Abstract
Introduction: Colchicine-binding site inhibitors are some of the most interesting ligands belonging to the wider family of microtubule-destabilising agents.
Results: A novel series of 4′-fluoro-substituted ligands (5–13) was synthesised. The antiproliferative activity assays resulted in nM values for the new benzotriazole-acrylonitrile derivatives. Compound 5, the hit compound, showed an evident blockade of HeLa cell cycle in the G2-M phase, but also a pro-apoptotic potential, and an increase of early and late apoptotic cells in HeLa and MCF-7 cell cycle analysis. Confocal microscopy analysis showed a segmented shape and a collapse of the cytoskeleton, as well as a consistent cell shrinkage after administration of 5 at 100 nM. Derivative 5 was also proved to compete with colchicine at colchicine-binding site, lowering its activity against tubulin polymerisation. In addition, co-administration of 5 and doxorubicin in drug-resistant A375 melanoma cell line highlighted a synergic potential in terms of inhibition of cell viability.
Discussion: The 4′-fluoro substitution of benzotriazole-acrylonitrile scaffold brought us a step forward in the optimisation process to obtain compound 5 as promising MDA antiproliferative agent at nanomolar concentration.
Graphical Abstract

Introduction
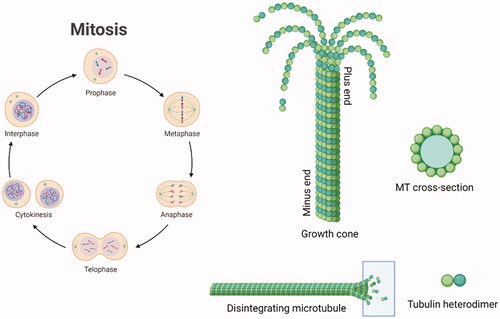
Cancer is one of the main clinical issues worldwide. Anticancer therapy is still a focus of academic and industrial research, which aims to improve the potency and safety of validated anticancer protocols and to create new ones.Citation1 Antitumor compounds targeting the microtubule (MT) structure affect the cell skeleton and the replication process but also act on apoptosis, therefore some of them have been approved for clinical cancer treatment.Citation2 Microtubules (MTs) have a fine-tuned dynamic mechanism of polymerisation and depolymerisation dealing with cell division. A single microtubule is constituted by heterodimers of α- and β-tubulin.Citation3–5 Tubulin polymerisation is crucial for the creation of microtubules. It is regulated by the hydrolysis of GTP (guanosine-5′-triphosphate) in the β-portion of tubulin dimer. GTP caps stabilise the formed microtubule ends.Citation6 The α- and β-hetero-polypeptides of tubulin have about 36–42% similarity to each other and each subunit consists of about 445 amino acids. The 3D structure of the α,β-tubulin heterodimer has been determined by X-ray diffraction (Protein Data Bank Identity [PDB ID]: 4O2B)Citation7 and both monomers were shown to surround a GTP molecule. MT growth occurs at the plus end and the shortening at the minus end.Citation8,Citation9 reports a simplified representation of the mitotic cycle and the microtubule depolymerisation at the plus end.
Figure 1. Schematic representation of the mitotic cycle. On the right, a picture of an MT growth cone, made of α,β-tubulin heterodimers. Created (and partly adapted from a template) with BioRender.com.
MT-targeting drugs are commonly divided into microtubule-destabilising agents (MDAs), e.g. colchicine or vinblastine, and microtubule-stabilising agents (MSAs), such as paclitaxel or epothilones. MDAs prevent tubulin polymerisation and the consequent MT assembly; differently, MSAs promote tubulin polymerisation.Citation10 Focussing on MDAs therapy, their clinical challenges include low therapeutic windows and innate or acquired drug resistance. Some tumours thought to have microtubule-independent trafficking of key oncogenic proteins seem not to respond to MDAs at all, such as renal cell carcinomas.Citation11 One of the most famous MDAs is colchicine (), an alkaloid mainly indicated as a treatment of inflammatory diseases, such as recurrent pericarditis treatment,Citation12 gout and familial Mediterranean fever.Citation13,Citation14 Colchicine interacts with tubulin heterodimer in the so-called colchicine-binding site (CBS), located at the interface between α- and β-tubulin.Citation15 Colchicine-tubulin interaction makes the microtubule polymerisation process energetically unfavourable and prevents the microtubule growth by sterically blocking further addition of tubulin dimers at the plus end.Citation16 Colchicine was also shown to alter the mitochondrial membrane potential and release pro-apoptotic factors like caspases, leading to apoptotic cell death.Citation17 Despite its multimodal activity, its therapeutic potential against cancer is restrained due to its low therapeutic index. Also, compound ZD6126, a water-soluble phosphate prodrug, initially selected as a gold alternative to colchicine (), did not succeeded further as an anticancer agent due to cardiotoxicity and other severe side effects.Citation18 Although colchicine failure, multiple efforts were made to develop clinically potent colchicine-binding site inhibitors (CBSIs). CBSIs are some of the most interesting MDAs as they target the β-subunit of tubulin in its curved and unassembled form and prevent it from adopting a microtubular straight structure.Citation19,Citation20 So far, combretastatin A-4 is arguably the most successful representative of the CBSI family.Citation21,Citation22
Figure 2. Chemical structures of colchicine and its derivative ZD6126, combretastatin A-4, our previous lead compounds 34 and 9a, and target compound 5 of the present work.
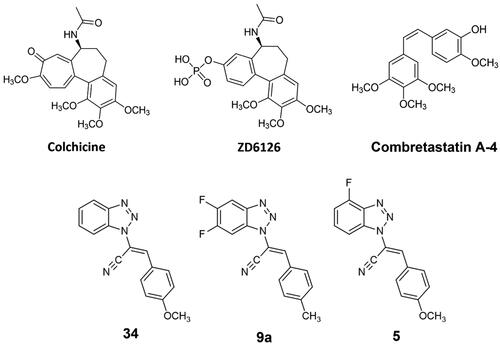
Many other CBISs have been synthesised in the last decades, including compound 34,Citation23 an acrylonitrile derivative developed by us and identified as CBSI through computational and experimental analysis.Citation23–29 Following this direction, we decided to explore the effect of a difluoro substitution on the benzotriazole scaffold, therefore a series of 5′,6′-difluoro-benzotriazole-acrylonitrile derivatives were designed, synthesised and screened, indicating the hit compound 9a as the most potent.Citation30 The two hit compounds 34 and 9a showed a consistent inhibition of cancer cell growth and a general blockage of the cancer cell cycle in the G2/M phase. Starting from this background, in the present work, we carried out a hit optimisation strategy on the heterocycle scaffold consisting of the mono-fluoro substitution in position C–4′ of the benzotriazole in place of the 5′6′-difluoro functionalisation previously reported, in order to reduce the molecular weight while maintaining the halogen interactions that were proved successful in the former series.
Materials and methods
Molecular docking
The academic program AutoDock Vina 1.1.2. was employed for the molecular docking investigation.Citation31 The tubulin-colchicine crystal structure (PDB ID: 4O2B; 2.30 Å of resolution) was downloaded from the Protein Data Bank.Citation32 The starting crystal structure of tubulin was processed: water molecules were removed, as well as GOL (glycerol), the buffer MES (2-(N-morpholino)-ethanesulfonic acid), Ca2+ and Mg2+ ions, GDP (guanosine-5′-diphosphate) and GTP. For a less time-consuming process, only the A and B chains were considered for the simulations. Protein Data Bank, Partial Charge, Atom Type (PDBQT) files of protein and ligands were obtained through PyMOL 2.3.4.Citation33 and AutoDockTools (ADT).Citation34 ADT Grid Box was set as 32–32-32 Å (x,y,z). Docking was performed by setting amino acids Serα178, Thrα179, Alaα180, Alaα181 flexible, considered crucial for the binding affinity of colchicine in its binding site on tubulin.Citation7 The exhaustiveness was set to 64 and 10 best-predicted poses were generated for each calculation. The range of binding energy was set as ≤ 2 kcal/mol, above that of the top-ranked pose. LigPlot + v.2.2.Citation35 generated 2D representations of protein-ligand interactions with a 5 Å cut-off from the ligand centre of mass. 3D figures were taken through PyMOL, with a 3000 dpi of rendering. A PC with an Intel® Core™ i7–9705H CPU @ 2.60 GHz with 8 GB RAM (operating system: Ubuntu 16.04) was employed, utilising 6 CPUs for each simulation.
Chemistry
Synthesis and compounds’ characterisation
As reported in Scheme 1, compound 1 was alkylated to yield the acetonitrile isomers 2–4. Final compounds 5–7, 8–10, and 11–13 were obtained via Knoevenagel condensation of intermediates 2–4 with aldehydes I–IV. Regarding the spectroscopic characterisation, the nuclear magnetic resonance (NMR) spectra were registered in a solution of deuterated dimethyl sulfoxide (DMSO-d6) and recorded with a Bruker Avance III 400 NanoBay (400 MHz). 1H NMR δs (chemical shifts) are reported in parts per million (ppm) downfield from tetramethylsilane (TMS), as the internal standard. Chemical shift values are reported in ppm and coupling constants (J) are reported in Hertz (Hz). In some cases, D2O was added to assign exchangeable protons (OH, NH). Signal splitting is represented as s (singlet), d (doublet), dd (doublet of doublets), ddd (doublet of doublets of doublets), t (triplet), dt (doublet of triplets), m (multiplet), and wm (wide multiplet). 13 C NMR chemical shifts are reported downfield from TMS, jmod (J-modulated spin-echo for X-nuclei coupled to H–1, to determine the number of attached protons) was employed. The assignment of resonances was evaluated based on relative intensity, chemical shift, and fine structure using assignment predictions by www.nmrdb.org. ESI–MS characterisation involved the preparation of each compound, dissolved in CH3CN for HPLC (1.0–2.0 ppm). Full mass spectra were recorded on a Q Exactive Plus Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific), in positive- and negative-ion mode. The solutions were infused into the ESI chamber at a flow rate of 5.00 μl/min. The spectra were recorded in the m/z range 150–800 at a resolution of 140,000 and accumulated for approximately 2 min to increase the signal-to-noise ratio. Positive-ion acquisitions were conducted with the following parameters: spray voltage 2300 V, capillary temperature 250 °C, sheath gas 10 (arbitrary units), auxiliary gas 3 (arbitrary units), sweep gas 0 (arbitrary units) and probe heater temperature 50 °C. Negative-ion measurements were done as follows: spray voltage −1900 V, capillary temperature 250 °C, sheath gas 20 (arbitrary units), auxiliary gas 5 (arbitrary units), sweep gas 0 (arbitrary units), probe heater temperature 50 °C. ESI–MS spectra analysis was made by using Thermo Xcalibur 3.0.63 software (Thermo Fisher Scientific), and the Xtract tool was used to extract the deconvoluted monoisotopic masses, as an averaged value. Commercially available chemicals were purchased from Carlo Erba Reagents and Sigma Aldrich. Evaporation was performed in vacuo (rotating evaporator). Sodium sulphate was always used as the drying agent. Celite® 545 was used as filter agent. A mixture of PS (petroleum spirit) and EA (ethyl acetate), ratio 8:2, was employed to calculate the retention factors (Rf), using Thin Layer Chromatographies (TLCs) on Merck F-254 plates. Melting points (m.p.) were measured in a Köfler hot stage in open capillaries or Digital Electrothermal melting point apparatus and were uncorrected.
Biology
NCI60 in vitro screening
A first NCI60 screening was performed to assess the in vitro anticancer activity through the Developmental Therapeutics Program of the National Cancer Institute (NCI, Bethesda, USA). All detailed NCI reports are depicted in Table S1 and Figures S29-44, and the experimental methods are explained on the NCI website (https://dtp.cancer.gov). To summarise, a first single compound dose of 10 µM was administered to the entire panel of NCI60 cancer cell lines. If the growth percentage, 50% of growth inhibition, tumour growth inhibition and 50% of lethal concentration profile of the molecule agrees with the selection criteria, the above-mentioned derivative is administered to the entire NCI60 panel of cancer cell lines in 5 × 10-fold dilutions, from 100 µM up to 0.01 µM.
Cell culture
HeLa, MCF-7, and A375 cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco). The medium was supplemented with 10% foetal bovine serum, 100 units/ml penicillin, 100 µg/ml streptomycin (Gibco), and 1% l-glutamine. Cells were incubated at 37 °C with 5% CO2 in humidified air. Every cell line was tested for mycoplasma contamination as previously described.Citation36 Doxorubicin (Sigma Aldrich) were resuspended in DMSO and administered for 24 h at 2 μM.
Proliferation assay (XTT)
Cells were seeded at a density of 1000 cells/well in a plate-based 96-well on different sizes, population doublings, and phenotypes. Twenty-four hours after seeding, cells were treated for 48 h with different concentrations of all compounds in Scheme 1 in a range from 0.125 μM to 2 μM in a final volume of 100 μl. DMSO and compound 34 were used as negative and positive controls, respectively. After 48 h, the XTT assay was performed using the Cell Proliferation Kit II (Roche, Basel, Switzerland). 0.5 μl of XTT electron-coupling reagent and 25 μl of labelling reagent were resuspended in 74.5 μl of medium (final volume 100 μl/well) and incubated for 4 h at 37 °C. After incubation, absorbance was measured at 490 nm with a spectrophotometric plate reader (SPECTRAMax 384 PLUS) and the results obtained were used to calculate IC50 values. Based on the IC50 value, compounds 5 and 12 were selected for subsequent experiments. All experiments were performed in triplicate.
Cell cycle analysis
Cells were seeded at 50% confluence on 6-cm dishes. After 24 h, cells were treated with 2 µM of compounds 5 and 12 for 24 h. Afterwards, floating (necrotic and in late apoptosis cells) and adherent cells were first centrifuged at 3000 rpm for 5 min; washed in PBS and then fixed with ice-cold 70% ethanol and incubated at −20 °C overnight. Fixed cells were washed twice with cold PBS and then resuspended with 150 μl of a solution of PBS and 10 μl/test of 7-aminoactinomycin D (7-AAD; Bioscience, San Diego, CA) and incubated for 20 min at room temperature. Cell cycle assay was performed by flow cytometry using BD FACS CANTO II and collecting approximately 20,000 events for each sample. Data were analysed by BD FACS DIVA software. All experiments were performed in triplicate.
Annexin V assay
Annexin V assay was performed using the FITC Annexin V Apoptosis Detection Kit II (BD Pharmingen, San Jose, CA), according to the manufacturer’s protocol. After 24 h, cells were treated at concentrations of 2 µM of compounds 5 and 12. After 24 h, floating and adherent cells were harvested, centrifuged, and washed twice with cold PBS. Annexin V 1x buffer was used to resuspend the cells. Five microliters of 7-AAD and 5 μl of V-fluorofluorine-annexin isothiocyanate were added to the cells and incubated in the dark for 15 min at room temperature. Subsequently, 200 μl of Annexin V 1× buffer was added in each sample. Flow cytometry was performed immediately using BD FACS CANTO II. 20,000 events were analysed using BD FACS DIVA software. All experiment were performed in triplicate.
Protein extraction and Western blot
Total proteins were extracted using lysis buffer (20 mM Tris HCl pH 8; 137 mM NaCl; 10% glycerol; 1% Nonidet P-40; 2 mM EDTA) with the addition of Protease Inhibitor Cocktail (Roche, Basel, Switzerland). Twenty micrograms of protein was dissolved in 8% SDS/PAA (Sodium Dodecyl Sulphate - PolyAcrylamide) and then transferred to a nitrocellulose membrane (GE, Healthcare, Whatman) for 1 h at 4 °C and 100 V. The nitrocellulose membrane was blocked with 5% bovine serum albumin for 1 h at room temperature. Blots were incubated overnight at 4 °C with primary anti-PARP (Cell Signalling, Boston, MA) and anti-GAPDH (Santa Cruz Biotechnology, CA) antibodies. Membranes were incubated with antibodies-conjugated secondary peroxidase and the signal was detected with Western Lightning Plus-ECL (PerkinElmer, Waltham, MA, USA).
Colchicine-5 competition assay
Tubulin polymerisation assay was carried out using the kit supplied by Cytoskeleton (BK006P, Cytoskeleton, Denver, CO). The assay is based on the principle that light is scattered by microtubules to an extent that fluorescence is proportional to the concentration of the microtubule polymer. The absorbance was recorded at 340 nm at 37 °C for 30 min with fixed acquisitions every 2 s. Colchicine-5 competition assay was performed incubating equimolar (1 μM) concentrations of colchicine and compound 5 with tubulin. After 30 min, fluorescence emitted by tubulin polymerisation was recorded by reading absorbance for 30 min.
High-content imaging of HeLa cells and IC50 calculation
Imaging experiments were performed using an Operetta CLS high-content imaging device (PerkinElmer, Hamburg, Germany), and analysed with Harmony 4.6 software (PerkinElmer). To assess the tubulin integrity, we acquired HeLa cells images using 63× magnification, taking 25 fields per sample in biological and technical triplicates. Data were analysed using the following building blocks: 1—Find Nuclei, 2—Find Cytoplasm (Tubulin+). IC50 was obtained by counting the number of nuclei in each field. Cell dimensions were assessed using the following building blocks: 1—Find Nuclei, 2—Find Cytoplasm (Tubulin+), 3—Calculate Morphology properties (Area), as previously described.Citation37–39 IC50s were calculated using the number of cells detected in every well as indexing parameter by Harmony 4.6 software as described here.Citation40
Experimental section
Materials
All starting materials and the different aldehydes (I–IV) were commercially purchased. Compound 1 was synthesised as previously reported.Citation41
General procedure for the synthesis of intermediates 2-(4-fluoro-1H-benzo[d][1,2,3]triazol-1-yl)acetonitrile (2), 2-(4-fluoro-2H-benzo [d][1,2,3]triazol-2-yl)acetonitrile (3) and 2-(7-fluoro-1H-benzo[d] [1,2,3]triazol-1-yl)acetonitrile (4)
Methods: Compound 1/chloroacetonitrile/KOH (1:1:1.1), CH3CN, reflux, overnight. The three isomers were separated by flash chromatography (PS/EA 9/1). C8H5FN4; MW: 176.15. Total yield of the three isomers was 47%. Compound 2: Yellow solid; 10% yield (0.18 g, 1.0 mmol); m.p. 73.4–75.6 °C. Rf 0.19. 1H NMR (DMSO-d6): δ 7.82 (1H, d, 1JH–H= 8.4 Hz, H–7), 7.69 (1H, dt, H–6), 7.34 (1H, dd, 1 J = 10.8 Hz, 2 J = 8.0 Hz, H–5), 6.22 (2H, s, CH2). 13 C NMR (DMSO-d6): δ 152.15 (C, d, 1JC–F= 254.0 Hz, C–F), 135.26 (C, d, 1JC–F= 7.0 Hz, C), 135.03 (C, d, 1JC–F = 19.0 Hz, C), 129.74 (C, d, 1JC–F= 7.0 Hz, CH), 114.67 (C≡N), 109.41 (C, d, 1JC–F= 16.0 Hz, CH), 106.87 (C, d, 1JC–F= 5.0 Hz, CH), 35.95 (CH2). ESI–MS m/z calcd for C8H5FN4 177.0571, found 176.8251 [M + H]+. Compound 3: Yellow solid; 23% yield (0.45 g, 2.6 mmol); m.p. 85.2–87.1 °C; Rf 0.58. 1H NMR (DMSO-d6): δ 7.92 (1H, d, 1JH–H= 8.4 Hz, H–7), 7.58 (1H, dt, H–6), 7.42 (1H, dd, 1JH–H= 5.0 Hz, 2JH–H= 2.0 Hz, H–5), 6.40 (2H, s, CH2). 13 C NMR (DMSO-d6): δ 151.24 (C, d, 1 J = 255.0 Hz, C–F), 146.69 (C, d, 1JC–F= 3.0 Hz, C), 135.17 (C, d, 1JC–F= 16.0 Hz, C), 127.91 (C, d, 1JC–F= 7.0 Hz, CH), 114.56 (C, d, 1JC–F= 5.0 Hz, CH), 114.09 (C≡N) 110.83 (C, d, 1JC–F= 16.0 Hz, CH), 44.18 (CH2). ESI–MS m/z calcd for C8H5FN4 177.0571, found 177.0913 [M + H]+. Compound 4: White solid; 14% yield (0.27 g, 1.5 mmol); m.p. 89.1–91.2 °C; Rf 0.38. 1H NMR (DMSO-d6): δ 7.89 (1H, d, 1 J = 8.4 Hz, H–7), 7.56 (1H, dt, H–6), 7.39 (1H, dd, 1 J = 4.0 Hz, 2 J = 1.0 Hz, H–5), 6.38 (2H, s, CH2). 13 C NMR (DMSO-d6): δ 148.54 (C), 146.99 (C, d, C–F), 127.91 (C, d, 1JC–F= 6.0 Hz, CH), 122.26 (C, d, 1JC–F= 14.0 Hz, C), 115.98 (C, d, 1JC–F= 5.0 Hz, CH), 110.83 (CH–CF), 114.83 (C≡N), 113.26 (C, d, 1JC–F= 16.0 Hz, CH), 37.44 (CH2). ESI–MS m/z calcd for C8H5FN4 177.0571, 178.0604, found 176.8253, 178.2626 [M + H]+.
General procedure for compounds (E)-2-(4-fluoro-1H-benzo[d] [1,2,3]triazol-1-yl)-3-(R)acrylonitrile (5–7), (E)-2-(4-fluoro-2H-benzo[d][1,2,3]triazol-2-yl)-3-(R)acrylonitrile (8–10), (E)-2-(7-fluoro-1H-benzo[d] [1,2,3]triazol-1-yl)-3-(R)acrylonitrile (11–13)
Compounds 5–7 were obtained via Knoevenagel condensation of 2 and the appropriate aldehydes I–IV. 8–10 were synthesised by reaction of 3 and I–IV, while intermediate 4 and the aldehydes I–IV led to compounds 11–13. Reaction conditions: (A) DMA (ratio 1:1.2), CH3CN for 9–10 and 12; (B) DIMCARB (ratio 1:1.2), CH3CN for 5, 6, 7; (C) Cs2CO3 (ratio 1:1.2), toluene for 8, 11, 13. Work up: (A) filtration; (B) crystallisation from EtOH; (C) flash chromatography (PS/EA 9/1).
(E)-2-(4-fluoro-1H-benzo[d][1,2,3]triazol-1-yl)-3-(4-methoxyphenyl) acrylonitrile (5)
Work-up procedure (B). Yellow solid; C16H11FN4O, MW: 294.28; 52% yield (0.23 g); m.p. 141.2–143.4 °C; Rf 0.35. 1H NMR (400 MHz, DMSO-d6): δ 8.23 (1H, s, =CH), 8.03 (2H, d, 1JH–H= 8.8 Hz, H–3″,5″), 7.86 (1H, d, 1JH–H= 8.4 Hz, H–7′), 7.75 (1H, dt, H–6′), 7.43 (1H, dd, 1 J = 10.4 Hz, 2 J = 8.0 Hz, H5′), 7.21 (2H, d, 1JH–H= 8.4 Hz, H–2″,6″), 3.89 (3H, s, OCH3). 13 C NMR (DMSO-d6): δ 162.48 (C), 152.27 (C, d, 1JC–F= 255.0 Hz, C–F), 143.01 (=CH), 135.17 (C, d, 1JC–F= 19.0 Hz, C), 134.57 (C, d, 1JC–F= 6.0 Hz, C), 131.96 (2CH), 132.43 (2CH), 130.95 (C, d, 1JC–F= 8.0 Hz, CH), 123.17 (C), 115.31 (2CH), 115.00 (C≡N), 105.42 (C, d, 1JC–F = 16.0 Hz, CH), 108.08 (C, d, 1JC–F = 5.0 Hz, CH), 102.99 (C), 56.12 (OCH3). ESI–MS m/z calcd for C16H11FN4O 295.0989, 296.1023, found 295.0989, 296.1023 [M + H]+.
(E)-2-(4-fluoro-1H-benzo[d][1,2,3]triazol-1-yl)-3-(p-tolyl)acrylonitrile (6)
Work-up procedure (C, PS/EA 7/3). Yellow solid; C16H11FN4, MW: 278.10; 50% yield (0.36 g); m.p. 128.1–130.3 °C; Rf 0.27. 1H NMR (DMSO-d6): δ 8.27 (1H, s, =CH), 7.93 (2H, d, 1JH–H= 8.0 Hz, H–3″,5″), 7.89 (1H, d, 1JH–H= 8.0 Hz, H–7′), 7.75 (1H, dt, H–6′), 7.45 (2H, d, 1JH–H= 8.0 Hz, H–2″,6″), 7.44 (1H, m, H5′), 2.43 (3H, s, CH3). 13 C NMR (DMSO-d6): δ 157.22 (C), 152.26 (C, d, 1JC–F= 255.0 Hz, C–F), 142.73 (=CH), 135.23 (C, d, 1JC–F= 19.0 Hz, C), 134.44 (C, d, 1JC–F= 6.0 Hz, C), 130.58 (C, d, 1JC–F= 7.0 Hz, CH), 129.90 (2CH), 129.74 (2CH), 127.65 (C), 114.11 (C≡N), 110.04 (C, d, 1JC–F = 16.0 Hz, CH), 107.71 (C, d, 1JC–F= 5.0 Hz, CH), 104.67 (C), 21.22 (OCH3). ESI–MS m/z calcd for C16H11FN4 279.1040, 280.1074, found 279.1032, 280.1187 [M + H]+.
(E)-3-(4-bromophenyl)-2-(4-fluoro-1H-benzo[d][1,2,3]triazol-1-yl)acrylonitrile (7)
Work-up procedure (B). Yellow solid; C15H8BrFN4, MW: 343.15; 35% yield (0.20 g); m.p. 158.6–160.3 °C; Rf 0.55. 1H NMR (DMSO-d6): δ 8.31 (1H, s, =CH), 7.94 (3H, m, H–3″,5″,7′), 7.86 (2H, d, 1JH–H= 8.8 Hz, H–2″,6″), 7.77 (1H, dt, H–6′), 7.45 (1H, dd, 1JH–F= 10.6 Hz, 2JH–F= 2.0 Hz, H–5′). 13 C NMR (DMSO-d6): δ 152.25 (C, d, 1JC–F= 255.0 Hz, C–F), 140.53 (CH), 135.29 (C, d, 1JC–F = 19.0 Hz, C), 134.24 (C, d, 1JC–F= 6.0 Hz, C), 132.33 (2CH), 131.47 (2CH), 130.70 (C, d, 1JC–F= 7.0 Hz, CH), 129.76 (C), 125.56 (C), 113.66 (C≡N), 110.18 (C, d, 1JC–F= 16.0 Hz, CH), 107.89 (C, d, 1JC–F= 4.0 Hz, CH), 106.67 (C). ESI–MS m/z calcd for C15H8BrFN4 342.9989, found 342.9972 [M + H]+.
(E)-2-(4-fluoro-2H-benzo[d][1,2,3]triazol-2-yl)-3-(4-methoxyphenyl)acrylonitrile (8)
Work-up procedure (C). Yellow powder; C16H11FN4O, MW: 294.28; 42% yield (0.23 g); m.p. 127.2–129.2 °C; Rf 0.69. 1H NMR (DMSO-d6): δ 8.72 (1H, s, =CH), 8.10 (2H, d, 1JH–H= 8.8 Hz, H–2″,6″), 7.89 (1H, d, 1JH–H= 8.4 Hz, H–7′), 7.55 (1H, dt, H–6′), 7.40 (1H, dd, 1JH–H= 11.0 Hz, 1JH–H= 8.0 Hz, H–5′), 7.20 (2H, d, 1JH–H= 8.8 Hz, H–3″,5″), 3.89 (3H, s, OCH3). 13 C NMR (DMSO-d6): δ 162.63 (C), 151.10 (C, d, 1JC–F= 256.0 Hz, C–F), 146.62 (C, d, 1JC–F= 3.0 Hz, C), 138.84 (CH), 135.29 (C, d, 1JC–F= 16.0 Hz, C), 132.37 (2CH), 128.53 (C, d, 1JC–F= 6.0 Hz, CH), 122.32 (C), 115.00 (2CH), 114.50 (C, d, 1JC–F= 5.0 Hz, CH), 113.48 (C≡N), 111.47 (C, d, 1JC–F= 15.0 Hz, CH), 108.70 (C), 55.66 (OCH3). ESI–MS m/z calcd for C16H11FN4O 295.0989, found 295.1803 [M + H]+.
(E)-2-(4-fluoro-2H-benzo[d][1,2,3]triazol-2-yl)-3-(p-tolyl)acrylonitrile (9)
Work-up procedure (A). Yellow solid; C15H8F2N4. MW: 282.07; 58% yield (0.28 g); m.p. 135.6–137.2 °C; Rf 0.70. 1H NMR (DMSO-d6): δ 8.75 (1H, s, =CH), 7.99 (2H, d, 1JH–H= 8.4 Hz, H–3″,5″), 7.90 (1H, d, 1JH–H= 8.8 Hz, H–7′), 7.57 (1H, dt, H–6′), 7.44 (2H, d, 1JH–H= 7.6 Hz, H–2″,6″), 7.41 (1H, m, H–5′), 2.42 (3H, s, CH3). 13 C NMR (DMSO-d6): δ 151.12 (C, d, 1JC–F= 256.0 Hz, C–F), 146.66 (C, d, 1JC–F= 4.0 Hz, C), 143.01 (C), 139.02 (CH), 135.40 (C, d, 1JC–F= 16.0 Hz, C), 130.10 (2CH), 129.97 (2CH), 128.71 (C, d, 1JC–F= 6.0 Hz, CH), 127.24 (C), 114.62 (C, d, 1JC–F= 5.0 Hz, CH), 113.09 (C≡N), 111.61 (C, d, 1JC–F= 16.0 Hz, CH), 110.46 (C), 21.24 (CH3). ESI–MS m/z calcd for C15H8F2N4 283.07898, 284.08233, found 283.9893, 284.1545 [M + H]+.
(E)-3-(4-bromophenyl)-2-(4-fluoro-2H-benzo[d][1,2,3]triazol-2-yl)acrylonitrile (10)
Work-up procedure (A). Light brown solid; C15H8BrFN4, MW: 343.15; 31% yield (0.18 g); m.p. 137.2–138.1 °C; Rf 0.71. 1H NMR (DMSO-d6): δ 8.79 (1H, s, =CH), 8.01 (2H, d, 1JH–H= 8.4 Hz, H–3″,5″), 7.91 (1H, d, 1JH–H= 8.8 Hz, H–7′), 7.85 (2H, d, 1JH–H= 8.4 Hz, H–2″,6″), 7.57 (1H, dt, H–6′), 7.42 (1H, dd, 1JH–H= 10.8 Hz, 2JH–H= 7.6 Hz, H–5′). 13 C NMR (DMSO-d6): δ 151.12 (C, d, 1JC–F= 256.0 Hz, C–F), 146.72 (C, d, 1JC–F= 3.0 Hz, C), 137.63 (CH), 135.53 (C, d, 1JC–F= 16.0 Hz, C), 132.39 (2CH), 131.77 (2CH), 129.33 (C), 128.93 (C, d, 1JC–F= 6.0 Hz, CH), 125.89 (C), 114.68 (C, d, 1JC–F= 6.0 Hz, CH), 112.71 (C≡N), 112.0 (C), 111.79 (C, d, 1JC–F= 16.0 Hz, CH). ESI–MS m/z calcd for C15H8BrFN4 344.9968, found 344.9965 [M + H]+.
(E)-2-(7-fluoro-1H-benzo[d][1,2,3]triazol-1-yl)-3-(4-methoxyphenyl)acrylonitrile (11)
Work-up procedure (C). Yellow powder; C16H11FN4O, MW: 294.28; 60% yield (0.25 g); m.p. 117.4–119.3 °C; Rf 0.35. 1H NMR (DMSO-d6): δ 8.28 (1H, s, =CH), 8.04 (2H, d, 1JH–H= 9.2 Hz, H–3″–5″), 7.64 (2H, m, H–5′,6′), 7.27 (2H, d, 1JH–H= 8.8 Hz, H–2″,6″), 3.95 (OCH3). 13 C NMR (DMSO-d6): δ 162.67 (C), 148.16 (C, d, 1JC–F= 16.0 Hz, C–F), 145.58 (CH), 131.90 (2CH), 126.11 (C, d, 1JC–F= 6.0 Hz, C), 122.39 (C), 122.09 (C, d, 1JC–F= 13.0 Hz, C), 116.26 (C, d, 1JC–F= 4.0 Hz, CH), 115.10 (2CH), 114.88 (C≡N), 114.33 (C, d, 1JC–F= 17.0 Hz, CH), 102.21 (C), 55.67 (OCH3). ESI–MS m/z calcd for C16H11FN4O 295.0989, found 295.0987 [M + H]+.
(E)-2-(7-fluoro-1H-benzo[d][1,2,3]triazol-1-yl)-3-(p-tolyl)acrylonitrile (12)
Work-up procedure (A). White powder; C16H11FN4, MW: 278.10; 38% yield (0.18 g); m.p. 145.3–147.9 °C; Rf 0.50. 1H NMR (DMSO-d6): δ 8.26 (1H, s, =CH), 8.09 (1H, d, 1JH–H= 8.0 Hz, H–7′), 7.89 (2H, d, 1JH–H= 8.4 Hz, H–2″,6″), 7.60 (2H, m, H–5′,6′), 7.46 (2H, d, 1JH–H= 8.0 Hz, H–3″,5″), 2.43 (3H, s, CH3). 13 C NMR (DMSO-d6): δ 148.13 (C, d, 1JC–F= 19.0 Hz, C–F), 145.58 (=CH), 143.18 (C), 130.09 (2CH), 129.86 (C, d, 1JC–F= 12.0 Hz, CH), 129.64 (2CH), 27.28 (C), 126.24 (C, d, 1JC–F= 6.0 Hz, CH), 122.09 (C, d, 1JC–F= 13.0 Hz, C), 116.25 (C, d, 1JC–F= 9.0 Hz, CH), 114.43 (C, d, 1JC–F= 22.0 Hz, CH), 114.49 (C≡N), 104.25 (C), 21.22 (OCH3). ESI–MS m/z calcd for C16H11FN4 279.1040, found 279.1037 [M + H]+.
(E)-3-(4-bromophenyl)-2-(7-fluoro-1H-benzo[d][1,2,3]triazol-1-yl)acrylonitrile (13)
Work-up procedure (C). Beige powder; C15H8BrFN4; MW: 343.15; 33% yield: (0.19 g); m.p. 109.1–111.2 °C; Rf 0.45. 1H NMR (DMSO-d6): δ 8.30 (1H, s, =CH), 8.10 (1H, d, 1JH–H= 4.0 Hz, H–7′), 7.91 (2H, d, 1JH–H= 8.8 Hz, H–3″,5″), 7.87 (2H, d, 1JH–H= 7.6 Hz, H–2″,6″), 7.56 (2H, m, H–5′,6′). 13 C NMR (DMSO-d6): δ 148.15 (C, d, 1JC–F= 25.0 Hz, C–F), 145.53 (C), 143.77 (=CH), 132.57 (2CH), 131.34 (2CH), 129.36 (C), 126.31 (C, d, 1JC–F= 6.0 Hz, CH), 125.98 (C), 121.84 (C, d, 1JC–F= 13.0 Hz, C), 116.34 (C, d, 1JC–F= 5.0 Hz, CH), 114.60 (C, d, 1JC–F= 17.0 Hz, CH), 114.02 (C≡N), 106.08 (C). ESI–MS m/z calcd for C15H8BrFN4 340.9832, 342.9812, found 341.1957, 342.1990 [M – H]–.
Results and discussion
Molecular modelling and drug design
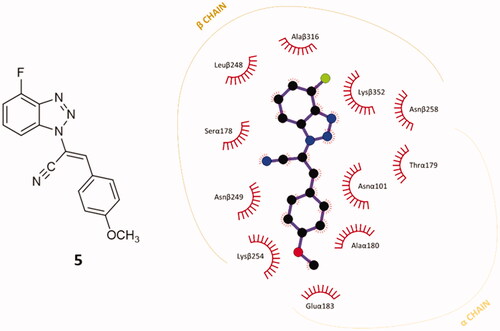
A preliminary docking study was built on compound 5, the 4′-fluoro parental compound of our former hit p-methoxy derivatives 34 and 9a in the previously validated CBS.Citation30 The docking simulation was conducted with AutoDock Vina.Citation31 The top-ranked conformational pose of compound 5 revealed an affinity energy of −9.3 kcal⋅mol−1. It shows also notable stability in the other nine poses generated from the simulation, with average affinity energy of circa −9.2 kcal⋅mol−1. The re-docked best-ranked pose of colchicine had affinity energy of −9.8 kcal⋅mol−1,Citation30 hence they have a similar predicted affinity for CBS on tubulin. Notably, it is also higher than the affinity energy of previous hits 34 and 9a, (–6.7 and −8.6 kcal⋅mol−1, respectively).Citation30 This result pushed us to investigate in depth the predicted interactions between 5 and the CBS. The best-predicted pose of compound 5 generates different non-covalent interactions with the exposed amino acids of the tubulin-binding site, generating a different pattern of polar and nonpolar contacts in comparison to compound 9a. The higher contribution to the protein-ligand affinity is the hydrophobic term. As shown in , each of the main moieties constituting the ligand backbone is involved in nonpolar contacts with amino acids on both α- and β-subunits of tubulin. The 4′-fluoro benzotriazole scaffold establishes connections mainly with β-chain amino acids: Leuβ248, Alaβ316, Lysβ352 and Asnβ258. The acrylonitrile linker in compound 5 interacts mainly with Serα178 (C≡N) and Asnα101 (=CH). The phenoxy moiety undertakes several hydrophobic interactions with aa located at the interface of the two subunits: Asnβ249, Lysβ254, Gluα183, Alaα180, Asnα101.
Figure 3. 2D-representation of the best-docked pose of compound 5 and its nonpolar interactions with the amino acids (here labelled with the three-letters denomination, the belonging chain and the number of the protein secondary structure) of the binding pocket at the interface of α,β-tubulin. Picture made with LigPlot+.Citation35 Hydrophobic interactions are depicted as hashes, for both protein and ligand atoms. Atoms are differently coloured: black (C), blue (N), green (F), and red (O).
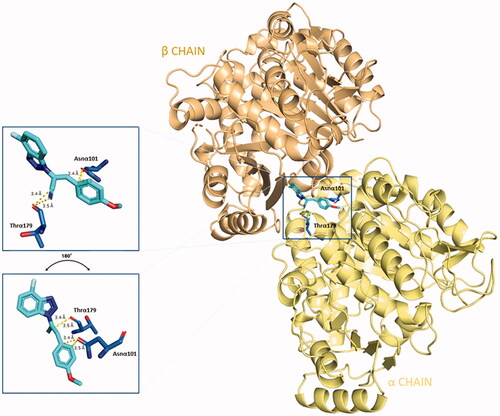
Apart from the consistent hydrophobic term, different polar contacts were recognised to contribute to the affinity, with α-subunit amino acids. A polar interaction is established between the cyan nitrogen of compound 5 and the carbonyl oxygen of Thrα179 (3.5 Å), while a polarised C⋅⋅⋅O interaction can be identified between the cyan carbon and the carbonyl oxygen of Thrα179. The lateral amide oxygen of Asnα101interact both with the C–1″ and C–2″ of the lateral phenoxy ring (∼ 3.5 Å both) through polar aryl π-heteroatom interactions, comparable to an amide-π stacking where the C = O points towards the phenyl ring (bond energy ∼ −2 kcal⋅mol−1).Citation42,Citation43 Polar interactions are depicted in . This docking prediction gives hints for an interesting mode of binding to the CBS on tubulin.
Figure 4. Three-dimensional representation of the binding conformation of compound 5 in CBS at the interface between α- and β-tubulin. Α-subunit is coloured in pale-yellow tint, β-one in light-orange tint. The ligand is in blue marine, interacting amino acids in blue. Yellow dashed lines consider the polar interactions, with a distance value in Å. Native 3D figures were produced via PyMOL.Citation33
Given the good predicted profile of binding of compound 5, our work proceeded with the design of different analogues bearing the benzotriazole scaffold, the linked acrylonitrile chain (to each of the three triazole nitrogen atoms), and various aromatic moieties selected based on the activity profile of our previous MTAs.Citation30 The final compounds are reported in . Introductory pharmacokinetic and drug-likeness properties were predicted for the newly designed series of compounds (5–13) that were subjected to a SwissADME prediction (http://www.swissadme.ch) also reported in and the full prediction output is listed in the SM.
Table 1. Chemical structure of the final compounds (5–13) and pharmacokinetic and drug-likeness properties obtained by SwissADME prediction. 
The designed derivatives have a molecular weight range of 278–343 g/mol, with a certain degree of rigidity (2–3 rotatable bonds). The designed molecules possess acceptor atoms for H-bonds, but not donors, as confirmed by the preliminary docking study. They showed a good profile of lipophilicity, with average LogP values of 2.85–3.54. All compounds are predicted as soluble or moderately soluble in water. As for the pharmacokinetic properties, in general, they are predicted to be highly absorbable from the gastrointestinal barrier and the blood–brain barrier. They seem also to be drug-like molecules, respecting Lipinski’s rule of five (0 violations). Figures S2–S4 in SM show the full SwissADME report for each compound. This hit optimisation process improved the predicted ADME properties. Comparing the ADME prediction of previous compound 9aCitation30 and new derivative 5 here reported, the latter were proved more drug-like than the parental compound. A lower predicted Log p values for ligand 5, 2.86 while 9a Log P was calculated at 3.53.Citation30 Compound 5 is predicted to be a lead-like compound, with a Log Po/w < 3, unlike the previous hit 9a.
Given the satisfying virtual results obtained by docking and ADME predictions, we proceeded in synthesising this promising series of compounds.
Chemistry
Scheme 1 depicts the synthetic route for the designed compounds 5–13. The benzotriazole derivative 1 was obtained as previously reported.Citation41 Compound 1 was used as a starting material to obtain three geometric isomers (2–4) bearing an acetonitrile chain on each of the three nitrogen atoms on the triazole ring. The reaction was conducted under basic conditions, by potassium hydroxide (KOH). Acetonitrile intermediates 2–4 were fully characterised, N-1 isomer (compound 2) was identified through NOESY NMR spectra, while isomers N-2 and N-3 were identified by chemical shift of the CH2 carbon at 13 C NMR spectra, see Figures S8 and S10 in the Supplemental Material. The stereochemistry of the double bond was identified as Z through 1H- and 13 C NMR spectra, as already established and described.Citation30 The chemical shift of the 1H and 13 C for the CH moiety clearly distinguish if the Z or E isomer is obtained.
Scheme 1. General procedure for final compounds (5–13). aReaction conditions: chloroacetonitrile/KOH (1:1.1), CH3CN, reflux, o/n. bReaction conditions: (A) DMA (ratio 1:1.2), CH3CN; (B) DIMCARB (ratio 1:1.2), CH3CN; (C) Cs2CO3 (ratio 1:1.2), toluene; yields:31–60%.
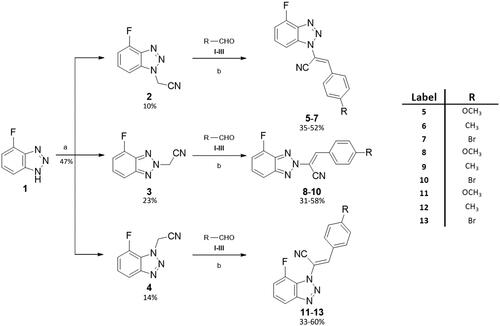
Following Knoevenagel condensation reactions with four different aromatic aldehydes (I–IV) gave final benzotriazolacrilonitrile compounds 5–7, 8–10, and 11–13 as single E isomer, identified by NMR, as previously described.Citation30 Derivatives 5–7 were obtained via Knoevenagel condensation of 2 and the proper aldehydes I–IV. Derivatives 8–10 were synthesised by reaction of 2 and I–IV, while intermediate 3 and the aldehydes I–IV led to compounds 11–13 through the same synthetic approach. Knoevenagel reactions were conducted in three different conditions: (1) compounds 9, 10 and 12 were yielded by using dimethylamine (DMA), as base and catalyst; (2) dimethylammonium dimethylcarbamate (DIMCARB) was employed as a catalyst to furnish derivatives 5 and 7, and (3) compounds 8, 11, and 13 were obtained utilising Cs2CO3 to alkalise the reaction environment.
Biology
Antiproliferative activity: NCI60 in vitro screening
As first antiproliferative activity evaluation, all synthesised compounds were subjected to the in vitro screening offered by the National Cancer Institute of Bethesda (USA, https://dtp.cancer.gov). The molecules were tested at the initial concentration of 10 µM on the complete panel of cancer cell lines which comprises 60 lines of solid and haematological tumours. All the derivatives with a remarkable antiproliferative activity at 10 µM, were further tested on the same panel, at five different dilutions ranging from 100 µM to 10 nM. Complete NCI60 results for compounds 5–13 are reported in Figures S29–S44 of the Supplemental Material section.
Structure-Activity relationships (SARs) report
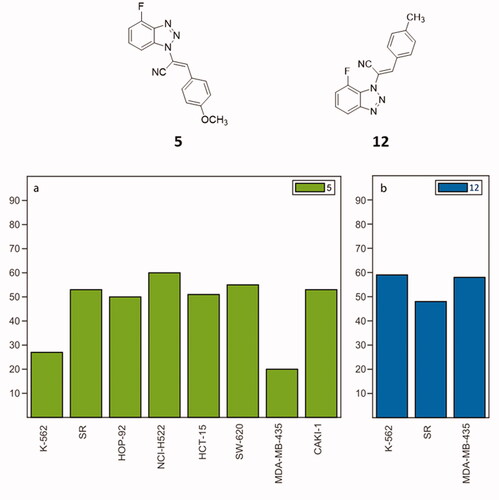
Among the tested compounds, all the N2-derivatives 8–10 showed a low activity on the growth inhibition of most of the NCI cancer cell lines. N3-substituted molecules 11–13 and N1-derivatives 5–7, showed appealing values of growth inhibition (GI). Considering the N3-derivatives, at 10 µM, compound 11 showed strong antiproliferative activity against all cancer cell lines, displaying GI ranging from 70 to 100%, and showing cytotoxicity against 21 of the 60 cancer cell lines. GI percentages were also calculated for nine cancer cell lines at 1 µM, showing cytotoxicity against three cell lines. Compound 13 elicited the most cytotoxic effect of this series at 10 µM with an intense cell growth inhibition on almost all the tested cell lines along with a strong cytotoxic activity on 57 out of 60 cell lines. Decreasing the concentration to 1 µM, compound 13 still inhibited twelve cancer cell lines (GI from 70 to 100%), also showing cytotoxicity against two of them. As for compound 12, it revealed an overall growth inhibition and cytotoxic activity comparable with parental compounds 11 and 13. At 1 µM concentration, it exerted the best antiproliferative effect against all leukaemia cell lines, while at 100 nM was proved strongly active against three cancer cell lines: K–562, SR (leukaemia) and MDA–MB–435 (melanoma) as shown in . As concerns N1-derivatives, compounds 5–7 displayed a general improved antiproliferative profile compared to the N2- and N3-derivatives. Compound 7 showed percentages of cancer cell growth inhibition around 80–100% at 10 µM, with a lethal effect on 48 of the 60 cancer cell lines. At 1 µM, derivative 7 displayed a cytostatic effect on 10 different cancer cell lines, while at 100 nM, the cytostatic effect resulted in a GI of 50% on the HOP–92 NSCLC cell line. When it comes to compound 6, at 10 µM, it displayed a GI of 80–100% involving 53 cancer cell lines of the 9 types of tumours, but also a cytotoxic effect on 24 cell lines. As for its activity at 1 µM, a < 50% reduction of GI can be identified against 52 cancer lines, particularly for leukaemia and melanoma and the cytotoxicity effect was proven against 8 cell lines. Compound 6 is also active at 100 nM on different tumour cell lines such as K-562 (leukaemia, growth percentage 40%), and MDA-MB-435 (melanoma, 20%). The p-OCH3 substituted 1-derivative 5 showed an overall proliferation inhibition ranging from 75 to 100%, with a limited cytotoxic effect. The antiproliferative effect is kept at a concentration of 1 µM: derivative 5 has a general detectable cytostatic effect on the entire NCI60 panel, also exhibiting cytotoxic effect against six lines. Remarkably, an 80% GI was registered against the K–562 leukaemia cell line when derivative 5 was administered at 100 nM concentration. The reduced GI on eight cancer cell lines induced by 5 at 100 nM, is charted in . Given these results, compounds 5 and 12 were selected as the most effective molecules and subjected to further biological assessment.
Antiproliferative activity of compounds against HeLa cells
Further analyses were carried out using the HeLa cell line, as previously validated.Citation30 To calculate the IC50 values for the newly synthesised compounds 5–13, an XTT assay was performed in triplicate on HeLa cells after 48 h of treatment, . Cells were treated with all compounds at different concentrations ranging from 2 to 5 μM, and DMSO was used as a control. Compounds 5 and 12 showed more pronounced antiproliferative activity and were used at concentrations of 0.125 to 2 μM, while compound 34 was used as an in-house positive control (). A colorimetric assay was performed after 48 h of treatment, absorbance data were obtained and processed to calculate the percentage viability of the cells. A 50% reduction in live cells was detected and IC50 values were calculated. Compounds 5 and 12 turned out as the most promising with IC50 values of 0.6 μM and 0.45 μM, respectively.
Figure 6. Inhibition of HeLa cell proliferation in vitro, as determined by the XTT assay. (A) HeLa cells were incubated with DMSO (vehicle control) and with compounds 34 (positive control) 5 and 12 at 2 and 5 μM concentrations. (B) HeLa cells were treated with compounds 34, 5 e 12 at different concentrations in a range between 0.125–2 μM. All experiments were performed in triplicate.
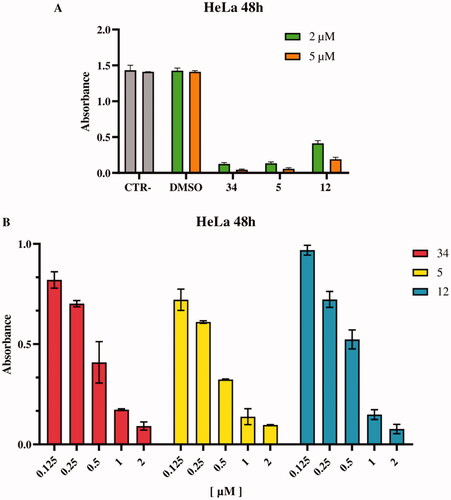
Table 2. Cytotoxic activities of the new compounds 5–13 as given by IC50 values in HeLa cells.
Mechanism of action of compounds 5 and 12 against HeLa and MCF-7 cells
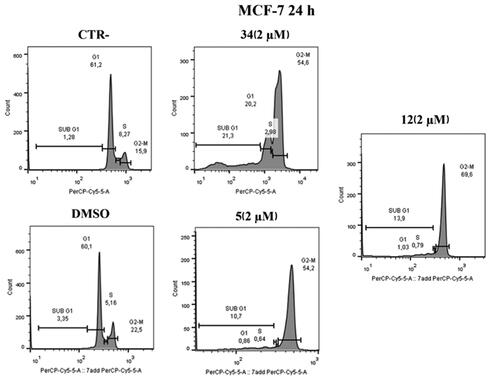
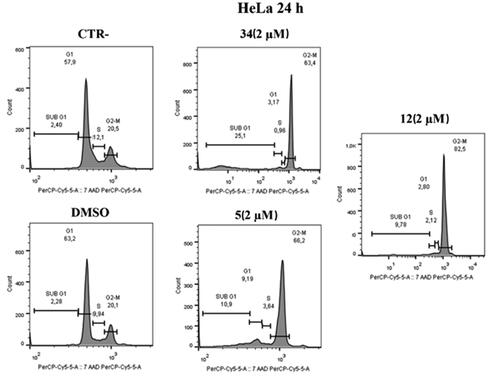
To prove that compounds 5 and 12 can hijack the cell cycle, flow cytometry was performed to assess changes in DNA content after treatment on HeLa and MCF-7 cells. As shown in , administration of compounds 5 and 12 at 2 μM over 24 h caused an increase in the number of cells in the G2/M phase in HeLa cells; our previous hit compound 34 was used as an in-house positive control. Specifically, an increase (82.5%) was observed in compound 12 treated cells compared to control (20.5%) and to DMSO (20.1%) at 2 µM concentrations. Furthermore, the behaviour is very similar to the positive control under the same conditions (63.4%). Similarly, in compound 5 treated cells, an increase (66.2%) in the G2/M phase was observed at a 2 µM concentration.
Figure 7. Distribution of HeLa cell in cell cycle assay. At 2 μM, both compounds 5 and 12 cause cell blockade in the G2/M phase compared to control and DMSO after 24 h. Compound 34 was used as a positive control.
Similar results were obtained on MCF-7 cell lines after 24 h at 2 µM. As shown in , in MCF-7 cell lines treated with compound 5 at the 2 µM concentration, an increase in G2/M phase (54.2%) was observed, similarly to the positive control. The same effect is evident in cells treated with compound 12 where an increase of 69.6% was observed.
Annexin V assay uncovers apoptosis activation in HeLa and MCF-7 cell lines after treatment with compounds 5 and 12
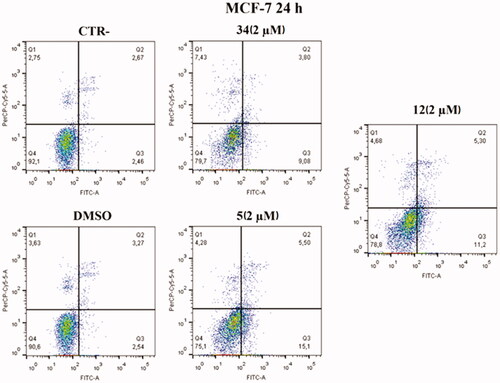
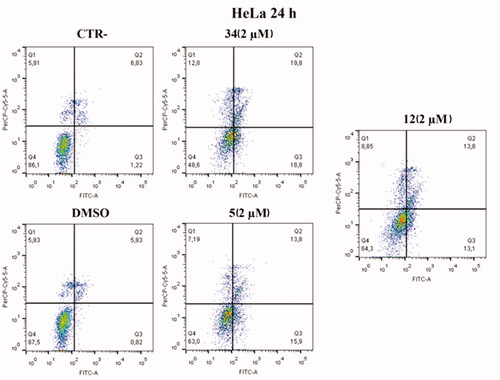
Based on cell cycle assay and XTT results, Annexin V–FITC/7-AAD double staining kit was used in flow cytofluorimetric analyses. HeLa cells were treated with compounds 5 and 12 at 2 µM concentration. After 24 h, cells were stained with both FITC and 7-AAD and analysed with BD FACS CANTO II. Compounds 5 and 12 activated apoptosis (). After 24 h of treatment with compound 5, the percentage of late apoptotic cells increased from 6.83% in control cells and 5.83% in cells with DMSO to 13.8% in treated cells. Besides, an increase in early apoptosis was noticeable, with compound 5 reaching rates of 15.9% compared to control and DMSO, 1.22% and 0.82%, respectively. Compound 12 caused late apoptosis in 13.8% of cells and early apoptosis in 13.1%. Both values approximate the percentage of cells in late apoptosis 19.8% and early apoptosis 18.8% caused by compound 34, which was used as an in-house positive control.
Figure 9. Annexin V analysis of HeLa cell lines after treatment with 2 µM of compounds 5 and 12 after 24 h. DMSO and compound 34 were used as controls. Apoptotic, necrotic, and live cells were stained with Annexin V FITC/7-AAD and then analysed by BD FACS DIVA software.
An increase in early apoptosis is observed in MCF-7 cell lines (). The percentage of cells in early apoptosis increased by 15.1% with compound 5 and 11.2% with compound 12 compared to control and DMSO, 2.46% and 2.54% respectively. Both values are higher than the percentage of early apoptosis caused by compound 34 (9.08%) used as an in-house positive control.
Compounds 5 and 12 cause PARP-1 cleavage in HeLa and MCF-7 cell lines
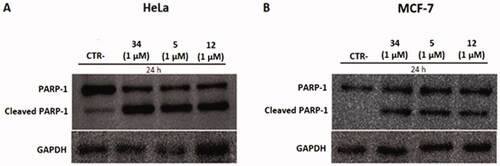
To investigate the mechanism of action of compounds 5 and 12 on HeLa and MCF-7 cells, the latter cells were treated with concentrations of 1 µM and the protein expression levels of PARP-1 and its cleaved form were measured. PARP-1 is a protein involved in the apoptotic cycle of a cell because it is the substrate of several “suicidal” proteases, such as caspases. The proteolysis of PARP-1 generates multiple cleavages in fragments, which are biomarkers to trace the proteases’ implication in cell death.Citation44 Twenty-four hours after treatment, pellets were collected, and western blotting was performed. As shown in , treatment with compounds 5 and 12 caused PARP-1 cleavage and generated an 89-kDa fragment on both cell lines, indicating the activation of the apoptotic pathway. The 89-kDa fragment of PARP-1 resulting from caspase cleavage is considered an early apoptosis hallmark, therefore these results confirm the pro-apoptotic activity of compounds 5 and 12 observed in the Annexin V assay.
Figure 11. Protein levels of PARP-1 and cleavage of PARP-1 in HeLa and MCF-7 cell lines after treatment with compounds 5 and 12 at 1 µM. Caspase-mediated cleavage of PARP1indicated activation of cell death by apoptosis in both cell lines. DMSO and compound 34 were used as negative and positive controls, respectively. All results were normalised using GAPDH.
When targeting tubulin and cell replication cycle, an antiproliferative but not cytotoxic activity may be desirable to target the fast-replicating tumour cells rather than normal cells. Given the comprehensively satisfying activity and the poorer cytotoxic effect, compound 5 was selected as the most promising compound and it was employed for further biological assessments to confirm the mechanism of action and to highlight its implication in different tumour pathways.
Compound 5 affects microtubule formation in HeLa cells
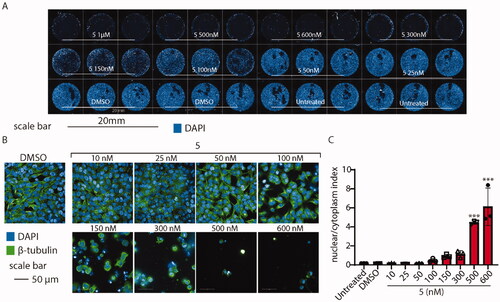
To probe whether microtubules might be affected by the administration of compound 5, we observed their structure on HeLa cells exposed to various concentrations of the drug for 24 h. Then, we analysed cells using high-content confocal microscopy. As shown in , compound 5 affects the shape and vitality of cells when administered above 100 nM. To better elucidate the cytoskeleton displacement, we tested whether compound 5 might change the ratio between the nuclear/cytoplasm dimension, segmenting the shape of every cell detected by the screening. As expected, the analysis revealed that, when compound 5 is administered above 100 nM, the cytoskeleton collapses leading to cell shrinkage, increasing the nuclear/cytoplasm ratio by ∼ 6 times ().
Figure 12. High-content confocal screening performed on HeLa cells. (A) Overview of an acquisition performed at 5×, nuclei are stained in blue (DAPI), scale bar 20 mm. (B) Exemplificative images of HeLa cells stained for β-tubulin (green) and nuclei (DAPI) treated or not with compound 5 at different concentrations. (C) Statistical analysis of nuclear/cytoplasm area of HeLa cells shown in B, performed using one-way ANOVA. Data are expressed as mean ± SD N = 3, α = 0.5 ***p < .0001.
Co-administration of compound 5-doxorubicin: compound 5 increases cytotoxicity of doxorubicin
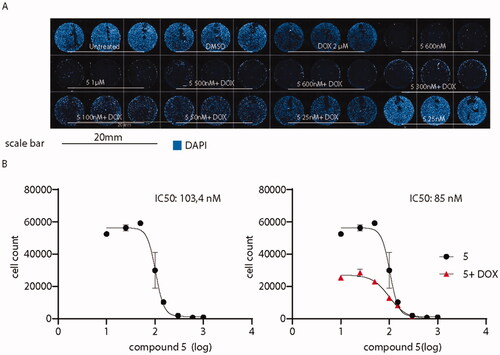
Then, it was determined to investigate whether the administration of compound 5 might enhance the antitumor activity of canonical antineoplastic drugs. Therefore, co-administration with doxorubicin was probed, as a well-established drug used to treat a wide variety of cancers including leukaemia, breast, lung, and ovary cancer. The co-administration of derivative 5 plus the standard drug were tested on A375, a cell line derived from an aggressive melanoma with an invasive phenotype resistant to clinical doxorubicin treatments.Citation37 A375 cells were exposed to doxorubicin alone at 2 µM for 24 h, a concentration that resembles the plasmatic concentration of the drug during high-dose chemotherapies.Citation45 As shown in , while doxorubicin alone did not affect A375 cells when administered alone, the co-administration with compound 5 dramatically affects A375 cell viability, decreasing compound 5 IC50 from 103.4 to 85 nM.
Figure 13. Administration of compound 5 decreases A375 melanoma cells viability. (A) Overview of high-content screening performed on A375 cells treated or not with compound 5 and doxorubicin. (B) IC50 of the sole compound 5 (black) or in association with doxorubicin (red). IC50 values were calculated by counting the number of nuclei in every experimental condition. Compound 5 IC50 when co-administered with doxorubicin is decreased from 103.4 to 85 nM.
Binding site confirmation
Compound 5 interferes with tubulin assembly and competes with colchicine binding
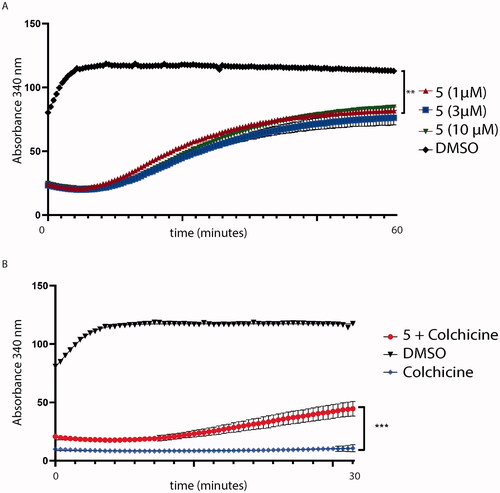
To confirm the mechanism of action, compound 5 was assessed to prove whether it might interfere with tubulin assembly decreasing tubulin polymerisation. As shown in , compound 5 affects tubulin polymerisation, if compared to tubulin treated with the sole DMSO. Then, to understand if compound 5 might bind tubulin in the CBS, we assessed a colchicine competition assay, as reported in . As previously predicted by docking simulation, compound 5, administered at equimolar concentration with colchicine, decreases colchicine effectiveness on tubulin polymerisation (p < .001 red curve, ). Colchicine and DMSO were used as control.
Figure 14. (A) Tubulin polymerisation assay. Tubulin was pre-incubated with compound 5 at different concentrations. Then, tubulin assembly was assessed by measuring absorbance every 2 s for 60 min at 340 nm. Compound 5 (red lines) decreases tubulin polymerisation at every concentration tested (1, 3 and 10 μM); (B) Colchicine-competition assay. Tubulin was pre-incubated with an equimolar concentration of colchicine with compound 5 at 1 μM. Then, tubulin polymerisation was assessed by measuring absorbance every 2 s for a total of 30 min. Statistical analyses were performed using a one-sample T test and expressed as mean ± SD, ***p ≤ .001.
Compound 5 is predicted to compete with colchicine at the interface of α- and β-tubulin
The binding conformation and interactions of compound 5 were already displayed before. Here, the crystal structure of colchicine in its binding pocket on tubulin (PDB ID: 4O2B), was superimposed with the best-predicted pose of compound 5 (), as described in Section 3.1, to highlight the different binding in the same spatial environment and to support the experimental results gained from the colchicine-compound 5 competition assay, previously reported.
Figure 15. Superimposition of the predicted conformation of compound 5 (coloured in blue marine) and the crystal structure of colchicine (in violet-purple, PDB ID: 4O2B).
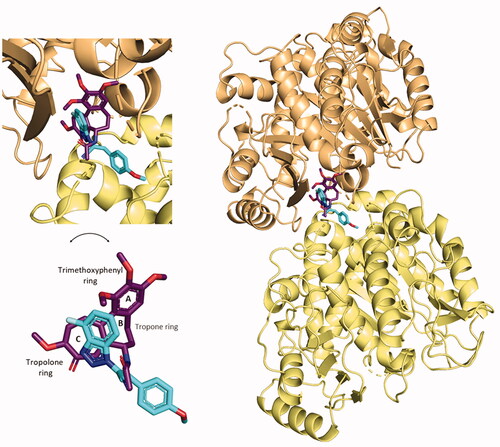
By the superimposition of the two structures, they clearly occupy the same specific portion at the interface of α- and β-tubulin. The 4′-fluoro benzotriazole moiety is predicted to sit in the specific space occupied by the crystal conformation of the tropolone ring of colchicine. The cyan group of the acrylonitrile linker directs in the same way as the amide group linked to the tropone ring. The p-OCH3 phenyl structure does not overlap with any moiety of colchicine, but points towards the interface-exposed part of the α-chain, interacting with its amino acids, as already shown.
This prediction shows the two structures bind with a good affinity profile, confirming the experimental competition between the two tubulin inhibitors and the binding to CBS on tubulin.
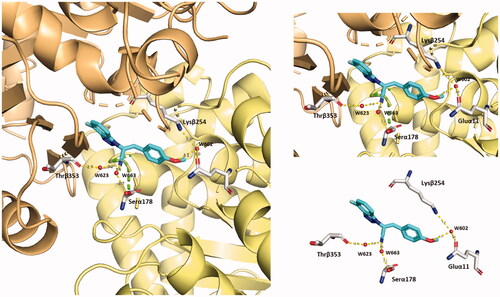
Compound 5 is predicted to stabilise its binding in CBS by solvent-mediated interactions
Compound 5 is predicted to establish several polar interactions with the exposed amino acids of CBS on tubulin (from both α- and β-tubulin). Some of them are mediated by water molecules. Notably, the major contribution in terms of water bridges is provided by the C≡N group and the methoxy oxygen in the ligand, see . The cyan group contacts Thrβ353 and Serα178 via two water bridges. The methoxy oxygen reaches both Lysβ254 and Gluα11 with the W602 water molecule. These additional polar interactions contribute to building the predicted unique binding mode of compound 5 to α- and β-tubulin interface.
Conclusions
As a background for this work, CBSI compounds 34 and 9a emerged for their remarkable antiproliferative activity and their effect on the cancer cell. In this paper, a single fluorine atom was introduced in C–4′ of benzotriazole scaffold, a position not investigated before, and a series of fluorinate compounds bearing the acrylonitrile chain on N–1,2,3 on the benzotriazole ring (5–13) was designed, in silico validated and synthesised. The synthetised compounds were assessed through a preliminary in vitro screening on a panel of 60 cell lines of 9 tumours. Compounds 5 and 12 were selected for further biological investigations, as they inhibited cancer cell growth on several lines at 100 nM. Further proliferative assay performed on Hela cells, showed IC50 values of 0.6 and 0.45 µM for compounds 5 and 12, respectively. Flow cytometry assay on both HeLa and MCF-7 cells confirmed these molecules interfere with the cell cycle showing a consistent blockade of cells in the G2/M phase. Pro-apoptotic activity of 5 and 12 was proved through the Annexin V assay resulting in an increase of early and late apoptotic cells, both in HeLa and MCF-7 cell lines. The pro-apoptotic activity of compounds 5 and 12 was confirmed by WB-analysis of PARP-1 and cleaved PARP-1,. Globally, the collected results suggested compound 5 as the most promising compound. It was further investigate for cytoskeleton collapse leading to severe cell shrinkage observed through a high-content screening and also for the competition with colchicine at the CBS on tubulin, showing a decreases the effectiveness of colchicine on the inhibition of tubulin polymerisation proving the interaction with the same binding site. To validate the latter result, the best-docked pose of compound 5 was superimposed with the crystal structure of colchicine. They showed to bind in the same specific portion of the CBS, but they did not show overlapping chemical portions between themselves, suggesting that compound 5 may undertake a different and novel pattern of interactions. Finally, co-administration of 5 and doxorubicin was assessed on the A375 cell line, a doxorubicin-resistant melanoma cell line. As expected, doxorubicin is not effective alone, while the co-administration exerts a synergic activity lowering the 5 IC50 value from 103 to 85 nM. In conclusion, the 4′-fluoro substitution of benzotriazole-acrylonitrile scaffold brought us a step forward in the optimisation process on previous hits 34 and 9a; among the new derivatives synthesised, compound 5 was proved to be a good antiproliferative agent acting as promising MDA at nanomolar concentration.
Author contributions
Conceptualisation: FR, RI, SP, and AC; formal analysis: FR, SZ, RI, LS, ML, and VB; investigation: FR, RI, SZ, LS, and ML; resources: AC; data curation: FR, RI, SS, SZ, LS, ML, LB, and AC; writing/original draft preparation: FR, RI, and SS; writing/review and editing: all authors; supervision: LB and AC; funding acquisition: LB and AC. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download PDF (4.1 MB)Acknowledgments
We would like to acknowledge “Assessorato della Programmazione, Bilancio, Credito e Assetto del territorio” of “Regione Autonoma della Sardegna” (Sardinia, Italy) for the funding support with the grant named “Legge Regionale 7 agosto 2007:CRP1_574, 22/41 del 2017,” grant number RASSR01499. The authors would thank the Italian Ministry for University and Research for the funding named PRIN2020. For the in vitro antiproliferative screening, we thank NCI (National Cancer Institute, Developmental Therapeutics Program, Bethesda, USA; https://dtp.cancer.gov).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bérubé G. How to utilize academic research efforts in cancer drug discovery. Expert Opin Drug Discov 2019;14:331–4.
- Giannakakou P, Sackett D, Fojo T. Tubulin/microtubules: still a promising target for new chemotherapeutic agents. J Natl Cancer Inst 2000;92:182–3.
- Florian S, Mitchison TJ. Anti-microtubule drugs. Methods Mol Biol 2016;1413:403–21.
- Chandrasekaran G, Tátrai P, Gergely F. Hitting the brakes: targeting microtubule motors in cancer. Br J Cancer 2015;113:693–8.
- Mukhtar E, Adhami VM, Mukhtar H. Targeting microtubules by natural agents for cancer therapy. Mol Cancer Therapeut 2014;13:275–84.
- Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol 2015;16:711–26.
- Prota AE, Danel F, Bachmann F, et al. The novel microtubule-destabilizing drug BAL27862 binds to the colchicine site of tubulin with distinct effects on microtubule organization. J Mol Biol 2014;426:1848–60.
- Huzil JT, Chen K, Kurgan L, Tuszynski JA. The roles of β-tubulin mutations and isotype expression in acquired drug resistance. Cancer Inf 2007;3:159–81.
- Mollinedo F, Gajate C. Microtubules, microtubule-interfering agents and apoptosis. Apoptosis 2003;8:413–50.
- Field JJ, Díaz JF, Miller JH. The binding sites of microtubule-stabilizing agents. Chem Biol 2013;20:301–15.
- Field JJ, Kanakkanthara A, Miller JH. Microtubule-targeting agents are clinically successful due to both mitotic and interphase impairment of microtubule function. Bioorg Med Chem 2014;22:5050–9.
- Markel G, Imazio M, Brucato A, Adler Y. Prevention of recurrent pericarditis with colchicine in 2012. Clin Cardiol 2013;36:125–8.
- Ter Haar NM, Frenkel J. Treatment of hereditary autoinflammatory diseases. Curr Opin Rheumatol 2014;26:252–8.
- Cocco G, Chu DCC, Pandolfi S. Colchicine in clinical medicine. A guide for internists. Eur J Intern Med 2010;21:503–8.
- Andreu JM, Timasheff SN. Conformational states of tubulin liganded to colchicine, tropolone methyl ether, and podophyllotoxin. Biochemistry 1982;21:6465–76.
- Brossi A, Yeh HJC, Chrzanowska M, et al. Colchicine and its analogues: recent findings. Med Res Rev 1988;8:77–94.
- Volbracht C, Leist M, Kolb SA, Nicotera P. Apoptosis in caspase-inhibited neurons. Mol Med 2001;7:36–48.
- Lu Y, Chen J, Xiao M, et al. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm Res 2012;29:2943–71.
- Ravelli RBG, Gigant B, Curmi PA, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 2004;428:198–202.
- Dorléans A, Gigant B, Ravelli RBG, et al. Variations in the colchicine-binding domain provide insight into the structural switch of tubulin. Proc Natl Acad Sci U S A 2009;106:13775–9.
- Tron GC, Pirali T, Sorba G, et al. Medicinal chemistry of combretastatin A4: present and future directions. ChemInform 2006;49:3033–44.
- Seddigi ZS, Malik MS, Saraswati AP, et al. Recent advances in combretastatin based derivatives and prodrugs as antimitotic agents. Medchemcomm 2017;8:1592–603.
- Carta A, Sanna P, Palomba M, et al. Synthesis and antiproliferative activity of 3-aryl-2-(1H-benzotriazol-1-yl)acrylonitriles. Part III. Eur J Med Chem 2002;37:891–900.
- Sanna P, Carta A, Nikookar MER. Synthesis and antitubercular activity of 3-aryl substituted-2-(1H(2H) benzotriazol-1(2)-yl)acrylonitriles. Eur J Med Chem 2000;35:535–43.
- Sanna P, Carta A, Gherardini L, et al. Synthesis and antimycobacterial activity of 3-aryl-, 3-cyclohexyl- and 3-heteroaryl. Il Farmaco 2002;57:79–87.
- Carta A, Sanna P, Bacchi A. Synthesis of E/Z 3-(1H-benzotriazol-1-yl-3-(pyridin-4- yl)acrylonitriles and E/Z 2-(3-imino-2-benzofuran-1(3H)- ylidene)acetonitriles. An unusual case of displacement of the benzotriazole ring. Heterocycles 2002;57:1079–90.
- Carta A, Palomba M, Boatto G, et al. Synthesis and antiproliferative activity of 3-aryl-2-[1H(2H)-benzotriazol- 1(2)-yl]acrylonitriles variously substituted: part 4. Farmaco 2004;59:637–44.
- Carta A, Briguglio I, Piras S, et al. 3-Aryl-2-[1H-benzotriazol-1-yl]acrylonitriles: a novel class of potent tubulin inhibitors. Eur J Med Chem 2011;46:4151–67.
- Briguglio I, Laurini E, Pirisi MA, et al. Triazolopyridinyl-acrylonitrile derivatives as antimicrotubule agents: synthesis, in vitro and in silico characterization of antiproliferative activity, inhibition of tubulin polymerization and binding thermodynamics. Eur J Med Chem 2017;141:460–72.
- Riu F, Sanna L, Ibba R, et al. A comprehensive assessment of a new series of 5′,6′-difluorobenzotriazole-acrylonitrile derivatives as microtubule targeting agents (MTAs). Eur J Med Chem 2021;222:113590.
- Trott O, Olson AJ. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–61.
- Berman HM, Battistuz T, Bhat TN, et al. The protein data bank. Acta Crystallogr D Biol Crystallogr 2002;58:899–907.
- The PyMOL Molecular Graphics System, Version 2.0, Schrödinger, LLC., (n.d.).
- Morris GM, Huey R, Lindstrom W, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 2009;30:2785–91.
- Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 2011;51:2778–86.
- Lai M, Iacono E, Spezia PG, et al. A low-cost simple test for weekly detection of Mycoplasma hyorhinis and arginini contaminations in cell cultures and viral preparations. J Virol Methods 2022;299:114327.
- Lai M, Amato R, Rocca VL, et al. Acid ceramidase controls apoptosis and increases autophagy in human melanoma cells treated with doxorubicin. Sci Rep 2021;11:1–14.
- Lai M, Rocca VL, Amato R, et al. Ablation of acid ceramidase impairs autophagy and mitochondria activity in melanoma cells. Int J Mol Sci 2021;22:3247.
- Scalise V, Sanguinetti C, Neri T, et al. PCSK9 induces tissue factor expression by activation of TLR4/NFkB signaling. Int J Mol Sci 2021;22:12640.
- Borgogna C, Bruna R, Griffante G, et al. Patterns of neutralizing humoral response to SARS-CoV-2 infection among hematologic malignancy patients reveal a robust immune response in anti-cancer therapy-naive patients. Blood Cancer J 2022;12:8–9.
- Carta A, Bua A, Corona P, et al. Design, synthesis and antitubercular activity of 4-alkoxy-triazoloquinolones able to inhibit the M. tuberculosis DNA gyrase. Eur J Med Chem 2019;161:399–415.
- Perutz MF, Fermi G, Abraham DJ, et al. Hemoglobin as a drug receptor. J Am Chem Soc 1986;108:1064–78.
- Meyer EA, Castellano RK, Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed Engl 2003;42:1210–50.
- Chaitanya GV, Alexander JS, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun Signal 2010;8:31.
- Barpe DR, Rosa DD, Froehlich PE. Pharmacokinetic evaluation of doxorubicin plasma levels in normal and overweight patients with breast cancer and simulation of dose adjustment by different indexes of body mass. Eur J Pharm Sci 2010;41:458–63.