoxicity; compounds 8a–c showed IC50 values in the range of 0.031–0.408 µM against the three cell lines. Among the three derivatives 8a–c, the methyl derivative 8b displayed the highest anticancer activity against the three cell lines.
Evaluation of the anticancer activity of pyrrolo[3,2-d]pyrimidine 9a–c, revealed that the 4-chloro congener derivative 9c was the most active against HCT116 cell line among all the tested compounds (IC50 = 0.009 µM). While, the pyrrolo[3,2-e][1, 4]diazepine derivatives (compounds 11a–c) showed comparable activity to their fused pyrrolo[3,2-d]pyrimidine analogues (9a–c) with enhancement in activity of the 4-chloro 11c over 9c against MCF-7 cell line (IC50 = 0.364 versus 1.479 µM). Thus, the SAR study pointed out the significance of a single methyl group as it presents in the most active compounds 8b (N-p-tolylcarbamoyl derivative) and 9a,c; 2-methyl group on the fused pyrrolo[3,2-d]pyrimidine scaffold. However, the comparable activity among the three series (A–C scaffolds) is raising a query about the significance of the molecular modifications on the physicochemical properties, which could affect the cellular barriers positively.
2.2.2. EGFR and CDK-2 inhibitory activity
According to the anti-proliferative activity of the tested compounds, 8b showed a substantial broad-spectrum activity against Hep3B, HCT116 and MCF-7 cell lines (IC50 = 0.049, 0.031 and 0.043 µM, respectively). Thus, compound 8b was tested for its inhibitory activity against both EGFR and CDK-2.
The results revealed that compound 8b showed excellent activity against CDK2 with an inhibition value of 15% compared to the reference molecule, imatinib (2%). On the other hand, it showed less activity against EGFR (% Inhibition = −70%) compared to imatinib (% Inhibition = −13%) ().
Table 2. Inhibitory activity of compound 8b and imatinib against 20 kinases at 10 µMCitation66.
2.2.3. Kinase profiling test
The previous results encouraged us to test compound 8b against other different kinases (18 kinases) to reach a good insight into its kinases inhibitory profile. The result showed that compound 8b has good inhibitory activity against DYRK3 and GSK3 alpha kinases. For its activity against DYRK3, it showed an inhibition value of 23% compared to imatinib (% Inhibition = 10%). Regarding its activity against GSK3 alpha, it showed an inhibition value of 10% compared to imatinib (% Inhibition = 1%).
Additionally, compound 8b showed imatinib comparable activity against ALK1, AMPK (A1/B1/G1), GRK1, MSK1, p38 Alpha, PDK1, and PRKG1. On the other hand, it showed moderate to weak activity against CK1 Alpha 1, Aurora A, BLK, BRAF, ASK1, EPHA1, FLT1, NEK1, and SGK1 ().
2.2.4. Determination of cell cycle perturbations
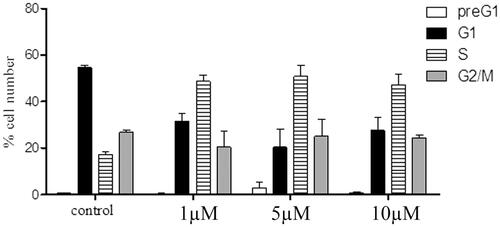
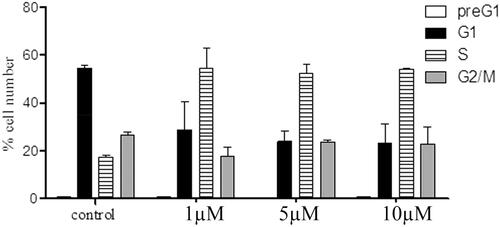
To study the mechanistic actions regarding compounds 6a and 8b, cell cycle analyses using flow cytometry (BC, FC500) were performed. The results of the cell cycle perturbation of the MCF-7 cell line treated with compounds 6a and 8b (72 h, separately) were presented in and , respectively. Each of the two compounds caused a three-fold increase of cells in the S phase at 1 µM compared to control. This increase in the S-phase cell population was accompanied by a concomitant decrease in the G1 cell population. However, there was no considerable change in the S phase cellular population at 5 and 10 µM, which means that the effect of compounds 6a and 8b are not dose-dependent.
2.3. In silico studies
2.3.1. Docking studies
A docking study was conducted in the hopes of learning more about how the synthesised compounds interact with their targetsCitation67–69. EGFR (PDB: 4HJO), CDK-2 (PDB: 6GUH), DYRK3 (PDB: 5Y86), and GSK3 (PDB: 5HLP) were employed as biological targets in docking investigations utilising MOE 14.0 software. The co-crystallised ligands (erlotinib and AZD5438) were utilised as anti-EGFR and anti-CDK-2 reference compounds respectively. When compared to the reference molecules, the docking results demonstrated. that the synthesised compounds have a high affinity for the two examined targets ().
Table 3. The binding free energies of the synthesised compounds against EGFR and CDK-2.
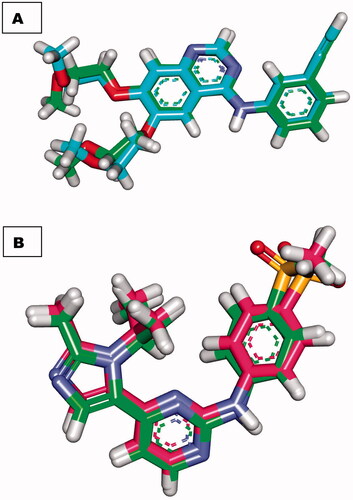
Redocking of co-crystallised ligands (Erlotinib and AZD5438) against EGFR and CDK-2, respectively, was used to validate docking experiments. Erlotinib and AZD5438 had RMSDs of 0.88 and 0.54 Å for docked and original ligands, respectively. The validity of the docking operation is shown by these values (.
Figure 6. (A) superimposition of the docked ligand of erlotinib (turquoise) and the original ligand (green) with an RMSD value of 0.88 Å. (B) superimposition of the docked ligand of AZD5438 (pink) and the original ligand (green) with RMSD value of 0.54 Å.
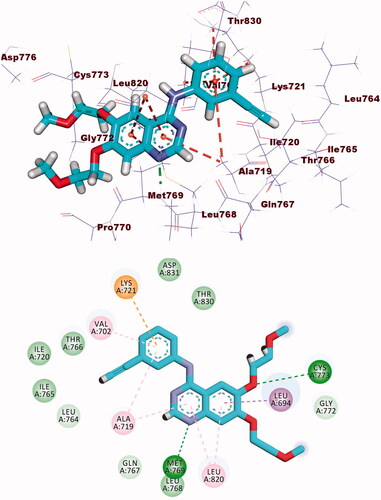
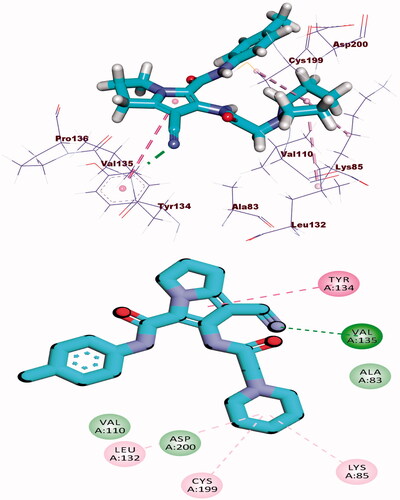
The binding energy of erlotinib was found to be −23.49 kcal/mol. The quinazoline molecule was buried in the adenine pocket, creating a hydrogen connection with Met769. Furthermore, Lue694, Ala719, and Leu820 established four hydrophobic contacts with quinazoline. The ethynylphenyl moiety was positioned in the hydrophobic pocket I, resulting in three hydrophobic interactions with Ala719, Val702, and Lys721. The 2-methoxyethoxy groups formed a hydrogen bond with Cys773 in the hydrophobic region II (.
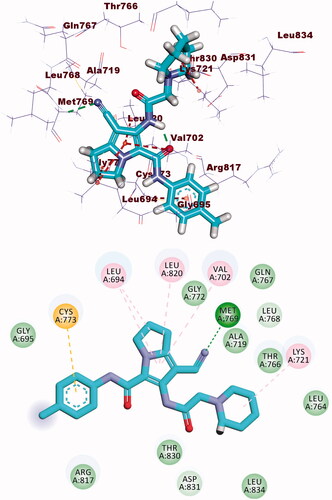
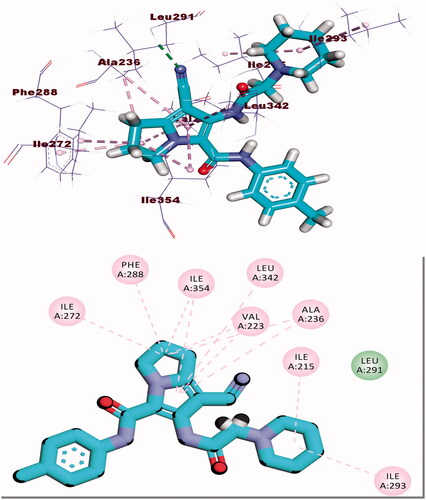
Compound 8b showed a binding mode like that of erlotinib with a binding energy of −22.46 kcal/mol. The 1H-pyrrole-3-carbonitrile moiety occupied the adenine pocket of the EGFR forming three hydrophobic interactions with Leu820, Leu694, and Val702. Piperidine moiety was oriented into the hydrophobic pocket I to form one hydrophobic interaction with Leu694. The pyrrolidine moiety occupied the hydrophobic pocket II forming one hydrophobic interaction with Lys721 in close contact with Thr766, Leu764, and Leu834. Moreover, p-tolyl moiety occupied the ribose binding pocket forming one electrostatic attraction with Cys773 (.
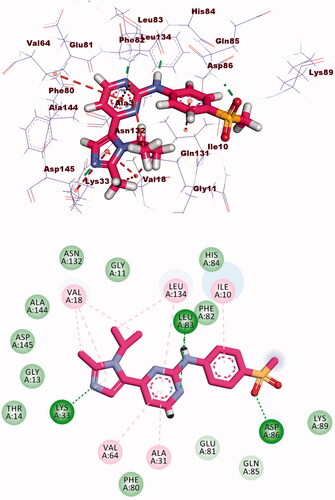
The co-crystallised ligand (AZD5438) showed binding energy of −21.66 kcal/mol against CDK-2. The pyrimidine moiety occupied the adenine pocket forming three hydrophobic bonds with Leu134, and Ala31, Val64. The NH-linker formed one hydrogen bond with Leu83. The terminal methylsulfonyl benzene moiety occupied the hydrophobic pocket forming a hydrogen bond with Asp86 and one hydrophobic interaction with Ile10. Additionally, the 1-isopropyl-2-methyl-1H-imidazole moiety occupied the other hydrophobic region forming a hydrogen bond with Lys33. Also, it formed four hydrophobic interactions with Leu134 and Val18 (.
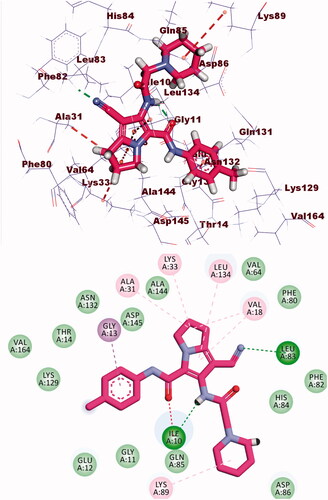
Compound 8b showed a binding mode like that of AZD543 with a binding energy of −24.52 kcal/mol. The 1H-pyrrole-3-carbonitrile moiety occupied the adenine pocket forming two hydrophobic interactions with Leu134 and Val18. The pyrrolidine moiety was oriented into the hydrophobic pocket forming three hydrophobic interactions with Val18, Lys33, and Ala31. The piperidine moiety occupied another hydrophobic pocket forming one hydrophobic interaction with Lys89 in close contact with Asp86 and Gln85. The two amide linkers formed one hydrogen bond and one electrostatic interaction with Ile10. Moreover, p-tolyl moiety occupied the ribose binding pocket forming one hydrophobic interaction with Gly13 (.
Docking calculations between compound 8b and DYRK3 and GSK3 revealed that it could occupy the active sites and interact with the key amino acids of each enzyme ( and ) with the binding energy of −24.54 and −16.85 kcal/mol, respectively. The binding free energy of the co-crystallised ligands of DYRK3 (HRM) and GSK3 (65 A) was calculated to be −20.80 and −16.41 kcal/mol, respectively and the binding modes were presented in Supplementary data.
2.3.2. In silico toxicity prediction
In this work, eight toxicity parameters were estimated computationally depending on the constructed toxicity models in Discovery studio softwareCitation70–72. The results revealed that the calculated toxicity potential of the synthesised compound was low. Except for compound 6a, all compounds were non-carcinogenic for mouse females based on the FDA rodent carcinogenicity model. In addition, compounds 6a, 8a, 8b, 9a, and 11a showed carcinogenic potency TD50 values ranging from 11.774 to 71.541 mg/kg body weight/day, which were higher than that of erlotinib (8.057 mg/kg body weight/day). All compounds had rat maximum tolerated dose values less than that of erlotinib (0.083 g/kg body weight) except compound 8c (0.107 g/kg body weight). For the rat oral LD50 model, except compounds 9a and 9c, all compounds showed oral LD50 values higher than that of erlotinib (0.662 mg/kg body weight/day). In addition, all compounds exhibited rat chronic LOAEL values higher than that of erlotinib (0.036 mg/kg body weight/day) except compounds 8c and 11c. Moreover, all the tested compounds were predicted to be mild irritants in the ocular irritancy model, and non-irritants in the skin irritancy model ().
Table 4. Toxicity properties of the synthesised compounds.
2.3.3. ADMET studies
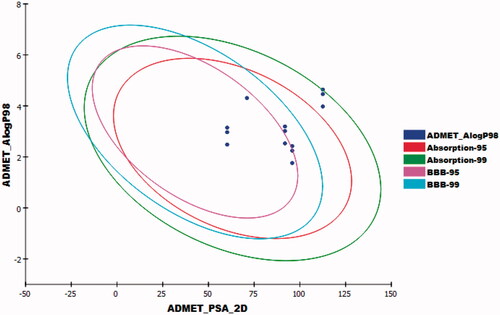
ADMET parameters were predicted using discovery studio softwareCitation69,Citation73–75. Erlotinib was used as a reference compound. The predicted ADMET parameters were listed in . The results revealed that compounds 6a–c had very low Blood Brain Barrier penetration power. Compounds 8a–c and 11a–c were anticipated to have low levels of BBB penetration. On the other hand, compound 9a–c was predicted to have medium levels of BBB penetration. Accordingly, the synthesised compounds were expected to be safe for CNS. The predicted aqueous solubility of the synthesised compounds ranged from good to low. All the synthesised compounds showed good absorption levels except compounds 6a and 6b which showed moderate absorption levels. All the synthesised members were predicted as non-inhibitors of CYP2D6. Additionally, all of them were expected to bind plasma protein by more than 90% (.
Table 5. Predicted ADMET profile for the synthesised compounds
3. Conclusion
In this work new compounds having the essential pharmacophoric features of EGFR inhibitors were designed and synthesised. Three series of 1H-pyrrole, pyrrolo[3,2-d]pyrimidine and pyrrolo[3,2-e][1, 4]diazepine derivatives were obtained. Compound 9c, the fused pyrimido[4,5-b]pyrrolizine derivative, showed selectivity towards the HCT116 colon cancer cell line with activity comparable with that of DOX (IC50 = 0.009 and 0.008 µM, respectively). While compound 8b, the 1H-pyrrole derivative featuring the extended 2-(piperidin-1-yl)acetamido moiety at C-6, showed a broad-spectrum inhibition against Hep3B, HCT116 and MCF-7cell lines (IC50 = 0.049, 0.031 and 0.043 µM, respectively). Regarding compound 8b, the kinase profiling evaluations revealed its inhibitory activity against three types of kinases: CDK2/Cyclin A1, DYRK3 and GSK3 alpha. Therefore, this study reinforces the promising anticancer efficacy of 1H-pyrrole pyrrolo[3,2-d]pyrimidine derivatives based on their multi-targets activity, which were supported by molecular modelling characteristics. Add to that, their superior predicted safety and predicted pharmacokinetic properties as drug-like or lead-like molecules.
4. Experimental protocol
4.1. Chemistry
All details of chemical reagents and apparatus were described in Supplementary Data. Compounds 2Citation61, 4a–cCitation63, 5a–c,Citation63 7a–c and 10a–cCitation76 were prepared using the previously reported methods.
4.1.1. General method for preparation of compounds (6a–c)
To a solution of 4-nitrobenzaldehyde (3.75 mmol) in absolute ethanol (20 ml), the pyrrolizines 5a–c (3.75 mmol) and 0.5 ml of glacial acetic acid were added. The reaction mixture was refluxed for 4 h. The reaction mixture was left to cool. The products 6a–c were crystallised out as orange crystals and recrystallized from ethanol
4.1.2. (EZ)-7-Cyano-6-((4-nitrobenzylidene)amino)-N-phenyl)-2,3-dihydro-1H-pyrrolizine-5-carboxamide (6a)
The title compound was obtained from compound 5a as orange crystals, m.p. 278–80 °C, yield 76%, IRʋmax/cm−13279 (NH), 3076 (C-H aromatic), 2935, 2852 (CH2), 2210 (CN), 1658 (CO), 1601 (C = N), 1548, 1469 (C = C, NH), 1412, 1337 (C-N, C-O). 1H NMR (CDCl3, 400 MHz): δ 2.60 (m, 2H, CH2-2), 3.07 (t, 2H, J = 7.4 Hz, CH2-1), 4.57 (t, 2H, J = 7.4 Hz, CH2-3), 7.17 (t, 1H, J = 8.2 Hz, CH-4′), 7.41 (t, 2H, J = 8.2 Hz, CH-3′, −5′), 7.62 (d, 2H, J = 8.2 Hz, CH-2′, −6′), 8.1 and 8.41 (two d, 4 H, J = 8 Hz, CH-2′', −3′', −4′', −5′', −6′'), 9.28 (s, H, N=CH) and 10.40 (s, 1H, NH, disappeared on deuteration). 13 C NMR (CDCl3, 100 MHz): δ 24.5, 25.4, 50.4, 76.7, 116.0, 119.1, 119.6, 124.4, 124.4, 129.1, 129.3, 137.8, 137.9, 140.9, 148.9, 149.7, 156.4, 158. MS (EI): m/z (%) 400 (M++1, 13), 399 (M+, 35), 307 (100). Anal. Calcd. for C22H17N5O3 (399.40):C, 66.16; H, 4.29; N, 17.53. Found: C, 68.35; H, 4.15; N, 17.23.
4.1.3. (EZ)-7-Cyano-6-((4-nitrobenzylidene)amino)-N-(4-tolyl)-2,3-dihydro-1H-pyrrolizine-5-carboxamide (6b)
The title compound was obtained from compound 5b as orange crystals, m.p. 291–3 °C, yield 79%, IRʋmax/cm−13280 (NH), 3069 (C-H aromatic), 2918 (CH3, CH2), 2210 (CN), 1657 (CO), 1601 (C = N), 1544, 1519 (C = C,N-H), 1411, 1338 (C-N, C-O). 1H NMR (CDCl3, 400 MHz): δ 2.37 (s, 3H, CH3Ph), 2.60 (m, 2H, CH2-2), 3.10 (t, 2H, J = 7.4 Hz, CH2-1), 4.59 (t, 2H, J = 7.4 Hz, CH2-3), 7.21 (d, 2H,J = 8.4 Hz, CH-3′, −5′), 7.53 (d, 2H,J = 8.4 Hz, CH-2′, −6′), 8.10 (d, 2H,J = 8.4 Hz, CH-2′', −6′'), 8.39 (d, 2H,J = 8.4 Hz, CH-3′', −5′'), 9.28 (s, H, N=CH) and 10.37 (s, 1H, NH, disappeared on deuteration). 13 C NMR (CDCl3, 100 MHz): δ 20.9, 24.5, 25.5, 50.3, 76.7, 116.1, 119.3, 119.6, 124.4, 129.1, 129.8, 134.1, 135.4, 137.7, 140.9, 148.7, 149.6, 156.3, 157.9.MS (EI): m/z (%) 414 (M + 1, 15), 413 (M+, 51), 307 (100). Anal. Calcd. for C23H19N5O3 (413.43): C, 66.82; H, 4.63; N, 16.94. Found: C, 67.01; H, 4.58; N, 16.91.
4.1.4. (EZ)-N-(4-Chlorophenyl)-7-cyano-6-((4-nitrobenzylidene)amino)-2,3-dihydro-1H-pyrrolizine-5-carboxamide (6c)
The title compound was obtained from compound 5c as yellow crystals, m.p. 283–5 °C, yield 83%, IRʋmax/cm−13245 (NH), 3111 (C-H aromatic), 2968 (CH2), 2214 (CN), 1680 (CO), 1597 (C = N), 1541, 1482 (C = C, NH), 1412, 1344 (C-N, C-O), 836, 754 (C-Cl). 1H NMR (CDCl3, 400 MHz): δ 2.63 (m, 2H, CH2-2), 3.11 (t, 2H, J = 7.4 Hz, CH2-1), 4.58 (t, 2H, J = 7.4 Hz, CH2-3), 7.38 (d, 2H,J = 8.4 Hz, CH-3′, −5′), 7.60 (d, 2H,J = 8.4 Hz, CH-2′, −6′), 8.10 (d, 2H,J = 8.4 Hz, CH-2′', −6′'), 8.42 (d, 2H, J = 8.4 Hz, CH-3′', −5′'), 9.31 (s, H, N=CH) and 10.45 (s, 1H, NH, disappeared on deuteration). 13 C NMR (CDCl3, 100 MHz): δ 24.5, 25.4, 50.4, 76.7, 115.9, 119.2, 119.8, 124.4, 129.1, 129.3, 135.4, 137.6, 137.7 140.9, 148.9, 149.7, 156.4, 157.9 MS (EI): m/z (%) 435 (M + 2, 74), 433 (M+, 76), 405 (100). Anal. Calcd. for C22H16ClN5O3 (433.85):C, 60.91; H, 3.72; N, 16.14. Found: C, 61.10; H, 3.68; N, 16.11.
4.1.5. General method for preparation of compounds (8a–c)
A mixture of compounds 7a–c (2.91 mmol), piperidine (0.5 g, 5.9 mmol) and dry sodium bicarbonate (0.5 g, 5.9 mmol) in absolute ethanol (10 ml) was refluxed for 8 h. Then, the reaction mixture was filtered while hot, and the produced white crystals upon concentration were collected, dried, and recrystallized from ethanol.
4.1.5.1. 7-Cyano-N-phenyl-6–(2-(piperidin-1-yl)acetamido)-2,3-dihydro-1H-pyrrolizine-5-carboxamide (8a)
The title compound was obtained from compound 7a as white crystals, m.p. 205–7 °C, yield 72%, IRʋmax/cm−13243, 3130 (NHs), 3060 (C-H aromatic), 2935, 2852 (CH2), 2220 (CN), 1660 (COs), 1601, 1557, 1493 (C = C, NH), 1451, 1390, 1319 (C-N, C-O). 1H NMR (CDCl3, 400 MHz):δ 1.5 (m, 2H, CH2-4′'), 1.7 (m, 4H, CH2-3′', −5′'), 2.54 (m, 2H, CH2-2), 2.69 (t, 4H,J = 5.2 Hz, CH2-2′', −6′'), 3.02 (t, 2H, J = 7.6 Hz, CH2-1), 3.27 (s, 2H, COCH2), 4.40 (t, 2H, J = 7.2 Hz, CH2-3), 7.12 (t, 1H, J = 8 Hz, CH-4′), 7.34 (t, 2H, J = 8 Hz, CH-3′, −5′), 7.6 (d, 2H, J = 8 Hz, CH-2′, CH-6′), 9.44 (s, H, NHCOCH2, which disappeared on deuteration) and 9.80 (s, H, CONH phenyl, which disappeared on deuteration). 13 C NMR (CDCl3, 100 MHz): δ 23.5, 25, 25.7, 26.1, 49.6, 55.1, 62.1, 83.9, 114, 119.5, 120.3, 124.1, 124.4, 129, 138.3, 145.7, 157.6, 173.2. MS (EI): m/z (%) 392 (M++1, 57), 391 (M+, 67), 138 (100).Anal. Calcd. forC22H25N5O2 (391.47): C, 67.50; H, 6.44; N, 17.89. Found: C, 67.80; H, 6.72; N, 17.67.
4.1.5.2. 7-Cyano-6–(2-(piperidin-1-yl)acetamido)-N-(4-tolyl)-2,3-dihydro-1H-pyrrolizine-5-carboxamide (8b)
The title compound was obtained from compound 7b as white crystals, m.p. 178–80 °C, yield 72%, IRʋmax/cm−13240, 3186 (NHs), 3064 (C-H aromatic), 2983, 2930, 2853 (CH3, CH2), 2222 (CN), 1656 (COs), 1604, 1547, 1513 (C = C, NH), 1455, 1385, 1321 (C-N, C-O). 1H NMR (CDCl3, 400 MHz): δ 1.51 (m, 2H, CH2-4′'), 1.72 (m, 4H, CH2-3′' −5′'), 2.33 (s, 3H,CH3Ph), 2.54 (m, 2H, CH2-2), 2.70 (t, 4H,J = 5.2 Hz, CH2-2′' −6′'), 3.02 (t, 2H, J = 7.4 Hz, CH2-1), 3.28 (s, 2H, COCH2), 4.39 (t, 2H, J = 7.2 Hz, CH2-3),7.13 (d, 2H, J = 8 Hz, CH-3′, −5′), 7.49 (d, 2H, J = 8 Hz, CH-2′, −6′), 9.42 (s, H, NHCOCH2, disappeared on deuteration) and 9.69 (s, H, CONH phenyl, disappeared on deuteration). 13 C NMR (CDCl3, 100 MHz): δ 20.9, 23.5, 25.1, 25.7, 26.1, 49.5, 55.1, 62, 83.9, 114.1, 119.5, 120.4, 124.2, 129.5, 133.8, 135.8, 145.6, 157.5, 173.2. MS (EI): m/z (%) 407 (M++2, 69), 406 (M++1, 82), 405 (M+, 68) 148 (100).Anal. Calcd. for C23H27N5O2(405.49): C, 68.13; H, 6.71; N, 17.27. Found: C, 68.33; H, 6.59; N, 17.50.
4.1.5.3. N-(4-Chlorophenyl)-7-cyano-6–(2-(piperidin-1-yl)acetamido)-2,3-dihydro-1H-pyrrolizine-5-carboxamide (8c)
The title compound was obtained from compound 7c as white crystals, m.p. 210–2 °C, yield 74%, IRʋmax/cm−13233, 3179 (NHs), 3053 (C-H aromatic), 2986, 2932 (CH2), 2222 (CN), 1658 (COs), 1602, 1546 (C = C, NH), 1489, 1456, 1387 (C-N, C-O), 829, 792 (C-Cl). 1H NMR (CDCl3, 400 MHz):δ 1.51 (m, 2H, CH2-4′'), 1.74 (m, 4H, CH2-3′' −5′'), 2.55 (m, 2H, CH2-2), 2.74 (t, 4H, J = 5.2 Hz, CH2-2′' −6′'), 3.02 (t, 2H, J = 7.4 Hz, CH2-1), 3.32 (s, 2H, COCH2), 4.39 (t, 2H, J = 7.4 Hz, CH2-3), 7.28–7.58 (4H, aromatic protons), 9.41 (s, H, NHCOCH2, disappeared on deuteration) and 9.93 (s, H, CONH phenyl, disappeared on deuteration). 13 C NMR (CDCl3, 100 MHz): δ23.4, 25, 25.6, 26.1, 49.5, 55, 62, 83.9, 113.9, 120, 120.7, 124.4, 129, 131.6, 136.9, 145.8, 157.6, 173.3. MS (EI): m/z (%) 426 (M++1, 0.9), 98 (100). Anal. Calcd. for C22H24ClN5O2 (425.91): C, 62.04; H, 5.68; N, 16.44. Found: C, 62.30; H, 5.90; N, 16.15.
4.1.6. General method for preparation of compounds (9a–c)
A mixture of 6-amino-7-cyano-N-phenyl-2,3-dihydro-1H-pyrrolizine-5-carboxamide 5a–c (3.75 mmol) and excess triethyl orthoformate was refluxed for 12 h. The solvent was then removed using a rotary evaporator and the obtained residue was crystallised from ethanol.
4.1.6.1. Methyl-4-oxo-3-phenyl-4,6,7,8-tetrahydro-3H-pyrimido[4,5-b]pyrrolizine-9-carbonitrile (9a)
The title compound was obtained from compound 5a as white crystals, m.p. 243–5 °C, yield 55%, IRʋmax/cm−13061 (C-H aromatic), 2966, 2918 (CH3, CH2), 2214 (CN), 1693 (CO), 1524 (C = C), 1421, 1304 (C-N, C-O).1H NMR (CDCl3, 400 MHz): δ 2.26 (s, 3H, CH3C = N), 2.67 (m, 2H, CH2-7), 3.19 (t, 2H, J = 7.4 Hz, CH2-8), 4.40 (t, 2H, J = 7.2 Hz, CH2-6),7.25 (d, 2H, J = 8 Hz, CH-2′, −6′) and 7.51–7.59 (m, 3H, CH-3′, −4′, −5′). 13 C NMR (CDCl3, 100 MHz): δ 24.3, 25.1, 26.2, 48, 81.1, 113.4, 113.9, 128, 129.4, 130, 137.3, 148.4, 152.3, 154.1, 154.3. MS (EI): m/z (%) 291 (M++1, 3), 290 (M+, 15), 85 (100). Anal. Calcd. for C17H14N4O (290.32): C, 70.33; H, 4.86; N, 19.30. Found: C, 70.12; H, 4.90; N, 19.41.
4.1.6.2. 2-Methyl-4-oxo-3–(4-tolyl)-4,6,7,8-tetrahydro-3H-pyrimido[4,5-b]pyrrolizine-9-carbonitrile (9b)
The title compound was obtained from compound 5b as white crystals, m.p. 251–3 °C, yield 59%, IRʋmax/cm−13073 (C-H aromatic), 2924 (CH3, CH2), 2208 (CN), 1694 (CO), 1591 (C = C), 1427, 1302 (C-N, C-O). 1H NMR (CDCl3, 400 MHz): δ 2.27 (s, 3H, CH3C = N), 2.46 (s, 3H, CH3Ph), 2.66 (m, 2H, CH2-7), 3.18 (t, 2H, J = 7.6 Hz, CH2-8), 4.41 (t, 2H, J = 7.2 Hz, CH2-6), 7.12 and 7.37 (two d, 4 H, J = 8 Hz, p-substituted phenyl ring). 13 C NMR (CDCl3, 100 MHz): δ21, 24.3, 25.1, 26.2, 48, 81.1, 113.5, 114, 127.9, 130.5, 137.2, 139.4, 148.5, 152.3, 154.1, 154.3. MS (EI): m/z (%) 305 (M++1, 7), 304 (M+, 51), 122 (100). Anal. Calcd. for C18H16N4O (304.35): C, 71.04; H, 5.30; N, 18.41. Found: C, 70.78; H, 5.50;N, 18.30.
4.1.6.3. 3–(4-Chlorophenyl)-2-methyl-4-oxo-4,6,7,8-tetrahydro-3H-pyrimido[4,5-b]pyrrolizine-9-carbonitrile (9c)
The title compound was obtained from compound 5c as white crystals, m.p. 268–70 °C, yield 60%, IRʋmax/cm−13060 (C-H aromatic), 2924 (CH3, CH2), 2223 (CN), 1689 (CO), 1533 (C = C), 1419, 1300 (C-N, C-O), 840, 774 (C-Cl). 1H NMR (CDCl3, 400 MHz): δ 2.25 (s, 3H, CH3C = N), 2.67 (m, 2H, CH2-7), 3.19 (t, 2H, J = 7.2 Hz, CH2-8), 4.39 (t, 2H, J = 7 Hz, CH2-6), 7.19 and 7.54 (two d, 4 H, J = 8 Hz, p-substituted phenyl ring). 13 C NMR (CDCl3, 100 MHz): δ 24.4, 25.2, 26.2, 48.1, 81.3, 113.3, 113.8, 129.5, 130.3, 135.6, 135.8, 148.4, 152.6, 153.7, 154.2. MS (EI): m/z (%) 324 (M+, 1), 91 (100).Anal. Calcd. for C17H13ClN4O (324.76): C, 62.87; H, 4.03; N, 17.25. Found: C, 62.69; H, 3.97; N, 17.55.
4.1.7. General method for preparation of compounds (11a–c)
To a solution of diazepino[5,6-b]pyrrolizine-10-carbonitrile derivative 10a–c (3.26 mmol) in acetone, ethyl chloroacetate (0.4 g, 3.26 mmol) and anhydrous potassium carbonate (0.45 g, 3.26 mmol) was added. After refluxing for 6 h, the mixture was filtered and left to cool. The obtained white crystals were collected and recrystallized from ethanol.
4.1.7.1. Ethyl-2–(10-cyano-2,5-dioxo-4-phenyl-2,3,4,5,8,9-hexahydro-[1, 4]diazepino[5,6-b]pyrrolizin-1(7H)-yl)acetate (11a)
The title compound was obtained from compound 10a as white crystals, m.p. 179–81 °C, yield 75%, IRʋmax/cm−13059 (C-H aromatic), 2977 (CH2), 2220 (CN), 1740, 1693, 1643 (COs), 1549 (C = C), 1461, 1369, 1306 (C-N, C-O). 1H NMR (CDCl3, 400 MHz): δ 1.29 (t, 3H, J = 7.2 Hz, CH3CH2),2.59 (m, 2H, CH2-8), 3.08 (t, 2H, J = 7.4 Hz, CH2-9), 4.24 (q, 2H, J = 7.2 Hz, CH2CH3), 4.39 (t, 2H, J = 7.2 Hz, CH2-7), 4.49 (s, 2H, CH2-3), 4.76 (s, 2H, NCH2CO) and 7.29–7.46 (m, 5H, aromatic protons). 13 C NMR (CDCl3, 100 MHz): δ 14, 25.1, 25.4, 48.3, 49.3, 54.5, 61.9, 79.1, 114.2, 114.5, 125.7, 127.2, 129.2, 134.2, 141.6, 148.4, 159.6, 166.5, 167.5. MS (EI): m/z (%) 393 (M + 1, 1), 392 (M+, 1), 106 (100).Anal. Calcd. for C21H20N4O4(392.41): C, 64.28; H, 5.14; N, 14.28. Found: C, 64.58; H, 5.36; N, 14.25.
4.1.7.2. Ethyl-2–(10-cyano-2,5-dioxo-4–(4-tolyl)-2,3,4,5,8,9-hexahydro-[1, 4]diazepino[5,6-b]pyrrolizin-1(7H)-yl)acetate (11b)
The title compound was obtained from compound 10b as white crystals, m.p. 213–5 °C, yield 71%, IRʋmax/cm−13077 (C-H aromatic), 2970, 2924 (CH3, CH2), 2220 (CN), 1741, 1690, 1642 (COs), 1548, 1465 (C = C), 1371, 1307 (C-N, C-O). 1H NMR (CDCl3, 400 MHz): δ 1.28 (t, 3H, J = 7.2 Hz, CH3CH2), 2.37 (s, 3H, CH3Ph),2.58 (m, 2H, CH2-8), 3.07 (t, 2H, J = 7.4 Hz, CH2-9), 4.24 (q, 2H, J = 7.2 Hz, CH2CH3), 4.40 (t, 2H, J = 7.2 Hz, CH2-7), 4.46 (s, 2H, CH2-3), 4.75 (s, 2H, NCH2CO), 7.21 and 7.30 (two d, 4 H, J = 8.4 Hz, p-substituted phenyl ring). 13 C NMR (CDCl3, 100 MHz): δ 14, 21, 25.1, 25.4, 48.3, 49.3, 54.6, 61.9, 79.1, 114.2, 114.6, 125.5, 129.8, 134.2, 137.2, 139, 148.2, 159.7, 166.5, 167.6.MS (EI): m/z (%) 407 (M++1, 14), 406 (M+, 52), 186 (100). Anal. Calcd. for C22H22N4O4(406.43): C, 65.01; H, 5.46; N, 13.78. Found: C, 65.30; H, 5.71; N, 13.77.
4.1.7.3. Ethyl-2–(4-(4-Chlorophenyl)-10-cyano-2,5-dioxo-2,3,4,5,8,9-hexahydro-[1, 4]diazepino[5,6-b]pyrrolizin-1(7H)-yl)acetate (11c)
The title compound was obtained from compound 10c as white crystals, m.p. 220–1 °C, yield 78%, IRʋmax/cm−13092 (C-H aromatic), 2970 (CH2), 2222 (CN), 1749, 1686, 1647 (COs), 1549 (C = C), 1490, 1471, 1373, 1306 (C-N, C-O), 838, 772 (C-Cl). 1H NMR (CDCl3, 400 MHz): δ 1.28 (t, 3H, J = 7.2 Hz, CH3CH2), 2.60 (m, 2H, CH2-8), 3.09 (t, 2H, J = 7.6 Hz, CH2-9), 4.24 (q, 2H, J = 7.2 Hz, CH2CH3), 4.39 (t, 2H, J = 7.2 Hz, CH2-7), 4.49 (s, 2H, CH2-3), 4.75 (s, 2H, NCH2CO), 7.38 and 7.43 (two d, 4 H, J = 8.4 Hz, p-substituted phenyl ring). 13 C NMR (CDCl3, 100 MHz): δ 14.1, 25.2, 25.5, 48.4, 49.4, 54.4, 62, 79.3, 114.1, 114.4, 126.9, 129.4, 132.8, 134.4, 140, 148.6, 159.6, 166.4, 167.5. MS (EI): m/z (%) 428 (M++2, 23), 426 (M+, 64), 186 (100). Anal. Calcd. for C21H19ClN4O4(426.85): C, 59.09; H, 4.49; N, 13.13. Found: C, 59.32; H, 4.39; N, 13.10.
4.2. Biological evaluation
4.2.1. In vitro anticancer screening
Cytotoxic activities evaluation of the synthesised compounds were done using the sulforhodamine B (SRB) method following the previous reported methodCitation65 described in Supplementary data.
Citation4.Citation2.Citation2. Kinase profiling assay
Compound 8b was selected to evaluate its inhibitory activity against 20 kinases according to the reported method.Citation66 The kinases inhibition assay was done by KINEXUS Corporation, Vancouver, BC, Canada, using the radiolabeled ATP determination method. Imatinib was used as a reference drug and blank control was set up and the corrected activity for the protein kinase target was determined. The results were presented as % inhibition, .
4.2.3. Perturbation of cell cycle analysis
Cell cycle distribution analysis was performed for compounds 6a and 8b based on the previously described method in Supplementary dataCitation77.
4.3. In silico studies
4.3.1. Docking studies
Crystallographic structures of EGFR CDK-2, and DYRK3, and GSK3 were retrieved from Protein Data Bank [PDB ID: 4HJO, resolution 2.75 Å and PDB ID: 6GUH, resolution 1.50 Å, PDB: 5Y86, resolution 1.90 Å, PDB: 5HLP, resolution 2.45 Å respectively] (http://www.pdb.org), and considered as targets for docking simulations. The docking analysis was performed using MOE softwareCitation78–80 to evaluate the free energies and binding mode of the designed molecules against EGFR, CDK-2 and DYRK3, and GSK3. At first, the crystal structures of EGFR, CDK-2 and DYRK3, and GSK3 were prepared by removing water molecules and retaining only one chain and their co-crystallised ligands, erlotinib and AZD5438, HRM, and 65 A, respectively. Then, the protein structures were protonated, and the hydrogen atoms were hidden. Next, the energy was minimised, and the binding pockets of the protein were defined.
The 2D structures of the synthesised compounds and the co-crystallised ligands, erlotinib and AZD5438, HRM, and 65 A were sketched using ChemBioDraw Ultra 14.0 and saved as MDL-SD format. Then, the saved files were opened using MOE and 3 D structures were protonated. Next, energy minimisation was applied. Before docking the synthesised compounds, validation of the docking protocol was carried out by running the simulation only using the co-crystallised ligands and low RMSD between docked and crystal conformations. The molecular docking of the synthesised compounds and the co-crystallised ligands was performed using a default protocol. MOE docking parameters include Triangle Matcher Algorithm with two rescoring functions London dG and GBVI/WSA dG were utilised to generate 10 poses of each compound. As a result, mdb output files were generated enclosing all docking results with scoring and multiple conformations of ligands. Results were finally inspected to determine the most promising compound by visualising various interactions of ligands within the binding pocket. The output from MOE was further analysed and visualised using Discovery Studio 4.0 software. The output from MOE was further analysed and visualised using Discovery Studio 4.0 softwareCitation47,Citation48,Citation56,Citation60. Mutations, missing regions and active/inactive states of the receptors were presented in Supplementary data.
4.3.2. In silico toxicity prediction
The toxicity parameters of the synthesised compounds were calculated using Discovery studio 4.0 as described in Supplementary dataCitation1,Citation57.
4.3.3. In silico ADMET studies
ADMET descriptors (absorption, distribution, metabolism, excretion, and toxicity) of the synthesised compounds were determined using Discovery studio 4.0 as described in Supplementary data.
Supplemental Material
Download PDF (312.8 KB)Disclosure statement
All authors of the above manuscript have not declared any conflict of interest.
Additional information
Funding
References
- El-Metwally SA, Abou-El-Regal MM, Eissa IH, et al. Discovery of thieno [2, 3-d] pyrimidine-based derivatives as potent VEGFR-2 kinase inhibitors and anti-cancer agents. Bioorg Chem 2021;112:104947.
- World Health Organization, Cancer. Available from: https://www.who.int/news-room/fact-sheets/detail/cancer [last accessed 1 March 2020].
- El-Zahabi MA, Sakr H, El-Adl K, et al. Design, synthesis, and biological evaluation of new challenging thalidomide analogs as potential anticancer immunomodulatory agents. Bioorg Chem 2020;104:104218.
- Chabner BA, Roberts TG. Chemotherapy and the war on cancer. Nat Rev Cancer 2005;5:65–72.
- G, Krauss N, Schönbrunner J, Cooper Biochemistry of signal transduction and regulation. Weinheim: Wiley Online Library; 2003.
- Nguyen K-SH, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non–small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer 2009;10:281–9.
- Abbass EM, Khalil AK, Mohamed MM, et al. Design, efficient synthesis, docking studies, and anticancer evaluation of new quinoxalines as potential intercalative Topo II inhibitors and apoptosis inducers. Bioorg Chem 2020;104:104255.
- Al-Sanea MM, Elkamhawy A, Paik S, et al. Synthesis and biological evaluation of novel 3-(quinolin-4-ylamino) benzenesulfonamides as carbonic anhydrase isoforms I and II inhibitors. J Enzyme Inhib Med Chem 2019;34:1457–64.
- Eldehna WM, Al-Rashood ST, Al-Warhi T, et al. Novel oxindole/benzofuran hybrids as potential dual CDK2/GSK-3β inhibitors targeting breast cancer: design, synthesis, biological evaluation, and in silico studies. J Enzyme Inhib Med Chem 2021;36:271–86.
- Shaldam M, Eldehna WM, Nocentini A, et al. Development of novel benzofuran-based SLC-0111 analogs as selective cancer-associated carbonic anhydrase isoform IX inhibitors. Eur J Med Chem 2021;216:113283.
- Grant S. Therapeutic protein kinase inhibitors. Cell. Mol. Life Sci 2009;66:1163–77.
- Normanno N, De Luca A, Bianco C, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene 2006;366:2–16.
- Sitohy B, Nagy JA, Dvorak HF. Anti-VEGF/VEGFR therapy for cancer: reassessing the target. Cancer Res 2012;72:1909–14.
- Touat M, Ileana E, Postel-Vinay S, et al. Targeting FGFR signaling in cancer. Clin Cancer Res 2015;21:2684–94.
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 1997;13:261–91.
- Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell 1996;85:149–58.
- Huang S-M, Harari PM. Epidermal growth factor receptor inhibition in cancer therapy: biology, rationale and preliminary clinical results. Invest New Drugs 1999;17:259–69.
- Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol 1995;19:183–232.
- Nicholson R, gee JM, harper M. egfR and cancer prognosis. Eur J Cancer 2001;37:9–15.
- Zhang H-Q, Gong F-H, Ye J-Q, et al. Design and discovery of 4-anilinoquinazoline-urea derivatives as dual TK inhibitors of EGFR and VEGFR-2. Eur J Med Chem 2017;125:245–54.
- Song Z, Huang S, Yu H, et al. Synthesis and biological evaluation of morpholine-substituted diphenylpyrimidine derivatives (Mor-DPPYs) as potent EGFR T790M inhibitors with improved activity toward the gefitinib-resistant non-small cell lung cancers (NSCLC). Eur J Med Chem 2017;133:329–39.
- Zhang Y, Chen L, Xu H, et al. 6, 7-Dimorpholinoalkoxy quinazoline derivatives as potent EGFR inhibitors with enhanced antiproliferative activities against tumor cells. Eur J Med Chem 2018;147:77–89.
- Belal A. Synthesis, molecular docking and antitumor activity of novel pyrrolizines with potential as EGFR-TK inhibitors. Bioorg Chem 2015;59:124–9.
- Jaiash DA, Belal A, Abdelgawad MA, Abdellatif KR. Design, synthesis and biological evaluation of new pyrroloazepines with potential and selective antitumor activity. Acta Poloniae Pharmaceutica 2016;73:359–68.
- Ding L, Cao J, Lin W, et al. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci 2020;21:1960.
- Belal A. 3D-pharmacophore modeling, molecular docking, and virtual screening for discovery of novel CDK4/6 selective inhibitors. Russian J Bioorg Chem 2021;47:317–33.
- Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov 2015;14:130–46
- Ahn YM, Vogeti L, Liu C-J, et al. Design, synthesis, and antiproliferative and CDK2-cyclin a inhibitory activity of novel flavopiridol analogues. Bioorg Med Chem 2007;15:702–13.
- Peng C, Zeng W, Su J, et al. Cyclin-dependent kinase 2 (CDK2) is a key mediator for EGF-induced cell transformation mediated through the ELK4/c-Fos signaling pathway. Oncogene 2016;35:1170–9.
- Yang A-l, Wu Q, Hu Z-d, et al. A network pharmacology approach to investigate the anticancer mechanism of cinobufagin against hepatocellular carcinoma via downregulation of EGFR-CDK2 signaling. Toxicol Appl Pharmacol 2021;431:115739.
- Joshi A, Bhojwani H, Wagal O, et al. Evaluation of benzamide-chalcone derivatives as EGFR/CDK2 inhibitor: synthesis, in-vitro inhibition, and molecular modeling studies, anti-cancer agents in medicinal chemistry (formerly current). Anti-cancer Agents Med Chem 2022;22:328–43.
- Mediani L, Antoniani F, Galli V, et al. Hsp90‐mediated regulation of DYRK3 couples stress granule disassembly and growth via mTORC1 signaling. EMBO Reports 2021;22:e51740.
- Wippich F, Bodenmiller B, Trajkovska MG, et al. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013;152:791–805.
- Papadopoli D, Pollak M, Topisirovic I. The role of GSK3 in metabolic pathway perturbations in cancer. Biochim Biophys Acta (BBA)-Mol Cell Res 2021;1868:119059.
- Quintayo MA, Munro AF, Thomas J, et al. GSK3β and cyclin D1 expression predicts outcome in early breast cancer patients. Breast Cancer Res Treat 2012;136:161–8.
- Patel P, Woodgett JR. Glycogen synthase kinase 3: a kinase for all pathways? Curr Top Dev Biol 2017;123:277–302.
- Bonomi P. Erlotinib: a new therapeutic approach for non-small cell lung cancer. Expert Opin Investig Drugs 2003;12:1395–401.
- Ou S-HI. Second-generation irreversible epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs): a better mousetrap? A review of the clinical evidence. Crit Rev Oncol Hematol 2012;83:407–21.
- Walter AO, Sjin RTT, Haringsma HJ, et al. Discovery of a mutant-selective covalent inhibitor of EGFR that overcomes T790M-mediated resistance in NSCLC. Cancer Discovery 2013;3:1404–15.
- Sequist LV, Besse B, Lynch TJ, et al. Neratinib, an irreversible pan-ErbB receptor tyrosine kinase inhibitor: results of a phase II trial in patients with advanced non–small-cell lung cancer. J Clin Oncol 2010;28:3076–83.
- Kim Y, Ko J, Cui Z, et al. The EGFR T790M mutation in acquired resistance to an irreversible second-generation EGFR inhibitor. Mol Cancer Therap 2012;11:784–91.
- Zhao Z, Wu H, Wang L, et al. Exploration of type II binding mode: a privileged approach for kinase inhibitor focused drug discovery? ACS Chem Biol 2014;9:1230–41.
- Furet P, Caravatti G, Lydon N, et al. Modelling study of protein kinase inhibitors: binding mode of staurosporine and origin of the selectivity of CGP 52411. J Comput Aided Mol Des 1995;9:465–72.
- Gandin V, Ferrarese A, Dalla Via M, et al. Targeting kinases with anilinopyrimidines: discovery of N-phenyl-N’-[4-(pyrimidin-4-ylamino) phenyl] urea derivatives as selective inhibitors of class III receptor tyrosine kinase subfamily. Sci Rep 2015;5:16750.
- Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol 2006;2:358–64.
- Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 2009;9:28–39.
- Elmetwally SA, Saied KF, Eissa IH, Elkaeed EB. Design, synthesis and anticancer evaluation of thieno [2, 3-d] pyrimidine derivatives as dual EGFR/HER2 inhibitors and apoptosis inducers. Bioorg Chem 2019;88:102944.
- Nasser AA, Eissa IH, Oun MR, et al. Discovery of new pyrimidine-5-carbonitrile derivatives as anticancer agents targeting EGFR WT and EGFR T790M. Org Biomol Chem 2020;18:7608–34.
- Traxler P, Furet P. Strategies toward the design of novel and selective protein tyrosine kinase inhibitors. Pharmacol Therap 1999;82:195–206.
- Malki Y, Martinez J, Masurier N. 1, 3‐Diazepine: a privileged scaffold in medicinal chemistry. Med Res Rev 2021;41:2247–315.
- Xu T, Zhang L, Xu S, et al. Pyrimido [4, 5‐d] pyrimidin‐4 (1H)‐one derivatives as selective inhibitors of EGFR threonine790 to methionine790 (T790M) mutants. Angewandte Chemie 2013;125:8545–8.
- Ibrahim M, Taghour M, Metwaly A, et al. Design, synthesis, molecular modeling and anti-proliferative evaluation of novel quinoxaline derivatives as potential DNA intercalators and topoisomerase II inhibitors. Eur J Med Chem 2018;155:117–34.
- Mahdy HA, Ibrahim MK, Metwaly AM, et al. Design, synthesis, molecular modeling, in vivo studies and anticancer evaluation of quinazolin-4 (3H)-one derivatives as potential VEGFR-2 inhibitors and apoptosis inducers. Bioorg Chem 2020;94:103422.
- El-Naggar AM, Eissa IH, Belal A, El-Sayed AA. Design, eco-friendly synthesis, molecular modeling and anticancer evaluation of thiazol-5 (4 H)-ones as potential tubulin polymerization inhibitors targeting the colchicine binding site. RSC Adv 2020;10:2791–811.
- Eissa IH, Metwaly AM, Belal A, et al. Discovery and antiproliferative evaluation of new quinoxalines as potential DNA intercalators and topoisomerase II inhibitors. Archiv Der Pharmazie 2019;352:1900123.
- Eissa IH, Ibrahim MK, Metwaly AM, et al. Design, molecular docking, in vitro, and in vivo studies of new quinazolin-4 (3H)-ones as VEGFR-2 inhibitors with potential activity against hepatocellular carcinoma. Bioorg Chem 2021;107:104532.
- Alanazi MM, Eissa IH, Alsaif NA, et al. Design, synthesis, docking, ADMET studies, and anticancer evaluation of new 3-methylquinoxaline derivatives as VEGFR-2 inhibitors and apoptosis inducers. J Enzyme Inhib Med Chem 2021;36:1760–82.
- Hagras M, El Deeb MA, Elzahabi HS, et al. Discovery of new quinolines as potent colchicine binding site inhibitors: design, synthesis, docking studies, and anti-proliferative evaluation. J Enzyme Inhib Med Chem 2021;36:640–58.
- Alanazi MM, Elwan A, Alsaif NA, et al. Discovery of new 3-methylquinoxalines as potential anti-cancer agents and apoptosis inducers targeting VEGFR-2: design, synthesis, and in silico studies. J Enzyme Inhib Med Chem 2021;36:1732–50.
- Gaber AA, Bayoumi AH, El-Morsy AM, et al. Design, synthesis and anticancer evaluation of 1H-pyrazolo [3, 4-d] pyrimidine derivatives as potent EGFRWT and EGFRT790M inhibitors and apoptosis inducers. Bioorg Chem 2018;80:375–95.
- El-Moghazy SM, Hanna MM, Farag AE, Belal A. Synthesis and biological evaluation of some novel substituted pyrrolizines and pyrimidopyrrolizines as Chemotherapeutic agents. Der Chemica Sinica 2017;8(1):102–16.
- Shaaban HG. Synthesis of some heterocyclic compounds derived from 2-chloro-Np-tolylacetamide. Al-Mustansiriyah J Sci 2017;27. DOI:10.23851/mjs.v27i4.26
- Gouda AM, Abdelazeem AH, Arafa E-SA, Abdellatif KR. Design, synthesis and pharmacological evaluation of novel pyrrolizine derivatives as potential anticancer agents. Bioorg Chem 2014;53:1–7.
- Khademi Z, Nikoofar K. Applications of alkyl orthoesters as valuable substrates in organic transformations, focusing on reaction media. RSC Adv 2020;10:30314–97.
- Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. JNCI: J Natl Cancer Inst 1990;82:1107–12.
- Elsayed MS, El-Araby ME, Serya RA, et al. Structure-based design and synthesis of novel pseudosaccharine derivatives as antiproliferative agents and kinase inhibitors. Eur J Med Chem 2013;61:122–31.
- El-Helby A-GA, Ayyad RR, El-Adl K, Elkady H. Phthalazine-1, 4-dione derivatives as non-competitive AMPA receptor antagonists: design, synthesis, anticonvulsant evaluation, ADMET profile and molecular docking. Mol Divers 2019;23:283–98.
- El‐Helby AGA, Ayyad RR, Zayed MF, et al. Design, synthesis, in silico ADMET profile and GABA‐A docking of novel phthalazines as potent anticonvulsants. Archiv Der Pharmazie 2019;352:1800387.
- Abdallah AE, Alesawy MS, Eissa SI, et al. Design and synthesis of new 4-(2-nitrophenoxy) benzamide derivatives as potential antiviral agents: molecular modeling and in vitro antiviral screening. N J Chem 2021;45:16557–71.
- Xia X, Maliski EG, Gallant P, Rogers D. Classification of kinase inhibitors using a Bayesian model. J Med Chem 2004;47:4463–70.
- BIOVIA, QSAR, ADMET and predictive toxicology. https://www.3dsbiovia.com/products/collaborative-science/biovia-discovery-studio/qsar-admet-and-predictive-toxicology.html [last accessed May 2020].
- Alanazi MM, Elkady H, Alsaif NA, et al. Discovery of new quinoxaline-based derivatives as anticancer agents and potent VEGFR-2 inhibitors: design, synthesis, and in silico study. J Mol Struct 2022;1253:132220.
- Alsaif NA, Taghour MS, Alanazi MM, et al. Discovery of new VEGFR-2 inhibitors based on bis ([1, 2, 4] triazolo)[4, 3-a: 3', 4'-c] quinoxaline derivatives as anticancer agents and apoptosis inducers. J Enzyme Inhib Med Chem 2021;36:1093–114.
- Alanazi MM, Elkady H, Alsaif NA, et al. New quinoxaline-based VEGFR-2 inhibitors: design, synthesis, and antiproliferative evaluation with in silico docking, ADMET, toxicity, and DFT studies. RSC Adv 2021;11:30315–28.
- Alsaif NA, Dahab MA, Alanazi MM, et al. New quinoxaline derivatives as VEGFR-2 inhibitors with anticancer and apoptotic activity: design, molecular modeling, and synthesis. Bioorg Chem 2021;110:104807.
- Abbas SE, Awadallah FM, Ibrahim NA, et al. Design, synthesis and preliminary evaluation of some novel [1, 4] diazepino [5, 6-b] pyrrolizine and 6-(2-oxopyrrolidino)-1H-pyrrolizine derivatives as anticonvulsant agents. Med. Chem. Res 2011;20:1015–23.
- Nicoletti I, Migliorati G, Pagliacci M, et al. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods 1991;139:271–9.
- Hammouda MM, Abo Elmaaty A, Nafie MS, et al. Design and synthesis of novel benzoazoninone derivatives as potential CBSIs and apoptotic inducers: In Vitro, in Vivo, molecular docking, molecular dynamics, and SAR studies. Bioorg Chem 2022;127:105995.
- Belal A, Elanany MA, Santali EY, et al. Screening a panel of topical ophthalmic medications against MMP-2 and MMP-9 to investigate their potential in keratoconus management. Molecules 2022;27:3584.
- Chemical Computing Group, Molecular Operating Environment (MOE). http://www.chemcomp.com/ [last accessed May 2017].