Abstract
Ceramide has a key role in the regulation of cellular senescence and apoptosis. As Ceramide levels are lowered by the action of acid ceramidase (AC), abnormally expressed in various cancers, the identification of AC inhibitors has attracted increasing interest. However, this finding has been mainly hampered by the lack of formats suitable for the screening of large libraries. We have overcome this drawback by adapting a fluorogenic assay to a 384-well plate format. The performance of this optimised platform has been proven by the screening a library of 4100 compounds. Our results show that the miniaturised platform is well suited for screening purposes and it led to the identification of several hits, that belong to different chemical classes and display potency ranges of 2–25 µM. The inhibitors also show selectivity over neutral ceramidase and retain activity in cells and can therefore serve as a basis for further chemical optimisation.
Introduction
Sphingolipids (SLs) are a major class of cellular lipids. Besides playing a structural role in cellular membranes, several members of the SLs family are also involved in the regulation of a variety of cellular processes. The metabolic hub in SLs biosynthesis and catabolism is ceramide, a bioactive lipid intimately involved in the regulation of stress responseCitation1, inflammationCitation2, apoptosisCitation3 and cancer cell deathCitation4.
In the recent years, there is more and more evidence that maintaining a tight regulation of ceramide levels is key to cells and strongly contributes to cell fate decisions. Moreover, altered ceramide levels are a hallmark in the manifestation of several pathological processes such as Alzheimer diseaseCitation5, metabolic disordersCitation6 or cancerCitation7, in which lower levels of this lipid are inversely correlated with the degree of malignant progressionCitation8. Consequently, tremendous efforts have been devoted to identifying small molecules targeting the enzymes involved in ceramide biosynthesis and degradation.
Ceramide can be generated de novo from serine and palmitate, by the degradation of sphingomyelin catalysed by sphingomyelinases and by the acylation of sphingosine in the salvage pathway. Ceramide degradation is in turn mediated by the actions of different ceramidases that are distinguished by the pH required for optimal activity, i.e. acid ceramidase (AC, ASAH1), neutral ceramidase (NC, ASAH2) and alkaline ceramidases 1, 2 and 3 (ACER1, ACER2 and ACER3)Citation9. Different functions and cellular roles, probably defined by their intracellular localisation and substrate specificity, have been suggested for these ceramidases. Hence, NC overexpression has been related to colon carcinogenesisCitation10, whereas ACER3 has been reported to contribute to hepatocellular carcinomaCitation11 and to acute myeloid leukaemia pathogenesisCitation12.
AC is one of the better-characterized ceramidases and its role in cancer initiation and progression has been largely studied. Abnormally elevated AC expression has been reported in various type of cancer including prostate cancerCitation13, colon adenocarcinomaCitation14, head and neck cancerCitation15, glioblastomaCitation16 and melanomaCitation17. Moreover, whereas AC overexpression renders the cells more resistant to chemo and radiotherapyCitation18, inhibition of the enzyme sensitises the cell to treatmentCitation19, thereby suggesting a role of AC in drug resistance associated to therapy. As a result, AC inhibition has emerged as an attractive target to improve the efficacy and lower the resistance to cancer treatments, and the identification of novel and selective AC inhibitors has gained increasing interest. Tremendous efforts have been done during the last two decades to develop AC modulators. However, most of the reported inhibitors are structurally related to ceramide, which has a negative impact on their selectivity, potency and drug-like propertiesCitation20. Thus, the discovery of ceramide-unrelated hits would be highly desirable. Some potent and structurally unrelated inhibitors of AC have already been described. Representative examples of this class of compounds are carmofur, identified after the screening of a commercial library using a LC/MS-based assayCitation14, and the related dioxypyrimidine and benzoxazolone carboxamidesCitation21–23. Despite these relevant examples, there is still a great need for the identification of novel molecules that can expand the toolbox for AC inhibitors.
One of the best alternatives to identify structurally diverse inhibitors is through the screening of large compound libraries. However, this approach requires a powerful, robust and cost-effective HTS assay, allowing the rapid and reliable testing of large number of compounds. Recently, we described a flow-cytometry-based assay that uses a deoxyceramide analog to monitor AC activity in intact cells. This assay could be potentially useful in the future for screening purposes, but it would require proper optimisation and the use of high-throughput flow cytometry platformsCitation24. Herein, we report a fluorescence-based AC assay that has been adapted to a 384-well format. Once validated, it has been employed to evaluate a 4100 compound library leading to the identification of novel compound classes targeting AC activity. Remarkably, the identified inhibitors also show selectivity over NC and retain activity in cellular studies.
Results and discussion
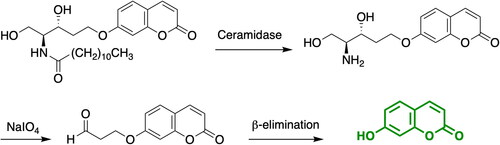
The screening for specific and potent AC inhibitors requires the availability of a high-throughput screening assay (HTS) capable of examining relatively large number of compounds simultaneously. A fluorescent-based assay in a 96-well format was previously set up in our group and applied to measure acid ceramidase activity using cell lysates and intact cells as a source of enzyme. The assay is based on the coumarinic substrate RBM14-C12Citation25, that shows a high affinity and specificity for AC over neutral and alkaline ceramidases. Briefly, hydrolysis of the amide bond of RBM14-C12 by AC yields an aminodiol that can be then oxidised upon treatment with sodium periodate. The resulting aldehyde undergoes a β-elimination reaction to release the fluorescent product umbelliferone (Scheme 1). The assay has been largely used by our group and others to identify novel ceramidase inhibitorsCitation21,Citation26,Citation27. However, assays with increased capacity are required to allow the rapid high-throughput screening (HTS) of large chemical libraries, thereby accelerating the drug discovery process as well as reducing the associated costs. Remarkably, recent advances have been performed in this area of research. Thus, a HTS screening assay for the identification of neutral ceramidase inhibitors was recently established by Spicer et al. using an analogous substrate RBM14-C16, developed in our groupCitation28. Moreover, Granier et al. synthesised a suite of doubly fluorophore-modified ceramides as turn-on probes for the direct FRET-based analysis of ceramidase activity in real-time. Although ACER3 was able to hydrolyse one of the probes, no synthetic substrates were hydrolysed by ACCitation29. Therefore, although the Granier’s method has the advantage of monitoring ceramidase activity in real-time, it cannot be applied to AC until suitable FRET ceramide substrates are discovered for this enzyme. Herein, we report the miniaturisation of an AC assay to a 384-well format and we employ it for the identification of new acid ceramidase inhibitors. The robustness and assay performance of the miniaturised assay were validated and assessed by the screening of a 4100 compound library, leading to the identification of novel AC inhibitors ().
Optimisation and miniaturisation of the AC HTS assay
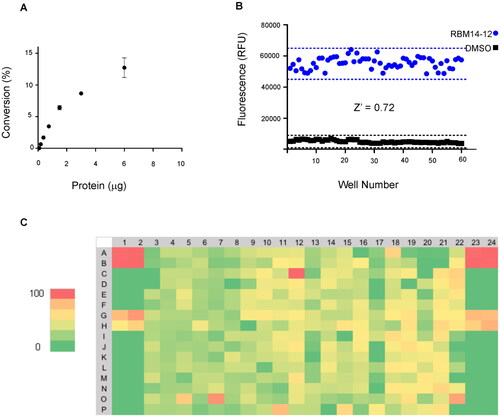
To start with, the final reaction volume was adapted from 100 µL (96-well-format) to 32 µL (384 well-format) with a final concentration of substrate of 20 µM. Next, protein concentration was adjusted using cell lysates of AC-overexpressing A375 melanoma cells. Ideally, the optimal amount of protein is the one that ensures reaction linearity over a period of time together with less than 10% of substrate depletion, in order to assume that the enzyme operates at steady-state conditions. Thus, different amounts of cell lysates were mixed with the substrate and product formation was measured over 1 h. displays the reaction progress curve for the hydrolysis of RBM14-C12 by AC obtained at a range of protein concentrations. Finally, an amount of 0.4 µg of protein was chosen, as higher amounts of proteins were not in the linear portion whereas lower amounts would compromise the signal window. The optimal incubation time of the reaction mixture was next explored. Although a higher signal was witnessed at 2 h reaction time, a final reaction time of 1 h was chosen since it exhibited an excellent signal window. As a first validation step, we performed a control run consisting of 60 wells representing AC activity and 60 wells representing the signal in absence of the enzyme. depicts the results and reveals excellent separation between high and low control wells resulting in a signal-to-background ratio >8 and a Z ’ factor of 0.72. The calculated coefficients of variation (CV) for both the high and low controls were 6.08% and 17.1% respectively. These data demonstrated that the assay was very robust, stable and possessed minimal well-to-well variability and therefore it is appropriate for HTS to identify AC inhibitors.
Figure 2. (A) Percentage of substrate conversion obtained using different amounts of AC (measured as μg of total protein). (B) Determination of the Z’-factor (Z’) of the AC assay in the 384-well plate format. Solid black squares and solid blue circles represent negative (without protein) and positive reaction, respectively. (C) Representative results for one 384-well plate. Each plate contained 8 negative controls, 16 positive controls and 8 samples with the known AC inhibitor SOCLAC. A colour gradient heat map, with red to green colours indicating high to low percentage of inhibition has been applied to the well values.
Screening of a compound library
After having successfully established the optimal reaction conditions in a 384-well format, we employed this newly miniaturised AC assay in a pilot screen against a library of 4100 compounds. The screen was carried out at a single dose of 20 µM for each library compound in 6% DMSO (v/v) and in duplicate. The acid ceramidase inhibitor SOCLAC was used as a pharmacological control for the assayCitation26. The Z’-factor per plate were consistent with those obtained during the initial validation. A total of 116 compounds of the 4100 screened at 20 µM met the active criterion, based on the hit cut-off at % inhibition >40, in the single-point primary screen, what resulted in a hit rate of 2.8%. An example hit map from one screening plate is shown (). The inhibitory activity of these hits was then validated in an additional assay performed at a single-point concentration in triplicates.
To confirm the results from the primary screening, top hits were cherry-picked and tested in a concentration-dependent response assay with triplicates per sample. Thus, 101 compounds were then subjected to 8-points two-fold dilution series starting at a maximum concentration of 200 micromolar. Among them, nine exhibited dose-dependent inhibition of AC with IC50 values in the range of 6 and 49 µM (, ), whereas 92 compounds showed a higher IC50 value or did not yield a concentration-dependent inhibition (structures not disclosed).
Figure 3. Structure of the identified inhibitors and their concentration-response curves. Active compounds were dose-tritiated to establish the corresponding IC50 values (N = 3 well per replicate point, errors bars are shown).
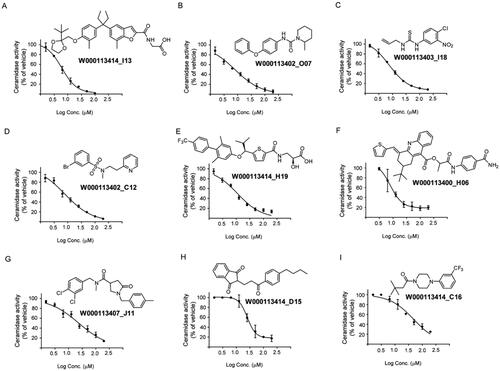
Table 1. The hit molecules identified through the primary assay performed at a single-point concentration of 20 μM were dose-titrated to establish their IC50 values (95% confidence interval).
Inhibition of neutral ceramidase
As mentioned above, hydrolysis of ceramides occurs by the action of ceramidases which are encoded by five known genes and are distinguished by the pH required for optimal activityCitation9. A common problem with AC inhibitors, especially those one with a scaffold related to the structure of ceramide, is the lack of selectivity over other ceramidases. Thus, to discard a selectivity issue, the activity of nine detected hits against neutral ceramidase was next explored at 50 µM. Activity over NC was tested using recombinant human NC (rhNC) and a specific NC substrate bearing a nervonic acid amide (RBM14-C24:1)Citation30. Remarkably, none of the molecules elicited a significant inhibitory activity, thereby confirming the selective inhibition of AC over NC (Supplemental Table 1).
Cellular inhibition of acid ceramidase
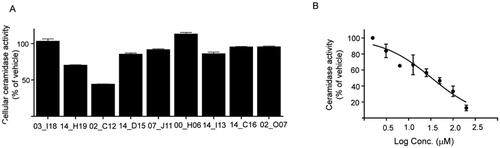
Cell-based assays allow evaluating biological activity in a more physiologically relevant system that also considers additional factors that might have a positive impact on inhibitory potency such as permeabilization through cellular membranes, metabolisation or concentration by intracellular compartmentalisation. Thus, the activity of the nine selected hits was next examined in intact cells using AC overexpressing A375 cells and the fluorogenic substrate RBM14-C12. First, compounds were tested in a primary screening assay. Measurements were performed in triplicated at a single-point concentration of 20 µM. The results showed that only two of the 9 tested compounds exhibited a significant inhibition of AC activity at this concentration: W000113402_C12 with a 53% inhibition and W000113414_H19 with a weaker 32% inhibition, whereas slight effects were observed for the other hits (). Dose-dependent inhibition could be also confirmed for W000113402_C12 in the cell-based assay displaying an IC50 of value of 32 µM (24.3–44.7) ().
Figure 4. (A) Cellular validation of hit compounds on acid ceramidase. The hit molecules identified during the primary validation were investigated using a cellular assay. Values shown are mean of three replicates and results are expressed as percentage of activity compared to vehicle control. Compounds are identified with the last 5 letters/numbers of their unique code (B) Concentration-response curve of the most potent hit molecule W000113402_C12 in a cellular assay.
Summary and conclusions
HTS is still the primary hit-finding strategy both in academia and industry. However, the identification of novel AC inhibitors has been hampered by the unavailability of appropriate screening platforms. Herein, we report a robust and cost-effective assay for the determination of AC activity that enables the rapid profile of large compound libraries. The screening platform has been employed to evaluate a 4100 compound library leading to the identification of 9 novel compound classes targeting AC activity with low micromolar IC50. Dose-dependent inhibition was confirmed for the primary hits identified in the screening campaign and now they can serve as a basis for hit-to-lead optimisation through chemical modifications, thereby opening new venues in the field of AC inhibition. Moreover, the reported technique can be considered an attractive drug-screening platform with a great potential to identify novel AC modulators. To further validate hits obtained in HTS campaigns of large libraries, an orthogonal assay using a different detection method (e.g. C12-Ceramide Bodipy and HPLC-based detection of substrate and reaction product) could be applied to discard potential interference of compounds on the fluorescence-based assayCitation31. As several diseases are linked to altered AC activity, novel compounds modulating its activity should allow progress in drug discovery and expand our knowledge in the essential role of this enzyme.
Materials and methods
Compound library
A library containing 4100 compounds was obtained from Eli Lilly. Compounds were distributed in triplicates in 384-well microtiter plates at a 10 mM concentration in DMSO (0.4 µL). Compounds were identified with a unique code. Plates were stored at −20 °C until use. Immediately prior to use, plates were withdrawn from −20 °C storage, thawed an ambient temperature and centrifuged.
Cell culture
The A375 cell line stably overexpressing ASAH1 under the control of a tetracycline/doxycycline-responsive promoter was kindly provided by Dr. Carmen Bedia and Prof. Thierry LevadeCitation17. The antibiotic selection of this cell line was performed with blasticidin (3 µg/mL) and hygromycin B (250 µg/mL). Ectopic expression of AC was induced with doxycycline at 1 µg/mL for 24 h before use. Cells were suspended in the appropriate volume of a 0.25 M saccharose solution with the proteases inhibitors aprotinin (1 mg/mL), leupeptin (1 mg/mmL) and PMSF (100 mM). The suspension was submitted to three cycles of a 5 s sonication (probe) at 10 watts/5 s resting on ice. The cell lysate was centrifuged at 600 g for 5 min. The supernatant was collected and protein concentration was determined with BSA as a standard using the bicinchoninic acid (BCA) protein determination kit (Thermo Scientific) according to the manufacturer’s instructions.
Acid ceramidase HTS assay
A previously described 96-well plate assayCitation25 was miniaturised into a 384-well plate format with a final reaction volume of 32 µL. Plated compounds were diluted with a mixture of DMSO/H2O (1.6 µL/18 µL), and after centrifugation, 3.2 µL were dispensed into a new 384-well plate, so that the final concentration of the compound in the final reaction volume of 32 µL was 20 µM and the DMSO content of the assay was 1%. Next, 20.8 µL of a substrate solution of RBM14-C12 in sodium acetate buffer (25 mM, pH 4.5) was added for a final concentration of 20 µM, followed by the addition of 8 µL of a 0.25 M sucrose solution of cell lysates from AC-overexpressing A375 melanoma cells containing 0.4 µg of protein The reaction was terminated after 60 min incubation at 37 °C by adding 8 µL of methanol. Oxidation was performed by treatment with 32 µL of a [2.5 mg/mL] solution of NaIO4 in 100 mM glycine-NaOH buffer (pH 10.6). The plates were incubated at 37 °C in the dark for another 1 h. Finally, 32 µL of 100 mM glycine-NaOH buffer (pH 10.6) were added and fluorescence was measured spectrophotometrically at excitation and emission wavelength of 355 and 460 nm, respectively. Blank reactions contained the same constituents as the test reactions except the cell lysates.
Neutral ceramidase assay
The NC assay was performed in 96-well plates at a final volume of 100 µL/well. Reaction buffers was 25 mM phosphate buffer 150 mM NaCl 1% (NaChol) pH 7.4. The reaction mixtures contained 25 µL/well of protein (5 ng recombinant NC R&D Systems, >95% pure), 70 µL/well of substrate (prepared from 4 mM stock solutions in ethanol) and 5 µL/well of inhibitor (prepared from 1 mM stock solutions in DMSO/H2O). Reaction mixtures were incubated at 37 °C for 1 h and reactions were stopped with 25 µL/well of MeOH followed by 100 µL/well of NaIO4 (2.5 mg/mL in 100 mM glycine-NaOH buffer, pH 10.6). After incubation at 37 °C for 1 h in the dark, 100 µL/well of 100 mM glycine-NaOH buffer (pH 10.6) was added and fluorescence was measured spectrophotometrically at excitation and emission wavelenght of 355 and 460 nm, respectively. The same reaction mixtures without enzymes were used as blanks.
Fluorogenic ceramidase activity assay in intact cells
To determine activity in intact cells, 2 × 104 cells/well were seeded in 96-well plates 24 h prior to the assay and maintained at 37 °C and 5% CO2. Overexpression of AC was induced with doxycycline at 1 µg/mL for 24 h. Medium was replaced by 100 µL of fresh medium (DMEM 10% FBS) containing 20 µM of the substrate and different concentrations of the indicated test compounds. Both substrates and test compounds were added simultaneously to the cell culture. The plate was incubated for 3 h at 37 °C in 5% CO2. The reaction was stopped with 25 µL/well of MeOH and then 100 µL/well of NaIO4 (2.5 mg/mL in glycine-NaOH buffer, pH 10.6) were added. After incubation at 37 °C for 1 h in the dark, 100 µL/well of 100 mM glycine-NaOH buffer (pH 10.6) were added and fluorescence was measured spectrophotometrically at excitation and emission wavelengths of 355 and 460 nm, respectively. The same reaction mixtures without cells were used as blanks.
Supplemental Material
Download PDF (63.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hannun YA, Obeid LM. The ceramide -centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J Biol Chem 2002;277:25847–50.
- MacEyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature 2014;510:58–67.
- Obeid LM, Linardic CM, Karolak L, Hannun YA. Programmed cell-death is mediated by ceramide. Clin Res 1993;41:A240.
- Nganga R, Oleinik N, Ogretmen B. Mechanisms of ceramide-dependent cancer cell death. Adv Cancer Res 2018;140:1–25.
- Filippov V, Song MA, Zhang K, et al. Increased ceramide in brains with Alzheimer’s and other neurodegenerative diseases. J Alzheimers Dis 2012;29:537–47.
- Turpin-Nolan SM, Brüning JC. The role of ceramides in metabolic disorders: when size and localization matters. Nat Rev Endocrinol 2020;16:224–33.
- Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer 2004;4:604–16.
- Riboni L, Campanella R, Bassi R, et al. Ceramide levels are inversely associated with malignant progression of human glial tumors. Glia 2002;39(2):105–13.
- Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta Mol Cell Biol Lipids 2008;1781:424–34.
- García-Barros M, Coant N, Kawamori T, et al. Role of neutral ceramidase in colon cancer. FASEB J 2016;30:4159–71.
- Yin Y, Xu M, Gao J, Li M. Alkaline ceramidase 3 promotes growth of hepatocellular carcinoma cells via regulating S1P/S1PR2/PI3K/AKT signaling. Pathol Res Pract 2018;214:1381–7.
- Chen C, Yin Y, Li C, et al. ACER3 supports development of acute myeloid leukemia. Biochem Biophys Res Commun 2016;478:33–8.
- Saad AF, Meacham WD, Bai A, et al. The functional effects of acid ceramidase overexpression in prostate cancer progression and resistance to chemotherapy. Cancer Biol Ther 2007;6:1455–60.
- Realini N, Solorzano C, Pagliuca C, et al. Discovery of highly potent acid ceramidase inhibitors with in vitro tumor chemosensitizing activity. Sci Rep 2013;3:1035.
- Mehta S, Blackinton D, Omar I, et al. Combined cytotoxic action of paclitaxel and ceramide against the human Tu138 head and neck squamous carcinoma cell line. Cancer Chemother Pharmacol 2000;46:85–92.
- Doan NB, Alhajala H, Al-Gizawiy MM, et al. Acid ceramidase and its inhibitors: a de novo drug target and a new class of drugs for killing glioblastoma cancer stem cells with high efficiency. Oncotarget 2017;8:112662–74.
- Bedia C, Casas J, Andrieu-Abadie N, et al. Acid ceramidase expression modulates the sensitivity of A375 melanoma cells to dacarbazine. J Biol Chem 2011;286:28200–9.
- Mahdy AEM, Cheng JC, Li J, et al. Acid ceramidase upregulation in prostate cancer cells confers resistance to radiation: AC inhibition, a potential radiosensitizer. Mol Ther 2009;17:430–8.
- Gouazé-Andersson V, Flowers M, Karimi R, et al. Inhibition of acid ceramidase by a 2-substituted aminoethanol amide synergistically sensitizes prostate cancer cells to N-(4-hydroxyphenyl) retinamide. Prostate 2011;71:1064–73.
- Saied EM, Arenz C. Inhibitors of ceramidases. Chem Phys Lipids 2016;197:60–68.
- Pizzirani D, Bach A, Realini N, et al. Benzoxazolone carboxamides: potent and systemically active inhibitors of intracellular acid ceramidase. Angew Chemie - Int Ed. 2015;54:485–9.
- Bach A, Pizzirani D, Realini N, et al. Benzoxazolone carboxamides as potent acid ceramidase inhibitors: synthesis and structure-activity relationship (SAR) studies. J Med Chem. 2015;58:9258–72.
- Di Martino S, Tardia P, Cilibrasi V, et al. Lead optimization of benzoxazolone carboxamides as orally bioavailable and CNS penetrant acid ceramidase inhibitors. J Med Chem 2020;63:3634–64.
- Casasampere M, Izquierdo E, Casas J, et al. Click and count: specific detection of acid ceramidase activity in live cells. Chem Sci 2020;11:13044–51.
- Bedia C, Casas J, Garcia V, et al. Synthesis of a novel ceramide analogue and its use in a high-throughput fluorogenic assay for ceramidases. Chembiochem 2007;8:642–8.
- Bielsa N, Casasampere M, Aseeri M, et al. Discovery of deoxyceramide analogs as highly selective ACER3 inhibitors in live cells. Eur J Med Chem 2021;216:113296.
- Ordóñez YF, Abad JL, Aseeri M, et al. Activity-based imaging of acid ceramidase in living cells. J Am Chem Soc 2019;141:7736–42.
- Otsuka Y, Airola MV, Choi YM, et al. Identification of small-molecule inhibitors of neutral ceramidase (NCDase) via target-based high-throughput screening. SLAS Discov 2021;26:113–21.
- Healey RD, Saied EM, Cong X, et al. Discovery and mechanism of action of small molecule inhibitors of ceramidases**. Angew Chemie Int Ed 2022;61:e202109967.
- Casasampere M, Bielsa N, Riba D, et al. New fluorogenic probes for neutral and alkaline ceramidases. J Lipid Res 2019;60:1174–81.
- He X, Li C, Park J, Dagan A, et al. A fluorescent-based high performance liquid chromarographic assay to determine acid ceramidase activity. Anal Biochem 1999;274:264–9.