Abstract
The increasing resistance of fungi to conventional antifungal drugs has prompted worldwide the search for new compounds. In this work, we investigated the antifungal properties of acylated Temporin L derivatives, Pent-1B and Dec-1B, against Candida albicans, including the multidrug-resistant strains. Acylated peptides resulted to be active both on reference and clinical strains with MIC values ranging from 6.5 to 26 µM, and they did not show cytotoxicity on human keratinocytes. In addition, we also observed a synergistic or additive effect with voriconazole for peptides Dec-1B and Pent-1B through the checkerboard assay on voriconazole-resistant Candida strains. Moreover, fluorescence-based assays, NMR spectroscopy, and confocal microscopy elucidated a potential membrane-active mechanism, consisting of an initial electrostatic interaction of acylated peptides with fungal membrane, followed by aggregation and insertion into the lipid bilayer and causing membrane perturbation probably through a carpeting effect.
Introduction
Fungal diseases affect billions of individuals and represent a worldwide threat to human health. The significant increase of fungal infections is likely due to the increased use of immunosuppressive therapies in cancer or transplantation, as well as medical devices and the indiscriminate consumption of antibiotics. Antifungal treatments are essential for reducing comorbidities and mortalities, but resistance-related issues have prompted the search for more effective alternatives with reduced toxicity, improved pharmacodynamics and pharmacokinetics, and increased specificityCitation1. Candidiasis is the most notable infection caused by fungi of the Candida genus and represents a major concern for public healthCitation2. Species of Candida are the leading pathogens causing around 400,000 systemic fungal diseases worldwideCitation3 and are considered the fourth most common cause of nosocomial blood infections, even overcoming Gram-negative pathogensCitation4,Citation5. Although more than one hundred Candida species exist, only a few are implicated in clinical infections. Of these, C. albicans is certainly the most isolated species from clinical specimens and it is generally found in 90–100% of mucosal isolates and 50–70% of blood infection isolatesCitation6. Candida spp infections exhibit different clinical manifestations, from superficial mucocutaneous candidiasis affecting mucosal, nail and skin surfaces, to systemic infections affecting organsCitation7. These infections are commonly seen in immunocompromised patients or in those conditions in which yeast overgrowth is favoredCitation7. Candida species have developed mechanisms of resistance towards antifungal drugs commonly used for the treatment of systemic or topical infectionsCitation8,Citation9. Among them, azoles (e.g. fluconazole, voriconazole), polyenes (e.g. amphotericin B) and echinocandins (e.g. caspofungin) are the most efficacious as they specifically interfere with the ergosterol or cell wall biosynthesis, which are not present in mammalian cellsCitation8. For example, activities of the azoles are directed against lanosterol 14-α-demethylase (Erg11p), which is encoded by the ERG11 gene, involved in the ergosterol biosynthesis. Among resistant clinical isolates of C. albicans, ERG11 over-expression is frequently observedCitation10. The numerous toxic effects, including liver and kidney dysfunctions, local phlebitis, haemolysis and feverCitation11,Citation12 caused by these antifungal drugs prompted the focus of recent research on new agents with improved safety profile and broad-spectrum antimicrobial activity for use alone and/or in synergy with antifungals.
Emerging drug candidates attracting the attention of researchers are antimicrobial peptides (AMPs), host defence peptides that play an essential role in the innate immune responseCitation12. AMPs, mostly isolated from living organisms, are typically featured by a net positive charge at physiological pH due to the presence of basic residues (e.g. Arg and Lys) while the hydrophobic residues determine their amphipathic conformation. Both the cationic nature and the secondary structure facilitate the interaction with the negatively charged membrane of microorganismsCitation12,Citation13, leading to insertion, destabilisation and disruption of the cell membrane, although they can also act on intracellular targetsCitation14,Citation15. Since the cell membrane is the primary target of AMPs, microorganisms are less likely to develop a resistance mechanism, making AMPs promising drug candidatesCitation16.
Temporins are AMPs isolated from the skin of the frog Rana temporaria, and exhibit strong antimicrobial, anti-Candida and antiviral activitiesCitation17–19. Temporin L (TL) is the most active isoform on both Gram-negative and Gram-positive bacteria and yeasts, but it has high haemolytic activityCitation18,Citation20. Previous structure-activity relationship studies (SARs) carried out by our research group led to the discovery of synthetic peptides derived from Temporin L possessing a strong ability to kill both Gram-positive and Gram-negative bacteria, and yeasts as Candida sppCitation21. Interestingly, some of them have also demonstrated scarce or absent haemolytic activity at their antimicrobial concentrationsCitation21,Citation22. Our group has deeply worked on a promising molecule derived from Temporin L, [ProCitation3DLeuCitation9DLysCitation10] TL (namely 1B, sequence: Phe-Val-Pro-Trp-Phe-Ser-Lys-Phe-dLeu-dLys-Arg-Ile-Leu-CONH2), that has proved to be an efficient antimicrobial agent, also devoid of cytolytic effects in vitroCitation23. Interestingly, peptide 1B has demonstrated the ability to inhibit the growth of methicillin-resistant Staphylococcus pseudintermedius (MRSP), and Malassezia pachydermatis and the prolonged application of this peptide did not cause M. pachydermatis drug-resistance and increased the susceptibility of MRSP to oxacillinCitation24. In addition, 1B has shown anti-inflammatory activity in vivo in response to zymosan-induce peritonitisCitation25. This result is worthy of note since it supports a possible application of 1B as an anti-inflammatory and antimicrobial agent. These results have also been described in a patent which has granted on 7 May 2021 (Number: 102019000009489).
Approaches to reduce or eliminate the toxicity of conventional antifungal drugs and enhance their activity may include the use of drug delivery systems or novel strategies that likely improve the interaction with the target membranesCitation26–28.
To improve the antimicrobial profile of peptide 1B, we have previously applied a lipidation strategyCitation29 consisting of the addition of fatty acids in N- and C-terminus of the peptide, and discovered the analogue Pent-1B, bearing a pentyl chain in para position of PheCitation1 (peptide C in Bellavita et al.Citation29). This compound has shown a good activity against carbapenem resistant Klebsiella pneumoniae and a low cytotoxicity against both human erythrocytes and keratinocytesCitation29,Citation30.
Through detailed biophysical studies focussed on the mode of action of Pent-1B using bacterial biomembrane models, we demonstrated that Pent-1B has a low ability to aggregate in aqueous solution, while it has a significant ability to oligomerize in membrane mimetic environments. This causes an alteration of the membrane fluidity and, by creating pores in both Gram-positive and Gram-negative membranes, eventually leads to cell deathCitation29.
Based on the results until here obtained, herein, we were prompted to investigate the antifungal properties of Pent-1B against reference and clinical strains of C. albicans, and with the further aim to increase the affinity and interaction with fungal membrane, we designed and synthesised the peptide Dec-1B bearing a longer lipid tail of 10-carbon atoms. The antifungal properties of the peptide Dec-1B were investigated against C. albicans, while its cytotoxicity was assessed on human keratinocytes. The effect of Pent-1B and Dec-1B in combination with antifungal drug voriconazole was evaluated on voriconazole-resistant C. albicans strains. Moreover, the mechanism of action on fungal membrane models of peptides Pent-1B and Dec-1B was explored by fluorescence assays, including Thioflavin T and leakage assays, tryptophan quenching experiments, nuclear magnetic resonance (NMR) spectroscopy and confocal microscopy.
Materials and methods
Materials
The conventional Fmoc-amino acids and coupling reagents N,N,N’,N’-tetramethyl-O-(1H-benzotriazol-1-yl) uranium hexafluorophosphate (HBTU) and 1-hydroxybenzotriazole (HOBt), were commercially obtained by GL Biochem Ltd acquired from GL Biochem Ltd (Shanghai, China). The unconventional amino acid Boc-l-Phe(4-NO2)-OH, Rink amide resin (loading substitution of 0.70 mmol/g), N,N-diisopropylethylamine (DIEA), piperidine, triisopropylsilane (TIS) and trifluoroacetic acid (TFA) were purchased from Iris-Biotech GMBH. Tin(II) chloride anhydrous, valeric and decanoic acids were acquired from Sigma Aldrich-Merck. Moreover, peptide synthesis solvents, such as N,N-dimethylformamide (DMF), water and acetonitrile for HPLC, were reagent grade acquired from commercial sources (Sigma Aldrich-Merck and VWR) and used without further purification.
Phospholipids: 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (DOPC), L-α-phosphatidylethanolamine (PE), L-phosphatidylglycerol (PG), phosphatidylinositol (PI), 1-palmitoyl-2–(6,7-dibromo)stearoyl-sn-glycero-3-phosphocholine (6,7 Br-PC), 1-palmitoyl-2–(9,10-dibromo)stearoyl-sn-glycero-3-phosphocholine (9,10 Br-PC), and 1-palmitoyl-2–(11,12-dibromo)stearoyl-sn-glycero-3-phosphocholine (11,12 Br-PC) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). Ergosterol, 1,1,1,3,3,3-Hexafluoro-2-propanol, Nile Red (NR) and Thioflavin T were acquired from Sigma Aldrich-Merck. 8-aminonaphtalene-1,3,6-trisulfonic acid, disodium salt (ANTS) and p-xylene-bis-pyridinium bromide (DPX) were purchased from Molecular Probes. Deuterium oxide (D2O) of 99.9% isotopic purity and dimethyl sulfoxide (DMSO-d6) of 99.9% isotopic purity were purchased from Sigma Aldrich (Milan, Italy). Dodecylphosphorylcholine-d38 (DPC-d38) of 98% isotopic purity and 16-doxylstearic acid were purchased from Merck Life Science S.r.l. (Milan, Italy). Sodium-3-(trimethylsilyl) propionate 2,2,3,3-d4 (TSP) was from Cambridge Isotope Laboratories (CIL), Inc. (Andover, MA, USA). Antibiotics were purchased from Sigma-Aldrich (Milan, Italy).
Candida strains and culture conditions
The yeast strains evaluated in this study included C. albicans ATCC 90028 (reference strain) and C. albicans ATCC 10231 (azole-resistant strain) obtained from the American Type Culture Collection (Rockville, MD), and six azole-resistant clinical isolates (Ca1-Ca6) belonging to an anonymous collection of fungal strains established at the Department of Molecular Medicine and Medical Biotechnology, University of Naples Federico II (). Identification was performed by subcultures on chromogenic agar (Chromid agar) (Becton Dickinson) and confirmed by MS MALDI-TOF (Bruker). Susceptibility to amphotericin B, anidulafungin, micafungin, caspofungin, posaconazole, voriconazole, itraconazole, and fluconazole was assessed using the Sensititre Yeast One colorimetric microdilution method (Thermofisher). The results were interpreted according to the MIC breakpoints reported by Clinical and Laboratory Standards Institute (CLSI)Citation31 with the exception of the MICs of itraconazole, posaconazole and amphotericin B, which were interpreted according to European Committee on Antimicrobial Susceptibility Testing (Eucast)Citation32. All strains were stored as 15% (v/v) glycerol stocks at −80 °C. Prior to each experiment, cells were subcultured from the stocks onto Sabouraud dextrose agar (SDA) (Becton Dickinson) at 37 °C for 48 h.
Table 1. Antimycotic-susceptibility profiles of clinical C. albicans used in our study.
Acylated peptide synthesis
The acyl-peptides Pent-1B and Dec-1B were synthesised by using ultrasounds-assisted solid-phase peptide synthesis (US-SPPS)Citation33 and by employing the Rink amide resin as solid supportCitation29. In details, peptide assembly was obtained by performing repeated cycles of (i) Fmoc deprotections, by treatments with 20% of piperidine in DMF solution [0.5 + 1 min under ultrasonic irradiation), and (ii) coupling reactions, by treatment with a solution of Fmoc-AA(3 eq), HBTU (3 eq), HOBt (3 eq) and DIEA (6 eq) in DMF, and exposing the reagent mixture to the ultrasonic irradiation for 5 min. For the construction of lipidic tails, the amino acid Boc-l-Phe(4-NO2)-OH was used and coupled as the last residue to the peptide sequence. Therefore, prior conjugation to lipidic tails, the nitro group was reduced to primary amine by treatment with SnCl2 1 M solution in DMF, at rt for 16 h. Then, valeric or decanoic acid (3 eq), together with HBTU (3 eq), HOBt (3 eq) and DIEA (6 eq) in DMF/DCM (1:1, v/v), were added to the resin-bound peptides Pent-1B and Dec-1B, respectively, and reactor was agitated on an automated shaker for 2 h. Once the lipidic tail coupling was ascertained by LC-MS analysis, peptides were released from the resin and all protecting groups were simultaneously cleaved by treatment with a cleavage cocktail of TFA/TIS/H2O (95:2.5:2.5, v/v/v), for 3 h under stirring. Finally, lipidated peptides were purified by preparative RP-HPLC equipped with a Phenomenex Kinetex column (C18, 5 µm, 100 Å, 150 × 4.6 mm) and by using a linear gradient of MeCN (0.1% TFA) in water (0.1% TFA) from 10 to 90% over 30 min, and the correct molecular masses were confirmed through electrospray ionisation mass spectrometry (ESI-MS) analyses (Table S1).
Fluorescein-labelling of peptide dec-1B
The peptide Dec-1B-K(Fam) was labelled to 5(6)-carboxyfluorescein (Fam) on side chain of an additional Lys placed in C-terminus. Thus, Fmoc-l-Lys(Mtt)-OH was introduced during peptide elongation, and, after the construction of the target sequence, as above described, the fluorescein-labelling proceeded as follows. The Mtt group was selectively removed by treating the resin-bound peptide sequence with a cocktail of TFA/TIS/DCM (1:5:94, v/v/v), for 25 min at rt.Citation16 The procedure was repeated seven times, and the complete deprotection was ascertained by the Kaiser test. The 5(6)-carboxyfluorescein (2 eq) was coupled by using COMU (2 eq), Oxyma (2 eq) and DIPEA (4 eq), and reagent mixture was allowed to stir for 25 min. Then, the resin was washed with DMF (× 3) and the coupling was repeated for an additional 25 min. The fluorescein labelling was ascertained by the LC-MS analysis. Finally, the peptide was cleaved from the resin and purified by preparative RP-HPLC equipped with a Phenomenex Kinetex column (C18, 5 µm, 100 Å, 150 × 4.6 mm) and a linear gradient of MeCN (0.1% TFA) in water (0.1% TFA) from 10 to 90% over 30 min, and the correct molecular mass was confirmed by ESI-MS analysis (Table S1).
Determination of the minimum inhibitory concentration (MIC) and the minimum fungicidal concentration (MFC)
The antifungal activity of compounds was determined using a standardised broth microdilution method (Clinical and Laboratory Standards Institute (CLSI) document M27-A4) Citation34. Briefly, cell suspension was adjusted to 3 × 103 CFU/mL in RPMI 1640 medium (Sigma) supplemented with 0.2% (w/v) glucose. One hundred microliter aliquots of these cell suspensions were dispensed into 96-well microtiter plates. Compounds were serially diluted using RPMI 1640 medium and added to the wells at a final concentration ranging from 0.4 to 26 μM, and the plate was incubated for 24 h at 37 °C. Voriconazole (ranged from 8 to 60 μg/mL for ATCC 10231 and Ca3 strains; ranged from 0.03 μg/mL to 8 μg/mL for other strains) and Amphotericin B (ranged from 0.12 to 2 μg/mL) were chosen as positive controls.
The MIC was defined as the lowest concentration of the peptide that resulted in 100% growth inhibition after 24 h of incubation. The MFC was determined by transferring 50 μL aliquots of each sample, previously treated with concentrations equal to or higher than the MIC, onto SDA plates and incubating the plates at 37 °C for 24 h. The lowest peptide concentration that yielded no fungal growth on agar plates was defined as the MFC. All the tests were conducted at least three times using independent cell suspensions.
Checkerboard assay
The interaction between compounds and voriconazole against voriconazole-resistant C. albicans strains ATCC 10231 and Ca3 clinical strain was evaluated by the checkerboard method in 96-well microtiter plates. Briefly, compounds Pent-1B or Dec-1B and voriconazole were serially diluted along the y and x axes, respectively. The final concentration of voriconazole ranged from 0.03 to 0.5 µg/mL while the final concentration of Pent-1B or Dec-1B ranged from 0.8 to 13 μM for ATCC 10231 and from 0.8 to 26 μM for the clinical strain. The checkerboard plates were inoculated with yeast as previously reported and incubated at 37 °C for 24 h, following which yeast growth was assessed visually and the turbidity measured by microplate reader at 595 nm. The Fractional Inhibitory Concentration (FIC) index for each combination was calculated as follows: FIC index = FIC of peptide + FIC of voriconazole, where FIC of peptide (or voriconazole) was defined as the ratio of MIC of peptide (or voriconazole) in combination and MIC of peptide (or voriconazole) alone. The FIC index values were interpreted as follows: ≤0.5, synergistic; >0.5 to ≤1.0, additive; >1.0 to ≤2.0, indifferent; and >2.0, antagonistic effectsCitation9.
Peptide cytotoxicity on HaCat cells
To evaluate the effect of Dec-1B on HaCaT cell viability, cells were cultured in Petri culture dishes (100 × 20 mm) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FBS, 2 mM L-glutamine, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, 25 mM HEPES, (Corning) and 130 μg mL−1 Na pyruvate in a humidified 5% carbon dioxide atmosphere at 37 °C. Cell viability was examined as previously describedCitation29. Briefly, using a colorimetric assay based on the MTT labelling reagent, HaCaT cells (1 × 104 cells/well) were seeded in 96-well plates and, after overnight incubation, were treated with Dec-1B at concentrations of 3.25, 6.50, 13.0, 26.0 µMCitation35. As internal control, cells containing only dimethylsulphoxide (DMSO) (Sigma-Aldrich D8418-100 ml) as the solvent at the same concentration used in each test wells were selected. After 24 h, 10 µL of MTT solution (5 mg mL−1 in phosphate-buffered saline, PBS; pH 7.4) was added to each well and the plates were incubated for 3 h at 37 °C. Then, the medium was removed, and the obtained formazan crystals were dissolved in 150 μL of DMSO for 15 min. The spectrophotometric absorbance was measured using a microtiter enzyme-linked immunosorbent assay reader (Multiskan™ GO Microplate Spectrophotometer; Thermo Scientific™) at 540 nm. The percentage of viability of cells was determined by the following formula: (OD of treated cells/OD of control) × 100. Our in vitro cytotoxic examination revealed a secure profile for Dec-1B at all tested concentrations on HaCaT cells.
Critical aggregation concentration
The critical aggregation concentration (CAC) of Dec-1B was calculated using Nile Red (NR) as the fluorescent probe. This probe has poor solubility in water, while in presence of hydrophobic environments such as aggregated produces a hyper-hypsochromic effect consisting of a blue-shift and an intensity increaseCitation36. The peptide stock concentration of 400 µM was prepared dissolving Dec-1B in 1,1,1,3,3,3-Hexafluoro-2-propanol (HFIP), and then different peptide aliquots (0.8, 1, 5, 10, 15, 20, 30, 50, 100, 150, 200, 250, 300 µM) were prepared. The organic solvent was removed under nitrogen stream, 0.5 ml of water was added to each peptide aliquots, and then were sonicated for 15 min and freeze-driedCitation36. Then, each peptide solution was hydrated with NR solution of 500 nM for 1 h. The NR spectra were recorded at a fluorescence emission between 570 and 700 nm (slit width, 5 nm), and an excitation wavelength of 550 nm (slit width, 10 nm). All data were analysed by plotting the maximum emission fluorescence corresponding wavelength (y) as a function of the peptide concentration (×) using the sigmoidal Boltzmann equation as described previouslyCitation29.
Thioflavin T assay
Peptide aggregation in presence of LUVs mimicking fungal membrane was monitored by Thioflavin T (ThT) assayCitation37. Once lipid film [PE/PC/PI/Ergosterol (5:4:1:2, w/w/w/w)] at Cf = 100 μM was prepared as described above, it was hydrated with 100 mM NaCl, 10 mM Tris-HCl, 25 µM ThT buffer for 1 h and then was treated through the extrusion method to obtain LUVs. The LUVs were titrated with different peptide concentrations (5, 10, 20, 30 μM) and the ThT emission spectra were recorded before and after the peptide treatment at a fluorescence excitation at 450 nm (slit width, 10 nm) and a fluorescence emission at 482 nm (slit width, 5 nm).
Tryptophan quenching by Br-PC
Three different preparations of large unilamellar vesicles (LUVs) containing 25% of phospholipids 6,7 Br-PC, 9,10 Br-PC, 11,12 Br-PC, were prepared to perform tryptophan quenching experiment being the phospholipid Br-PC known as tryptophan quencherCitation38. Lipid films composed of PE/PC/Br-PC/PI/Ergosterol (5:3.7:0.3:1:2, w/w/w/w/w) were prepared at a final concentration of 125 μM. After the lyophilisation, the film was hydrated with water and LUVs were obtained as described above. The, LUVs were incubated with 5 μM of peptides Pent-1B and Dec-1B for 2 min and the tryptophan spectra emission was recorded setting the fluorescence excitation at 295 nm.
Tryptophan quenching by acrylamide
The tryptophan residue of peptides Pent-1B and Dec-1B was quenched using a 3.25 M solution of acrylamide in the absence and in presence of LUVs composed of PE/PC/PI/Ergosterol (5:4:1:2, w/w/w/w) at a final concentration of 250 μMCitation39. Once LUVs were prepared as described above, they were incubated with the peptide at 5 μM for 2 min, and then the tryptophan residue was quenched by acrylamide at different concentrations of 0.02, 0.04, 0.06, 0.08, 0.1, 0.12, 0.14, 0.16, 0.18, and 0.2 M. The same quenching experiment was performed for both peptides in water at a concentration of 5 μM. Fluorescence was measured with λex 295 nm and λem 340 nm. The data were analysed with the Stern-Volmer equation, F0/F = 1 + Ksv [Q], where F0 and F represent the fluorescence intensities in the absence and the presence of the quencher (Q), respectively. The Ksv is the Stern-Volmer quenching constant, which is correlated to the accessibility of the tryptophan residue to acrylamide. As acrylamide does not significantly partition into the membrane bilayer, Ksv is a reliable reflection of the bimolecular rate constant for collisional quenching of the aromatic residues present in the aqueous phase. Ksv is thus determined by the amount of non-lipid-associated free peptide as well as the peptide fraction located on the surface of the bilayer.
NMR spectroscopy
Appropriate amounts of peptides were dissolved in 0.600 ml H2O/D2O (90/10 v/v) for a peptide concentration of 0.4–0.8 mM to carry out the NMR analysis in pure water. Then, increasing volumes (μL) of a stock DPC-d38 solution (1.0 M in H2O/D2O 90/10 v/v, pH 6) were added until no modification was observed on the one-dimensional spectraCitation40. The NMR analysis in micellar environments was performed at DPC/peptide molar ratio R of 67, corresponding to DPC concentrations of 20–40 mM and peptide concentrations of 0.3–0.6 mM. Assuming a micelle aggregation number of ∼30, the micelle/peptide molar ratio R is ∼ 2.
NMR spectra were recorded on a Bruker 700 MHz Spectrometer, located at the Department of Pharmacy – University “Federico II” of Naples, and equipped with a z-gradient 5 mm triple-resonance cryoprobe. One-dimensional proton (1 D) and two-dimensional homonuclear (2 D) NMR spectra were recorded at a temperature of 298 K. The spectra were calibrated relative to TSP (0.00 ppm) as an internal standard. 2 D TOCSY (with different mixing times ranging from 25 to 70 ms), and NOESY (mixing times ranging from 100 to 300 ms) spectra were recorded in the phase-sensitive mode using the method from States, using 4096 data points in t2 and 512 equidistant t1 values. The water resonance was suppressed by use of gradients. NMR spectra were analysed by using CARA program (http://cara.nmr.ch/doku.php/home). Proton resonances were sequentially assigned by following the Wuthrich standard methodCitation41 and are reported in Tables S2–S5 of the Supplementary Material.
Table 2. MIC and MFC values of Pent-1B and Dec-1B against test C. albicans strains.
Spin-label experiments
Few microliters of a stock solution of 16-doxylstearic acid (16-DSA) in DMSO-d6 was added to the DPC peptide samples to obtain a ratio spin-label/micelle 1:1.
Structure determinations
The NOE-based distance restraints were obtained from NOESY spectra recorded with a mixing time of 200 ms. The NOE cross-peaks were integrated with CARA and were converted into upper distance bounds using CALIBA programCitation42 incorporated into CYANACitation43. Geminal protons were chosen as distance reference with 2.0 Å. Three dimensional structures were obtained by using inter-proton distances evaluated from NOEs as upper limits. Structure calculations started from 100 randomised conformers and used the standard CYANA simulated annealing schedule with 20,000 torsion angle dynamics steps per conformer. As the peptide sequences contain modified residues such as D-residues at positions 9 and 10 (d-LeuCitation9 and d-LysCitation10) and the N-terminal phenylalanine (Phe) bearing a long aliphatic chain in position para of phenyl ring i.e. pentanoic acid for Pent-1B and decanoic acid for Dec-1B, all these modifications have been inserted in the special.lib of CYANA suite before calculations. All the conformers showed a fairly agreement with experimental constraints showing no violations.
The best CYANA structures out of 100, in terms of agreement with experimental data, i.e. with lowest target function (TF) values (Table S6), were chosen to represent the peptide molecular structures in solution and visualised by PyMOL software (DeLano (2002) http://www.pymol.org).
Leakage assay
Large unilamellar vesicles (LUVs) mimicking fungal membrane, PE/PC/PI/Ergosterol (5:4:1:2, w/w/w/w), with ANTs and DPX encapsulated, were prepared as previously described.Citation21 Firstly, dry lipid film was prepared at a final concentration of 100 µM, and fluorophores ANTs and DPX dissolved in water were added to the film. Then, the lipid preparation was lyophilised overnight. The lipid film loaded with ANTs and DPX was hydrated with PBS 1× buffer (pH= 7.2) for 1 h under stirring and then was treated using the extrusion method to obtain LUVs. In particular, the lipid film with ANTs and DPX was freeze-thawed 6 times and extruded through 0.1-μm pore size polycarbonate filters 10 times. Then, nonencapsulated ANTs and DPX were removed from LUVs through gel filtration using a Sephadex G-50 column (1.5 cm × 10 cm). In the experiment, we measured the dequenching of ANTs by DPX after the treatment of LUVs with different peptide concentrations of 5, 10, 15, 20, 30, and 50 μM for 15 min. The ANTs fluorescence was recorded at a fluorescence excitation at 385 nm (slit width, 5 nm) and a fluorescence emission at 512 nm (slit width, 5 nm). The complete LUVs leakage and release of ANTs were measured after the treatment of LUVs with a solution of 10% Triton-X for 15 min. Liposomes leakage at different peptide concentrations was calculated as follows: %leakage= (Fi-F0)/(Ft-F0), where F0 is the ANTs fluorescence of the undamaged LUVs, Fi is the ANTs fluorescence after the peptide addition, and Ft is the ANTs fluorescence after the Triton-X treatment.
Plasma membrane permeabilization assay
The effect of Pent-1B and Dec-1B on plasma membrane of C. albicans cells was investigated by propidium iodide uptake using LIVE/DEAD FungaLight Yeast Viability Kit (Molecular Probes), according to the methodology by Thevissen et al. with some modificationsCitation43. A cell suspension of C. albicans ATCC 10231 (200 μL; 3.2 × 106 cells/mL of PBS) was incubated with test peptide at MIC value concentration and live/dead dyes (according to manufacturer instructions) for 30 min and 2 h in microcentrifuges tubes with constant agitation. After this time, the cells were washed, centrifugated (5 min at 10,000 × g) and suspended in 20 μL of PBS. Images were captured using an LSM 710 inverted confocal laser-scanning microscope (Zeiss) equipped with a 63× objective lens.
Cellular localisation of dec-1B
Confocal laser-scanning fluorescence microscopy was used to study the localisation of Dec-1B in C. albicans ATCC 10231 cells using a double stainingCitation44. A cell suspension (200 μL; 3.2 × 106 cells/mL of PBS) was incubated with 150 nM MitoTracker Orange (chloromethyl-H2-tetramethyl rosamine, Molecular Probes), a permanent mitochondrion-selective dye, for 15 min at 37 °C. The cells were washed with 200 μL of PBS and treated for 30 min and 2 h with 6.5 µM of 5(6)-carboxyfluorescein-labelled Dec-1B. The cells were centrifugated (5 min at 10,000 × g), suspended in 20 μL of PBS and examined using an LSM 710 confocal laser-scanning microscope with a 63X objective lens. The entire experiment was performed protected from light.
Results
Peptide design
Promoting the peptide self-assembly in fungal membrane and reducing the aggregation in aqueous solution through the lipidation strategy could favour an enhancement of the biological activity of AMPs. Different studies have already shown that the addition of fatty acids in N- or C-terminus of AMPs increases their antifungal and antibacterial activitiesCitation45,Citation46. Starting from our previous lipidation study, we identified the para position of PheCitation1 of peptide 1B as the best position for the incorporation of lipid tailsCitation29 because when we conjugated fatty acids of variable length in N-terminus or C-terminus of peptide 1B, we observed a loss of activity. In this previous study, we discovered a promising lipidated analogue namely Pent-1B bearing a lipid tail of 5-carbon atoms (C-5) in the para position of PheCitation1 (), which showed a good biological profile in terms of antimicrobial and cytotoxicity activities.
Figure 1. Representation of peptide sequences of Pent-1B and Dec-1B derived from the lipidation of the peptide 1B.
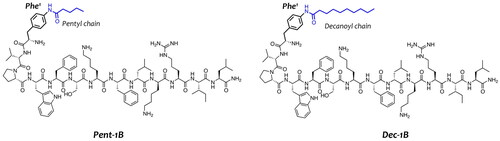
Herein, we explored the incorporation of a longer lipid tail of 10-carbon atoms (C-10) in the para position of PheCitation1 yielding the peptide Dec-1B (). The aim of this modification was to enhance the vehiculation and anchoring of the peptide within the fungal membrane, attempting to preserve the good biological profile of peptide Pent-1BCitation30. In our design, we kept unchanged the net positive charge of +4 and the amphipathic properties of the peptide Pent-1B and we did not explore the effect of further longer lipid tails to save the right hydrophobic/hydrophilic balance essential for the membrane interaction and to attain a good biological profile.
Antifungal activity of peptides against C. albicans strains
The antimicrobial activity of peptides Pent-1B and Dec-1B was evaluated against C. albicans ATCC 90028 and C. albicans ATCC 10231, and six clinical drug-resistant C. albicans strains. The antimycotic-susceptibility profiles of clinical C. albicans strains are reported in .
The antifungal activity of peptides, expressed as MIC and MFC values, is shown in . As reported, both peptides resulted active against both ATCC test strains with MIC values of 13 μM for Pent-1B and 6.5 μM for Dec-1B. The activity tested on clinical strains showed an inhibitory effect on the growth of 4/6 strains for the peptide Pent-1B (MIC values ranged from 26 to 6.5 μM) and of 3/6 strains for the peptide Dec-1B at the highest tested concentration of 26 μM. In addition, the activity of peptides was also investigated in terms of minimum fungicidal concentration. The MFC values were equal or two-fold higher than the MIC values for Pent-1B, and equal to the corresponding MICs for Dec-1B.
Synergistic effect with voriconazole of acyl-peptides
To further investigate the antifungal activity of Pent-1B and Dec-1B the synergistic interaction with voriconazole on voriconazole- resistant ATCC 10231 and voriconazole-resistant clinical Ca3 strain was determined using the checkerboard technique. After 24 h of incubation we analysed the results to determine the best combination of Pent-1B or Dec-1B with voriconazole. The total absence of growth of the ATCC 10231 strain was observed for 6.5 μM (1/2 MIC) of Pent-1B in combination with 0.06 μg/mL voriconazole (MIC of voriconazole alone was 30 μg/mL on C. albicans ATCC 10231). The FIC value of 0.502 was indicative of an additive effect for Pent-1B. Regarding Dec-1B, the total absence of growth was observed for the values of 3.25 μM (1/2 MIC) for Dec-1B in combination with 0.03 μg/mL voriconazole. The FIC value of 0.501 suggested an additive interaction as well. The best interaction on clinical strain Ca3 (voriconazole MIC = 60 μg/mL) was obtained at value of 13 μM (1/2 MIC) for Pent-1B in combination with 0.06 μg/mL voriconazole (FIC value = 0.501), resulting in an additive effect. Finally, peptide Dec-1B showed a synergistic effect at a concentration of 6.5 μM (1/4 MIC) in combination with 0.06 μM of voriconazole (FIC = 0.251) on clinical strain Ca3, thus being more effective than Pent-1B ().
Table 3. Fractional inhibitory concentration (FIC) index values of peptides on selected C. albicans strains.
Peptide effect on HaCaT cell viability
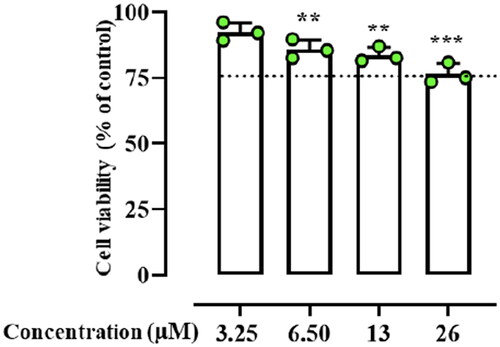
Based on the results from previous experiments, the cytotoxic effect of peptide Dec-1B was tested on human keratinocytes (HaCat cells) using the 3–(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay in a concentration range from 3.25 to 26 µM. As shown in , the peptide Dec-1B did not produce a significant cytotoxic effect on HaCat cells at its antimicrobial concentration of 6.50 µM. Only at the highest concentration of 26 µM, the incorporation of the lipid tails of C-10 caused a slight increase in the cytotoxicity of Dec-1B (75% of cell viability) in comparison with Pent-1B, which showed a negligible cytotoxic effect at highest concentration of 25 µM (86.7% of cell viability in Bellavita et al.)Citation29.
Figure 2. The cell viability of peptide treated HaCaT cells evaluated by MTT assay at 24 h. Dotted lines indicate 75 of cell viability as “cell viability limit”. Values are presented as means ± SD of 3 independent experiments **p ≤ 0.01; ***p ≤ 0.001 vs. Ctrl group. Statistical analysis performed by one way ANOVA followed by Bonferroni’s for multiple comparisons.
Lipid tails promote peptide aggregation in fungal membrane
Lipid tails promote peptide self-assembly in solution as well as in membrane environments. We calculated the critical aggregation concentration (CAC) of peptide Dec-1B in aqueous solution using Nile Red (NR) as fluorescent probe (, panel A). NR is not highly soluble in water, and it has a large preference to partition into hydrophobic environments producing a blue shift and hyperchromic effectCitation36. Despite the presence of the longer lipid tail (C-10), the peptide Dec-1B has the same CAC of 39.9 µM of the peptide Pent-1B, thus showing similarly a low ability to aggregate in aqueous solution at the concentration used for the antimicrobial assays.
Figure 3. Panel A reports the CAC of the peptide Dec-1B, while the CAC of Pent-1B was already calculated previouslyCitation29. The CAC was obtained plotting wavelength corresponding to the maximum fluorescence emission of Nile red as a function of the concentration of peptide. Panel B reports the peptide aggregation monitored by ThT fluorescence at different peptide concentrations of 5, 10, 20, and 30 µM.
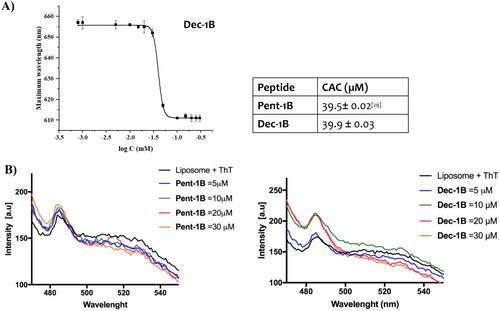
We also explored the ability of the peptides to oligomerize in membrane mimetic environments. The peptide Dec-1B, compared to peptide Pent-1B, clearly showed a stronger ability to oligomerize in fungal membranes (, panel B). Specifically, we studied peptide aggregation in LUVs composed of PE/PC/PI/Ergosterol (5:4:1:2, w/w/w/w) mimicking fungal membrane and Thioflavin T as fluorescent dye. LUVs were treated with different peptide concentrations (5, 10, 20, and 30 µM). A large enhancement of ThT was recorded at 10 µM for the peptide Dec-1B, revealing a complete peptide aggregation in LUVs. In contrast, for the peptide Pent-1B, the ThT fluorescence at different concentrations indicated a progressive phenomenon of aggregation in LUVs, obtaining an increase of ThT fluorescence at the higher concentration of 30 µM.
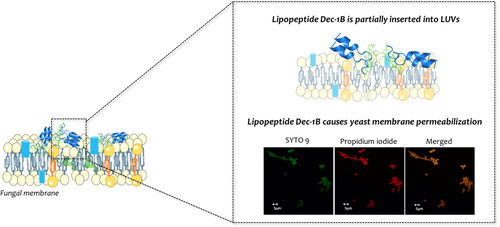
Lipopeptides pent-1B and dec-1B are partially inserted into LUVs
The depth of insertion of the peptide into the target membrane is key for activity and for understanding its mechanism of action. We monitored the insertion of lipopeptides Pent-1B and Dec-1B into LUVs mimicking fungal membrane [PE/PC/PI/Ergosterol (5:4:1:2, w/w/w/w)] recording the change of the fluorescence emission of tryptophan (Trp) present in position 4 in both peptides.
We compared the Trp fluorescence emission of peptides in presence of LUVs and in water; furthermore, the accessibility of the Trp residue was measured by quenching it with acrylamide at different concentrations, which is a water-soluble quencher and is unable to insert into the hydrophobic lipid bilayer. As shown in , when the peptides Pent-1B and Dec-1B are inserted into hydrophobic environments such as LUVs, the quantum yield of Trp residue changes determining an increase of the Trp fluorescence emission with respect to that recorded in water.
Figure 4. Tryptophan fluorescence spectra for the peptides Pent-1B (panel A) and Dec-1B (panel B) recorded in water (on the left) and in LUVs composed of PE/PC/PI/Ergosterol (5:4:1:2, w/w/w/w) (on the right).
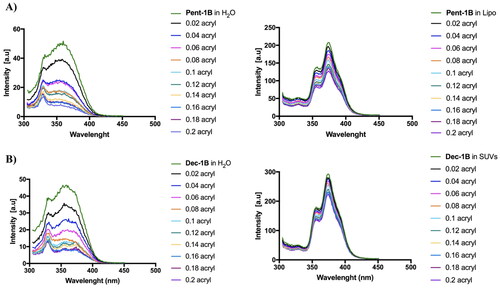
The emission spectra in indicated that both peptides (concentration of 10 µM) were buried in LUVs because the Trp residue resulted to be only partially accessible to the quenching by acrylamide at different concentrations. Being both peptides aggregated at the concentration of 10 µM in LUVs as shown in , it is likely that equilibrium phenomena take place between monomeric and oligomeric states which make Trp only partially accessible to quenching by acrylamide. In contrast, when the peptides are in aqueous solution, the Trp is highly accessible because both peptides are in monomeric state at 10 µM as indicated by CAC values ().
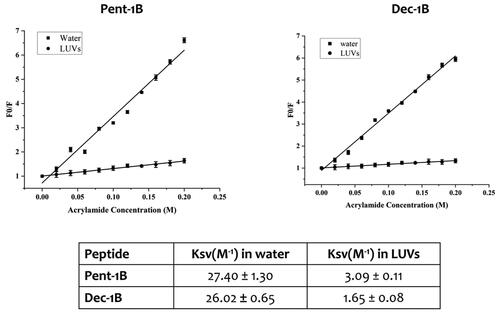
Stern-Volmer plots for the quenching of tryptophan by acrylamide are shown in . In the presence of LUVs, a great decrease in fluorescence intensity was evident, thus revealing that the tryptophan is less accessible to the quencher. In fact, the values for Ksv were lower () in LUVs, confirming that the tryptophan is more buried in the bilayer.
Figure 5. Quenching of tryptophan by acrylamide of peptides Pent-1B and Dec-1B and Stern-Volmer (Ksv) quenching constants of peptides both in water and LUVs mimicking fungal membrane.
In addition, the localisation of both peptides into LUVs mimicking the fungal membrane was investigated by measuring the quenching of the fluorescence of the Trp residue by the bromo probes bound to the lipid chains of the membrane (). We used LUVs composed of 3 different phospholipids bearing bromo quencher in different positions along the hydrocarbon chain. In particular, we used phospholipids 11,12-Br-PC, 9,10-Br-PC, and 6,7-Br-PC in our experiment. While in 6,7-Br-PC LUVs, the bromo quencher is located at the LUVs interface, in 11,12-Br-PC and 9,10-Br-PC LUVs, the bromo quencher is deeply buried into the lipid core.Citation38 As observed in , for both peptides, the largest quenching of Trp fluorescence was recorded in presence of 9,10-Br-PC and 11,12- Br-PC LUVs, while it was less in presence of 6,7- Br-PC LUVs.
Figure 6. Tryptophan fluorescence spectra for the peptides Pent-1B (panel A) and Dec-1B (panel B) recorded in the presence of the probes 11,12-Br-PC, 9,10-Br-PC, and 6,7-Br-PC.
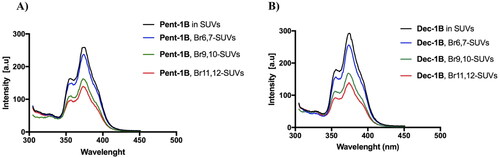
These results indicated that probably both peptides were inserted into the lipid bilayer and the Trp residue points towards the micellar interior.
NMR conformational analysis shows the localisation of lipopeptides into micelle mimicking membrane environment
We further investigated the interaction and structural properties of peptides Pent-1B and Dec-1B in fungal membranes by NMR spectroscopy using dodecyl-phosphorylcholine (DPC) micelles mimicking membrane environment. NMR studies of Pent-1B and Dec-1B peptides were performed in pure water, where they are perfectly soluble but poorly structured, and in DPC micelles, where they partially adopt helix structures.
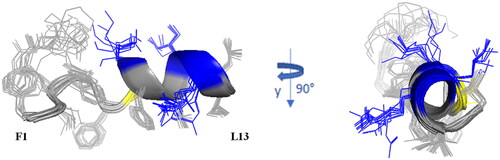
In aqueous medium, NMR data, such as the deviations of αCH proton chemical shift (Figure S1) from random coil valuesCitation47 and NOEs collected for Pent-1B and Dec-1B in H2O/D2O (90/10, v/v) (Figure S2A and 2C, and Table S6, respectively), indicate random coil conformations of both peptides. This structural diagnosis is confirmed by the molecular models computed by CYANA programCitation48 using NOE derived distances as upper limit (upl) of interproton distances sketched in Figure S3. The best ten CYANA structures in terms of agreement with experimental data, i.e. with lowest target function (TF) values, were chosen as representative of the conformational space accessible to the peptides and reveal a conformational flexibility of Pent-1B and Dec-1B in pure water, even though some hints of ordered structural elements, bends or turns, emerge in the middle region of peptides. 1 D 1H spectra of the peptides in water solution as well as the CYANA molecular models obtained for Pent-1B and Dec-1B are shown in .
The conformational behaviour of Pent-1B and Dec-1B was also tested in DPC/water mixture (20–40 mM DPC, pH 6, DPC/peptide molar ratio R of 67, DPC micelle/peptide ratio ca. 2). 1 D 1H spectra of Pent-1B and Dec-1B in DPC are shown in Figure S3A and S5B. In such environment, both peptides adopt a quite ordered structure. The negative αCH deviations from random coil values < −0.1 ppm (Figure S6) point to a helical conformation in the middle-C-terminal regions of the peptideCitation46. This structural diagnosis is consistent with the NOE pattern (Figure S2B, D).
Indeed, the presence of NOE effects of the type NHi–NHi+1, together with long range αi-Ni+2, αi-Ni+3 and αi-βi+3 contacts, indicates the occurrence of a helical structure in the central region of the peptides. Structural calculations of Pent-1B and Dec-1B were carried out by CYANA43 using as upper limit (upl) of inter-proton distances 191 NOE derived distances (127 intra-residues, 33 sequential, 31 long-range) for Pent-1B and 246 NOE derived distances (156 intra-residues, 45 sequential, 45 long-range) for Dec-1B. The statistic of the structural analysis is reported in Table S6. The best CYANA structures were chosen as representative of the conformational space accessible to the peptide ().
Figure 7. Cartoon representation of the best 15 NMR/CYANA structures of Pent-1B in DPC 20 mM. Side - chains are shown as sticks coloured by amino acid type: gray, hydrophobic; blue, basic; yellow, polar.
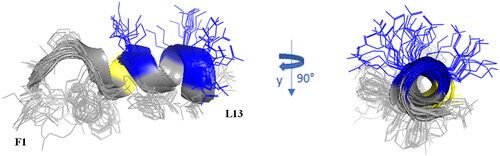
The backbone superimposition of the best 15 structures shows a well-defined helix arrangement in F5–I12 segment (RMSD value on the backbone atoms of 0.52 Å) that places positively charged and hydrophobic side chains on opposite sides. It is worth noticing that, in this arrangement, all the charged sidechains (KCitation7 KCitation10 R11) are iso-oriented as well as the aromatic side chains of N-terminal residues FCitation1 W4 and F.5 Similarly, for Dec-1B the NMR/CYANA analysis finds a helical arrangement in the segment 5–12 (RMSD value on the backbone atoms of 0.25 Å), with iso-orientation of the hydrophobic and charged side chains ().
Spin label studies
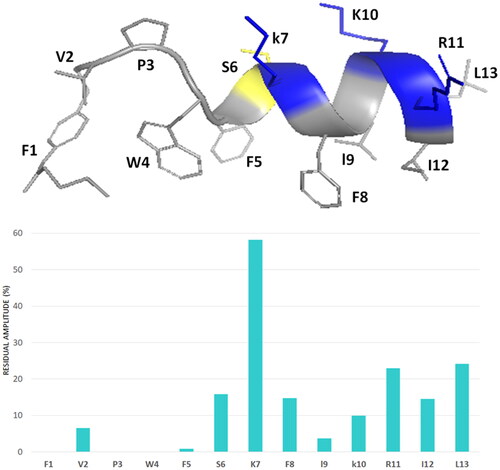
The spin label 16-doxyl stearic acid (16-DSA) was added to the peptide/DPC samples to examine the placement of peptides in DPC. 16-DSA is useful for identification of the membrane-embedded residues, as it solubilises into DPC micellesCitation49. This spin label contains one unpaired electron that induces paramagnetic relaxation resulting in line broadening of resonance signals in proximity to the spin label. A comparison of the cross-peak intensities in the TOCSY spectra of peptide/micelle systems with and without the spin label indicates the residues mostly interacting with micelles.
With a 16-DSA/DPC micelle ratio of 1:1 the cross-peak intensities of the N-terminal residues are strongly reduced, whilst the cross-peak intensities of the C-terminal residues show the same periodic medium or strong reduction for Pent-1B as well as for Dec-1B. The least affected residues are serine 6 and lysine 7 at the beginning of the α-helix structure.
These results suggest that helical domain lies parallel to the micelle surface, with the hydrophobic face towards the interior of the micelle, and the positively charged residues interacting with the negatively charged phosphate headgroups, anchoring the peptide just below the micelle surface ().
Lipopeptides pent-1B and dec-1B did not induce membrane leakage
Since different studies showed that temporins generally act causing the breakdown of bacterial membranesCitation22,Citation29, we investigated the mode of action of lipopeptides Pent-1B and evaluated the release of encapsulated ANTs probe in LUVs mimicking fungal membrane [PE/PC/PI/Ergosterol (5:4:1:2, w/w/w/w)]. In these conditions, we did not observe any ANTs release and liposome leakage at different peptide concentrations of 5, 10, 15, 20, 30, and 50 µM (data not shown). Thus, we hypothesised that the mode of action of these lipopeptides may involve an initial electrostatic interaction followed by aggregation and insertion in the lipid bilayer causing perturbation through a carpeting effect as already shown for some temporin analoguesCitation21 and other molecular mechanisms are surely underlying their antifungal activity.
Lipopeptides cause yeast membrane permeabilization
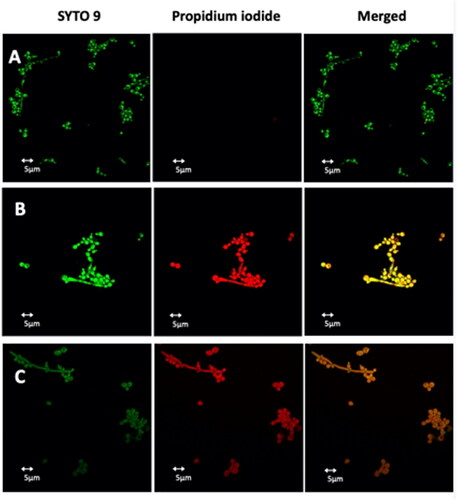
To evaluate the membrane permeabilization of peptides Pent-1B and Dec-1B, C. albicans cells were treated with the LIVE/DEAD FungaLight Yeast Viability kit consisting of labelling dyes such as SYTO 9 and propidium iodide (PI). SYTO 9 penetrates both viable and non-viable cells, while PI only penetrates cells with damaged membranes by turning off the fluorescence emitted by SYTO 9. Damaged or dead cells emit a yellow-red fluorescence, while viable cells emit only green fluorescence. The images were captured by an LSM710 inverted confocal laser scanning microscope (CLSM). All yeast cells showed yellow-red fluorescence after 30 min of treatment with Pent-1B or Dec-1B peptides (, panels B and C), while untreated cells showed green fluorescence (, panel A). The obtained results further support the data obtained by the other experiments and pointing on the ability of both peptides to act on the plasma membrane of these cells, causing a membrane perturbation and thus allowing the entry of the PI dye.
Figure 10. Detection of cell membrane permeabilisation in living C. albicans cells from control culture (A) and cultures treated with Pent-1B (B) or Dec-1B (C) by CLSM. The cells were stained with SYTO9 and Propidium iodide (PI). Fluorescence images of the same samples at 528 nm for SYTO9 (green fluorescence; left panels), 645 nm for PI (red fluorescence; middle panels) and merged images (right panels) are shown. SYTO 9 penetrates both viable and nonviable cells, while PI penetrates only cells with compromised membranes. Candida cells showing yellow-red fluorescence are considered as dead cells.
Dec-1B does not enter in yeast mitochondria
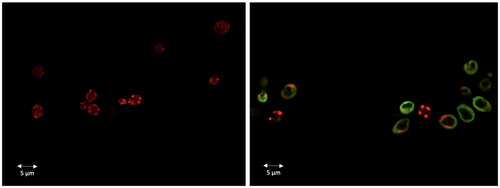
Dec-1B labelled with 5(6)-carboxyfluorescein was used to investigate the cellular localisation of the peptide in C. albicans cells using CLSM. A suspension of the ATCC 10231 strain was pre-incubated with MitoTracker Orange and then treated with the labelled peptide at the MIC value concentration. Confocal images showed that the green signal corresponding to the localisation of the fluorescent peptide was uniformly localised on the surface of the treated cells after 30 min. Since the peptide signal has not been shown to overlap the red signal (MitoTracker) of the mitochondria (), no localisation in mitochondria seems to occur. The green signal remained localised on yeast surface even after 2 h of treatment. The images therefore confirm that the peptide acts on the fungal wall/cell membranes.
Figure 11. Dec-1B localisation in C. albicans cells detected by CLSM. Dec-1B conjugated with 5(6)-carboxyfluorescein is coloured in green, while the mitochondria are visualised in red by MitoTracker. The images show C. albicans cells before treatment (left panel) and after treatment for 30 min with Dec-1B (right panel) with the green signal localised on the surface.
Discussions
The spread of Candida infections and the resistance issues to available antifungal drugs are rapidly increasing. In this scenario, AMPs represent a challenge to open novel avenues and hopes for overcoming the antifungal resistanceCitation14,Citation16. Previously, using several synthetic strategies on some Temporin L derivatives, including lipidation strategyCitation29 we developed different analogues which exhibited strong ability to kill both Gram-positive and Gram-negative bacteria, and yeasts as Candida sppCitation21. Our previous lipidation study, consisting in the conjugation of fatty acids at the N- and C-terminus of the most promising Temporin L analogue, peptide 1B, allowed to identify the para position of PheCitation1 as the best position for performing this functionalizationCitation29. In this lipidation screening, we discovered the acylated peptide Pent-1B bearing the acyl chain of C-5 on PheCitation1 which displayed a strong activity against carbapenem resistant Klebsiella pneumoniae (MIC = 6.25 µM) and a low cytotoxic profileCitation29,Citation30. Through a deep study on its potential mode of action, we observed that the presence of the chain of the C-5 in Pent-1B determined a monomeric state in aqueous solution (CAC= 39.5 µM) and a significant ability to oligomerize at a low concentration (∼10 µM) in LUVs mimicking Gram-positive and Gram-negative membranesCitation29. Following the oligomerization in bacterial membrane, Pent-1B induced the pore formation of the target membrane detected by a complete leakage of LUVs mimicking bacterial membrane.
Here, considering these encouraging results, we further investigated the activity of Pent-1B against reference and clinical strains of Candida and we designed and developed its analogue Dec-1B (), bearing a longer lipid tail of C-10 in para position of PheCitation1 to improve peptide anchoring within the fungal membrane.
In general, the interaction of AMPs with the target membranes is regulated by several features, including the length of the sequence, the number and density of positive charges, the secondary structure in aqueous environment and in membranes, and the hydrophobicityCitation50. The initial interaction involves the binding to the membrane by a purely non-specific electrostatic interaction between the positively charged residues of the peptide and the negatively charged membrane of bacteria or fungi. Whereas, the peptide hydrophobic domains interact with the hydrophobic core of the lipid bilayer, guiding the peptide deeper into the membraneCitation50,Citation51.
In our design, we kept the net positive charge of +4 of peptide Pent-1B due to Lys and Arg residues and added a longer lipid tail of C-10 to boost the peptide interaction and the internalisation into a complex membrane like that of fungi. The results of antifungal activity on C. albicans ATCC strains (ATCC 10231 and ATCC 90028) showed a significant antifungal activity for Pent-1B and Dec-1B with a MIC of 13 and 6.5 µM, respectively (). Both peptides were able to reduce the growth of C. albicans clinical strains, with MIC values ranging from 26 μM to 6.5 μM for peptide Pent-1B on Ca1, Ca2, Ca3 and Ca5 strains (), while the peptide Dec-1B worked at the highest tested concentration of 26 μM on 3 strains (Ca3, Ca4, Ca6). Although in most cases MIC values for clinical strains were higher than those obtained for ATCC strains, these results seem interesting as the peptides were effective against clinical strains with different resistance profiles, some of which very difficult to treat. In particular, Ca3 and Ca6 strains were resistant to all the azoles, which are widely used for the topical treatment of superficial candidiasis and the systemic therapy of invasive mycoses, while the Ca1 and Ca6 strains were resistant to both azoles and echinocandins, which frequently represent the therapeutic option for azole-resistant Candida strains in hospital settingsCitation52.
Currently, combination treatment of antifungal drugs with unconventional antifungal agents have been proposed to fight drug-resistant C. albicans infectionsCitation53. In our study, when combined with voriconazole, Dec-1B exhibited a synergistic effect on Ca3 strain, while the association had an additive effect on resistant ATCC strain (). On the other hand, the combination of Pent-1B with voriconazole was always additive. Noteworthy is the ability of both peptides to significantly reduce the MIC of voriconazole for both resistant strains ATCC 10231 and Ca3, thereby restoring their susceptibility to the antifungal drug. In this case, the combination approach seems particularly advantageous as it allows to recover the effectiveness of drugs such as azoles which have limited toxicity, are cheap and available for oral administration.
Based on these synergistic results, we hypothesised that both peptides may act with different mode of actions compared to conventional antifungal drugs, facilitating the entry of azoles due to their ability to enhance the cell membrane permeability. Clearly, we still cannot exclude that the peptides may also act on some specific molecular pathways and further studies will be necessary to demonstrate this latter mechanism.
We previously showed that Pent-1B acts through a membrane-active mechanism on bacteriaCitation29 thus, here, we explored the mode of action of peptides Pent-1B and Dec-1B through fluorescence assays using liposomes as model systems for mimicking fungal membrane and NMR studies to analyse their three-dimensional structure.
Both peptides were in a monomeric state in aqueous solution at their active antifungal concentrations (around 10 µM) detected by the value of CAC (around 39 µM). Whereas, both peptides showed the ability to oligomerize in liposomes mimicking the Candida membrane detected by the ThT experiment (). In particular, the peptide Dec-1B bearing the longer lipidic tails oligomerized at lower concentration of 10 µM than the peptide Pent-1B that is completely aggregated at 30 µM.
We also analysed through the Br-Trp quenching experiment the depth of internalisation of the peptides in LUVs and the results supported that the tryptophan of both peptides was strongly inserted in the bilayer (). The tryptophan is located at position 4 of the sequence from the N-terminus, which confirms the hypothesis that the flexible domain at the N-terminus is key for the insertion into the bilayer.
Since the other important feature that influences the mode of action of AMPs is three-dimensional structure, we studied the potential mode of action of both peptides by NMR spectroscopy. We observed that both peptides are disordered in water but partially organised in helix when interacting with membrane mimetic systems. The choice of the mimetic system is a critical point in this type of study. We opted for a micellar system instead of bicelles or other heavier mimics because micelles are generally light enough to allow the use of un-labelled (neither 15N and/or 13C labelling) cheaper peptides for the NMR interactions studies. The addition of DPC to each Pent-1B and Dec-1B in aqueous solution modifies the NMR spectra. The lines of all signals become larger and some amide protons belonging to the N-terminal segment vanish at all. These effects, by alone, demonstrate that the peptides bind the micelles. Indeed, both peptides change their roto-translation correlation times from those typical of the free forms to those, longer, of the micelle, with consequent higher relaxation rates and larger or vanished signals. Temporin analogue ability to interact with membrane mimics with consequent adoption of helix structure was previously demonstratedCitation21,Citation22,Citation29. Here, we showed that the pentanoic or decanoic acid covalently linked to the para position of PheCitation1 for Pent-1B and Dec-1B, respectively, favours the N-terminal anchorage of the peptides to the micelle.
For both peptides, the hydrophobic trait, TAIL-F1-V2-P3-W,4 deeply binds the micelle, while the trait F5-L13 adopts an α-helix structure. The helix arrangement is driven by the contact between the 5–13 trait, amphiphilic in nature, and micelle. The recourse to radical species like 16-doxyl stearic acid (16-DSA), makes it possible to catch the NMR imagine of Pent-1B and Dec-1B docked to the micelle surface. DSA, which solubilises into the micelle, accelerates the nuclear relaxation rates of the nuclei around it. That means that more the peptide side chains are sunk into the micelle, more their signal intensities are diminished. For both Pent-1B and Dec-1B, the nuclei with the most decreased intensities are those of the N-terminal lipophilic segments and those of the first 3–4 residues. The effect of the paramagnetic species on the intensity of the segment 5–13 suggests that the helix lies on the micelle surface, with the aromatic side chains of both Pent-1B () and Dec-1B iso-oriented towards the micelle interior, while the positively charged sidechains protrude on the opposite side. Thus, the picture emerging by NMR measurements points to peptides that are deeply anchored through the N-terminal segment, while the C-terminal trait lies on the micelle surface fastened by the hydrophobic side chains.
The Br-Trp quenching experiment in membranes mimicking the fungal membrane clearly showed a deeper insertion of the peptides compared to the NMR results obtained into micelles mimicking mammalian membranes, further supporting that the antifungal activity associated by the lack of toxicity to mammalian cells is associated to a different depth of penetration.
The results of the NMR studies in combination with biophysical data clearly showed that both peptides have the typical behaviour of AMPs, characterised by a random conformation in aqueous solution and the adoption of a helical conformation in membrane bilayers. Furthermore, the presence of the lipidic chain favours the peptide anchorage to the membrane and the formation of oligomers inside the membrane. In addition, the deep insertion and the absence of leakage are key to hypothesise a carpet mechanism for the perturbation of the membrane. This assumed mechanism of action of the peptides was further investigated by CLSM. Candida membrane permeabilisation assay showed that 30 min of treatment with Pent-1B or Dec-1B (separately) allowed the uptake of PI suggesting that wall/cell membrane damage and permeabilisation is a possible mode of action of these lipopeptides (). In agreement with these results, the fluorescent labelling of Dec-1B-K(Fam) showed that it remained uniformly localised at the surface of Candida cells and was not localised into mitochondria ().
These data support the hypothesis that Pent-1B and Dec-1B act by interacting and altering the fungal cell membrane as already reported for many antimicrobial peptides, although other mechanisms of action cannot be excluded.
Conclusions
In conclusion, this study elucidated the antifungal activity of our acylated Temporin L analogues Pent-1B and Dec-1B and explored their potential membrane active mechanism. The acylated peptides resulted to be efficacious on both reference and clinical isolates of C. albicans and they showed an additive or synergistic interaction with voriconazole. In addition, both peptides did not show cytotoxicity on human keratinocytes at their antimicrobial concentration. Through a deep study on their potential mechanism of action by using fluorescence-based assays, NMR studies and confocal microscopy, we hypothesised a membrane-active mechanism for peptides Pent-1B and Dec-1B, that consists in an internalisation and oligomerization into fungal membrane, causing a cell membrane damage by a carpeting effect.
These findings may provide insights into the possible therapeutic applications of Temporin analogues as antifungal agents or as enhancers of traditional antifungal drugs that are losing their efficacy due to the development of resistance.
Supplemental Material
Download PDF (368 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Domán M, Bányai K. COVID-19-associated fungal infections: an urgent need for alternative therapeutic approach.? Front Microbiol. 2022;13:919501.
- Ré ACS, Martins JF, Cunha-Filho M, Gelfuso GM, Aires CP, Gratieri T. New perspectives on the topical management of recurrent candidiasis. Drug Deliv Transl Res. 2021;11(4):1568–1585.
- Mukaremera L, Lee KK, Mora-Montes HM, Gow NAR. Candida albicans yeast, pseudohyphal, and hyphal morphogenesis differentially affects immune recognition. Front Immunol. 2017;8(1):629.
- de Cássia Orlandi Sardi J, de Souza Pitangui N, Gullo FG, Fusco-Almeida AM, Mendes-Giannini MJS. A mini review of Candida species in hospital infection: epidemiology, virulence factor and drugs resistance and prophylaxis. Trop. Med. Surg. 2013;1:5.
- Nucci M, Queiroz-Telles F, Tobón AM, Restrepo A, Colombo AL. Epidemiology of opportunistic fungal infections in Latin America. Clin Infect Dis. 2010;51(5):561–570.
- Talapko J, Juzbašić M, Matijević T, Pustijanac E, Bekić S, Kotris I, Škrlec I. Candida albicans-the virulence factors and clinical manifestations of infection. JoF. 2021;7(2):79.
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67(3):400–428, table of contents.
- Bhattacharya S, Sae-Tia S, Fries BC. Candidiasis and mechanisms of antifungal resistance. Antibiotics (Basel. 2020;9(6):312.
- Danby CS, Boikov D, Rautemaa-Richardson R, Sobel JD. Effect of pH on in vitro susceptibility of Candida glabrata and Candida albicans to 11 antifungal agents and implications for clinical use. Antimicrob Agents Chemother. 2012;56(3):1403–1406.
- Zhang MR, Zhao F, Wang S, Lv S, Mou Y, Yao CL, Zhou Y, Li FQ. Molecular mechanism of azoles resistant Candida albicans in a patient with chronic mucocutaneous candidiasis. BMC Infect. Dis. 2020;20:126.
- Tverdek FP, Kofteridis D, Kontoyiannis DP. Antifungal agents and liver toxicity: a complex interaction. Expert Rev anti Infect Ther. 2016;14(8):765–776.
- Buda De Cesare G, Cristy SA, Garsin DA, Lorenz MC. Antimicrobial peptides: a new frontier in antifungal therapy. mBio. 2020;11(6):20.
- Wang J, Dou X, Song J, Lyu Y, Zhu X, Xu L, Li W, Shan A. Antimicrobial peptides: promising alternatives in the post feeding antibiotic era. Med Res Rev. 2019;39(3):831–859.
- Falanga A, Maione A, La Pietra A, de Alteriis E, Vitale S, Bellavita R, Carotenuto R, Turrà D, Galdiero S, Galdiero E, et al. Competitiveness during dual-species biofilm formation of Fusarium oxysporum and Candida albicans and a novel treatment strategy. Pharmaceutics. 2022;14(6):1167.
- Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19(3):491–511,
- Maione A, Bellavita R, de Alteriis E. d, Galdiero S, Albarano L, La Pietra AL, Guida M, Parrilli E, D’Angelo C, Galdiero E, et al. A. WMR peptide as antifungal and antibiofilm against albicans and non-albicans Candida species: shreds of evidence on the mechanism of action. IJMS. 2022;23(4):2151.
- Silva ARP, Guimarães MS, Rabelo J, Belén LH, Perecin CJ, Farías JG, Santos JHPM, Rangel-Yagui CO. CO. Recent advances in the design of antimicrobial peptide conjugates. J Mater Chem B. 2022;10(19):3587–3600.
- Romero SM, Cardillo AB, Martínez Ceron MC, Camperi SA, Giudicessi SL. Temporins: an approach of potential pharmaceutic candidates. Surg Infect. 2020;21(4):309–322.
- Zannella C, Chianese A, Palomba L, Marcocci ME, Bellavita R, Merlino F, Grieco P, Folliero V, De Filippis A, Mangoni ML, et al. Broad-Spectrum antiviral activity of the amphibian antimicrobial peptide Temporin L and its analogs. IJMS. 2022;23(4):2060.
- Mangoni ML, Grazia AD, Cappiello F, Casciaro B, Luca V. Naturally occurring peptides from Rana temporaria: antimicrobial properties and more. Curr Top Med Chem. 2016;16(1):54–64.
- Bellavita R, Maione A, Merlino F, Siciliano A, Dardano P, De Stefano L, Galdiero S, Galdiero E, Grieco P, Falanga A. Antifungal and antibiofilm activity of cyclic Temporin L peptide analogues against albicans and non-albicans Candida species. Pharmaceutics. 2022;14(2):454.
- Bellavita R, Casciaro B, Di Maro S, Brancaccio D, Carotenuto A, Falanga A, Cappiello F, Buommino E, Galdiero S, Novellino E, et al. First-in-Class cyclic Temporin L analogue: design, synthesis, and antimicrobial assessment. J Med Chem. 2021;64(15):11675–11694.
- Merlino F, Carotenuto A, Casciaro B, Martora F, Loffredo MR, Di Grazia A, Yousif AM, Brancaccio D, Palomba L, Novellino E, et al. Glycine-replaced derivatives of [Pro3,Dleu9]TL, a temporin L analogue: evaluation of antimicrobial, cytotoxic and hemolytic activities. Eur J Med Chem. 2017;139:750–761.
- Bellavita R, Vollaro A, Catania MR, Merlino F, De Martino L, Nocera FP, Della Greca M, Lembo F, Grieco P, Buommino E. Novel antimicrobial peptide from Temporin L in the treatment of Staphylococcus Pseudointermedius and Malassezia Pachydermatis in polymicrobial inter-kingdom infection. Antibiotics (Basel. 2020;9(9):530.
- Bellavita R, Raucci F, Merlino F, Piccolo M, Ferraro MG, Irace C, Santamaria R, Iqbal AJ, Novellino E, Grieco P, et al. Temporin L-derived peptide as a regulator of the acute inflammatory response in zymosan-induced peritonitis. Biomed Pharmacother. 2020;123:109788–109788.
- Hamill RJ. Amphotericin B formulations: a comparative review of efficacy and toxicity. Drugs. 2013;73(9):919–934.
- de Alteriis E, Maione A, Falanga A, Bellavita R, Galdiero S, Albarano L, Salvatore MM, Galdiero E, Guida M. Activity of free and liposome-encapsulated essential oil from Lavandula angustifolia against persister-derived biofilm of Candida auris. Antibiotics. 2021;11(1):26.
- Allen TM, Cullis PR. Liposomal drug delivery systems: from concept to clinical applications. Adv Drug Deliv Rev. 2013;65(1):36–48.
- Bellavita R, Falanga A, Buommino E, Merlino F, Casciaro B, Cappiello F, Mangoni ML, Novellino E, Catania MR, Paolillo R, et al. Novel Temporin L antimicrobial peptides: promoting self-assembling by lipidic tags to tackle superbugs. J Enzyme Inhib Med Chem. 2020;35:751–1764.
- Roscetto E, Bellavita R, Paolillo R, Merlino F, Molfetta N, Grieco P, Buommino E, Catania MR. Antimicrobial activity of a lipidated Temporin L analogue against carbapenemase-producing Klebsiella pneumoniae clinical isolates. Antibiotics. 2021;10(11):1312.
- Clinical and Laboratory Standards Institute (CLSI). Performance standards for antifungal susceptibility testing of yeasts. M60 – 2nd Ed. Pennsylvania (US): Clinical and Laboratory Standards Institute; 2020.
- The European Committee on Antimicrobial Susceptibility Testing - EUCAST. Clinical breakpoints for fungi (Candida and Aspergillus species). Available from: https://www.eucast.org/astoffungi/clinicalbreakpointsforantifungals. v. 10.0, accessed on 4 April 2022.
- Merlino F, Tomassi S, Yousif AM, Messere A, Marinelli L, Grieco P, Novellino E, Cosconati S, Maro D. S. Boosting Fmoc solid-phase peptide synthesis by ultrasonication. Org Lett. 2019;21(16):6378–6382.
- Clinical and Laboratory Standards Institute. Reference method for broth dilution antifungal susceptibility testing of yeasts. M27 – 4th Ed. Pennsylvania (US): Clinical and Laboratory Standards Institute; 2017.
- Varma SR, Sivaprakasam TO, Mishra A, Prabhu S, M R, P R. Imiquimod-induced psoriasis-like inflammation in differentiated human keratinocytes: its evaluation using curcumin. Eur J Pharmacol. 2017;813:33–41.
- Del Genio V, Falanga A, Allard-Vannier E, Hervé-Aubert K, Leone M, Bellavita R, Uzbekov R, Chourpa I, Galdiero S. Design and validation of nanofibers made of self-assembled peptides to become multifunctional stimuli-sensitive nano-vectors of anticancer drug doxorubicin. Pharmaceutics. 2022;14(8):1544.
- Xue C, Lin TY, Chang D, Guo Z. Thioflavin T as an amyloid dye: fibril quantification, optimal concentration and effect on aggregation. R Soc Open Sci. 2017;4(1):160696.
- Bolen EJ, Holloway PW. Quenching of tryptophan fluorescence by brominated phospholipid. Biochemistry. 1990;29(41):9638–9643.
- De Kroon AI, Soekarjo MW, De Gier J, De Kruijff B. The role of charge and hydrophobicity in peptide-lipid interaction: a comparative study based on tryptophan fluorescence measurements combined with the use of aqueous and hydrophobic quenchers. Biochemistry. 1990;29(36):8229–8240.
- Yousif AM, Ingangi V, Merlino F, Brancaccio D, Minopoli M, Bellavita R, Novellino E, Carriero MV, Carotenuto A, Grieco P. Urokinase receptor derived peptides as potent inhibitors of the formyl peptide receptor type 1-triggered cell migration. Eur J Med Chem. 2018;143:348–360.
- Wuthrich K. NMR of proteins and nucleic acids. New York: Wiley; 1986.
- Güntert P, Qian YQ, Otting G, Müller M, Gehring W, Wüthrich K. Structure determination of the Antp (C39––S) homeodomain from nuclear magnetic resonance data in solution using a novel strategy for the structure calculation with the programs DIANA, CALIBA, HABAS and GLOMSA. J Mol Biol. 1991;217(3):531–540.
- Thevissen K, Terras FR, Broekaert WF. Permeabilization of fungal membranes by plant defensins inhibits fungal growth. Appl Environ Microbiol. 1999;65(12):5451–5458.
- Ruissen AL, Groenink J, Helmerhorst EJ, Walgreen-Weterings E, Van’t Hof W, Veerman EC, Nieuw Amerongen AV. Nieuw Amerongen AV. effects of histatin 5 and derived peptides on Candida albicans. Biochem J. 2001;356(Pt 2):361–368.
- Bugli F, Massaro F, Buonocore F, Saraceni PR, Borocci S, Ceccacci F, Bombelli C, Di Vito M, Marchitiello R, Mariotti M, et al. Design and characterization of myristoylated and non-myristoylated peptides effective against Candida spp. clinical isolates. IJMS. 2022;23(4):2164.
- Malina A, Shai Y. Conjugation of fatty acids with different lengths modulates the antibacterial and antifungal activity of a cationic biologically inactive peptide. Biochem J. 2005;390(Pt 3):695–702.
- Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J Mol Biol. 1991;222(2):311–333.
- Guntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378.
- Brown LR, Bosch C, Wuthrich K. Location and orientation relative to the micelle surface for glucagon in mixed micelles with dodecylphosphocholine: EPR and NMR studies. Biochim. Biophys. Acta. 1981;642(2):296–312.
- Hancock RE, Rozek A. Role of membranes in the activities of antimicrobial cationic peptides. FEMS Microbiol Lett. 2002;206(2):143–149.
- Shai Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by alpha-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim Biophys Acta. 1999;1462(1–2):55–70.
- Pristov KE, Ghannoum MA. Resistance of Candida to azoles and echinocandins worldwide. Clin Microbiol Infect. 2019;25(7):792–798.
- Liu S, Hou Y, Chen X, Gao Y, Li H, Sun S. Combination of fluconazole with non-antifungal agents: a promising approach to cope with resistant Candida albicans infections and insight into new antifungal agent discovery. Int J Antimicrob Agents. 2014;43(5):395–402.