Abstract
High aldehyde dehydrogenase (ALDH) activity is a metabolic feature of adult stem cells and various cancer stem cells (CSCs). The ALDEFLUOR system is currently the most commonly used method for evaluating ALDH enzyme activity in viable cells. This system is applied extensively in the isolation of normal stem cells and CSCs from heterogeneous cell populations. For many years, ALDH1A1 has been considered the most important subtype among the 19 ALDH family members in determining ALDEFLUOR activity. However, in recent years, studies of many types of normal and tumour tissues have demonstrated that other ALDH subtypes can also significantly influence ALDEFLUOR activity. In this article, we briefly review the relationships between various members of the ALDH family and ALDEFLUOR activity. The clinical significance of these ALDH isoforms in different cancers and possible directions for future studies are also summarised.
Introduction
The aldehyde dehydrogenase (ALDH) superfamily is the most important aldehyde-metabolizing enzyme system in the body and belongs to the group of NAD(P)+-dependent enzymes. The human ALDH superfamily includes 11 familiesCitation1,Citation2 and a total of 19 members: ALDH1s (1A1, 1A2, 1A3, 1B1, 1L1, and 1L2), ALDH2, ALDH3s (3A1, 3A2, 3B1, and 3B2), ALDH4A1, ALDH5A1, ALDH6A1, ALDH7A1, ALDH8A1, ALDH9A1, ALDH16A1, and ALDH18A1Citation1,Citation3. The genes encoding these enzymes are located on different chromosomal segments and are named based on their evolutionary relationships and on the amino acid homologies of the encoding proteinsCitation4. All enzyme subtypes in the ALDH family have similar structures, and most are homodimers or homotetramers. Each monomer contains 3 structural domains: a catalytic domain, a cofactor-binding domain, and an oligomerization domain. This family of enzymes plays important roles in embryo formation, development, cell proliferation, and differentiationCitation3. Its main functions include the followings: (1) Participation in retinoic acid (RA) synthesis: ALDH1As and ALDH8A1 participate in the second step of RA synthesis, in which all-trans or 9-cis retinal is oxidised to produce RACitation5. ALDH1A1 mainly oxidises all-trans and 9-cis retinal into RACitation6,Citation7, and ALDH1A3 mainly oxidises all-trans retinalCitation5,Citation8. The catalytic effects of ALDH1A1 on these 2 reactions are equivalentCitation6,Citation7; ALDH1A2 displays higher catalytic efficiency for all-trans retinalCitation9, and ALDH8A1 almost exclusively catalyses 9-cis retinalCitation5,Citation10,Citation11. (2) Detoxification: ALDHs can irreversibly oxidise endogenous and exogenous aldehydes into the corresponding acids with the participation of NADP+, thereby performing detoxification functions and helping maintain normal cell homeostasis and body functionsCitation4,Citation12. (3) Participation in resistance to chemotherapeutic drugs (especially drugs derived from aldehyde intermediates): ALDH oxidises cyclophosphamide intermediate products into non-toxic carboxyphosphamide, thereby increasing the resistance of tumour cells to cyclophosphamideCitation13,Citation14. (4) ALDH4A1 can also reduce cellular levels of reactive oxygen species (ROS), thereby protecting the cells from ROS injuryCitation15.
The ALDH family participates in physiological processes mainly involved in the development of normal tissues and in cell metabolismCitation16. For instance, ALDH1A1 is the most studied subtype and is extensively expressed in normal human tissues, including the brain, thyroid gland, lung, mammary gland, liver, pancreas, kidney, colon, bone, prostate, ovary, and cervix. Notably, ALDH1A1 expression was found to be closely associated with the occurrence, development, and therapeutic resistance of tumours and was considered a marker of cancer stem cells (CSCs)Citation16. Consequently, targeting CSCs through interference with ALDHs is considered a novel strategy for the chemical prevention and treatment of tumours.
History of ALDH activity detection
Traditional methods for the detection of ALDH subtypes include western blotting, reverse transcription–polymerase chain reaction (RT–PCR), spectrophotometry for the detection of enzyme activities, and immunohistochemistry (IHC)Citation17. However, none of the above methods can be used to measure ALDH activity in viable cellsCitation18,Citation19. By applying the ALDEFLUOR system, in which BODIPY-aminoacetaldehyde (BAAA) was used as the ALDH substrate and N,N-diethylaminobenzaldehyde (DEAB) as a negative control, researchers successfully isolated SSClo/ALDHbr cells from human umbilical cord blood, and 40–90% of the isolated cells were CD34+CD38lo/– haematopoietic stem cells (HSCs)Citation20. The ALDEFLUOR system has the following advantages: (1) it can detect human stem/progenitor cells of multiple lineages, including haematopoietic, mammary gland, endothelial, and neural stem/progenitor cells as well as mesenchymal stem cells; (2) it is applicable to the isolation of stem/progenitor cells of other species and human CSCs; and (3) it can be used with cryopreserved cell samples. Thus, ALDEFLUOR has become the most commonly used method for detecting ALDH enzyme activity in viable cells and for the isolation of stem/progenitor cells (http://www.stemcell.com).
Using ALDEFLUOR as a stem cell marker
ALDEFLUOR was originally used for the identification and sorting of haematopoietic stem/progenitor cellsCitation20,Citation21. Subsequently, this method was used to label stem cells in other normal tissues, including neural stem cells, adipose-derived adult stem cellsCitation22, and muscle precursor cellsCitation23. In addition, ALDEFLUOR was gradually extended to the isolation of stem cells from acute myeloid leukaemia (AML) and solid tumours, including breast cancerCitation24, oral squamous cell carcinoma (OSCC)Citation25, sarcomaCitation26, oesophageal squamous cell carcinomaCitation27,Citation28, oesophageal adenocarcinomaCitation29, gastric cancerCitation30,Citation31, colorectal cancerCitation32–34, head and neck squamous cell carcinomaCitation35, thyroid cancerCitation36, lung cancerCitation37–39, cholangiocarcinoma (CHOL)Citation40, hepatocellular carcinomaCitation41,Citation42, pancreatic cancerCitation43–45, osteosarcomaCitation46–48, prostate cancerCitation49,Citation50, bladder cancerCitation51–53, glioblastomaCitation54, melanomaCitation55, ovarian cancerCitation56–58, cervical cancerCitation59,Citation60 and multiple myelomaCitation61. Therefore, the ALDEFLUOR technique has become an important tool in CSCs research.
Pending controversy
Although the ALDEFLUOR detection and sorting system has been extensively applied in the field of stem cell/CSCs research, some important scientific questions remain to be answered. The most striking question is which enzyme or group of enzymes among the 19 members of the ALDH family determines the ALDEFLUOR-positive rate. Initially, the activity and positive rate of ALDEFLUOR were thought to be primarily determined by ALDH1A1Citation62. However, 9 of the 19 ALDH isoforms (ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B1, ALDH2, ALDH3A1, ALDH3A2, ALDH3B1, and ALDH5A1) were potentially involved in ALDEFLUOR activityCitation63. The results also implied that DEAB inhibits various ALDH isoforms to different extents except for ALDH3B1 and ALDH5A1Citation63,Citation64. Furthermore, the results of in-depth studies suggested that ALDH isoforms exhibited differential expression patterns in various cancer typesCitation63,Citation65,Citation66.
Haematopoietic system
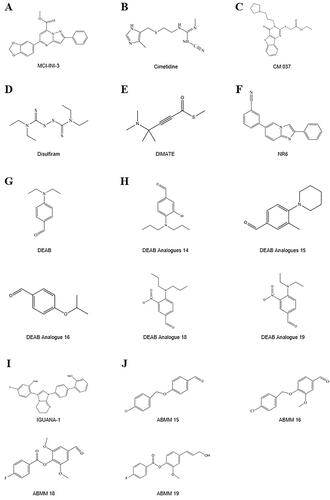
Accumulating evidence has indicated that ALDH might be a potential marker for HSCs and involved in leukaemia developmentCitation67. ALDH1A1 was found to be most highly expressed in both murine and human HSCs and immature progenitorsCitation68. Moreover, studies have demonstrated that ALDH1A1 and ALDH3A1 are involved in the metabolism of ROS and reactive aldehydes in HSCsCitation69,Citation70, which may play important roles in HSC biology and leukaemia transformationCitation67. With both ALDH1A1 and ALDH3A1 deletion, NUP98-HOXA10 homeodomain fusion protein can promote the development of leukaemia with B220+ and varied levels of CD11b, while NUP98-HOXA10 homeodomain fusion protein alone induces only in vitro expansion of HSCs without malignant transformationCitation71, suggesting the important role of ALDH1A1 and ALDH3A1 in leukaemia initiationCitation67,Citation72. Additional evidence indicated that ALDH1A1 expression induced by cytokines in bone marrow cells can result in increased resistance to 4-hydroperoxycyclophosphamide (4-HC) in AMLCitation73, and suppression of ALDH1A1 can sensitise leukaemia cells to 4-HCCitation74. Notably, inhibition of ALDH1A1 with all-trans RA was found to be a promising approach in AML by inducing the differentiation of leukaemia stem cells (LSCs)Citation75. Disulfiram an inhibitor of ALDH1A1 (), may offer a new treatment strategy by selectively eradicating LSCs through the simultaneous induction of ROS-JNK and inhibition of NF-kB and Nrf2Citation76,Citation77 and overcoming bortezomib and cytarabine resistance by inducing apoptosis and proteasome inhibition in AMLCitation78. The ALDH1A1 inhibitor dimethyl ampal thiolester (DIMATE) was also shown to be a drug candidate by specifically targeting LSCs in AMLCitation79 (). In a study of multiple myeloma, ALDH + cells isolated by ALDEFLUOR were found to have stem cell-like characteristics. With overexpression of ALDH1A1 and ALDH2 in NCI-H929 cells, although ALDH activity detected by ALDEFLUOR assay is associated with the expression of both ALDH1A1 and ALDH2, only ALDH1A1 plays a pivotal role in maintaining the stemness of ALDH+cellsCitation61. Taken together, these findings indicated the therapeutic potential of targeting ALDH1A1 in the clinical management of AML patientsCitation70.
Previous studies have indicated that ALDEFLUOR+ cells account for approximately 1% of the total number of cells in bone marrow, mobilised peripheral blood cells, and umbilical cord bloodCitation80,Citation81. These cells displayed the characteristics of HSCs, and the major enzyme subtype that determined the ALDEFLUOR-positive rate was ALDH1A1Citation81. However, recent studies have reported that ALDH1A1 is not the dominant expression subtype in mouse HSCs and that the expression levels of ALDH subtypes in mouse HSCs follow the order ALDH9A1 > ALDH2 > ALDH1A1 ≥ ALDH3A2 > ALDH1A7Citation62. In addition, upon knockout of ALDH1A1, ALDEFLUOR activity in mouse bone marrow stem cells and HSCs (CD150+CD41−CD48−Sca1+c-kit+) did not change significantly. However, if both ALDH1A1 and ALDH3A1 were knocked out in adult mouse bone marrow cells, the number of bone marrow stem cells and HSCs decreased significantlyCitation68,Citation69,Citation82. These results suggest that in addition to ALDH1A1, other ALDH subtypes, such as ALDH3A1, ALDH9A1, ADLH1A2, and ALDH7A1, might also influence ALDEFLUOR activity in HSCsCitation62.
Similarly, TaqMan low-density array analysis of ALDEFLUOR+ and ALDEFLUOR– populations of the K562 cell line reveal that in addition to the ALDH1 subfamily, K562 ALDEFLUOR+ cells also express the ALDH3, ALDH5, ALDH6, ALDH7, and ALDH8 subtypesCitation17.
Mammary gland
Recent efforts have suggested that ALDH1A1 is regulated at the posttranslational level by the Notch signalling pathway in breast cancer, which modulates ALDH1A1 acetylation through the induction of SIRT2 expression and therefore activates ALDH1A1 to promote tumorigenesis and tumour growthCitation83. High ALDH1A1 activity was associated with a poor prognosis for breast cancer patientsCitation24. Breast CSCs, which exhibit high ALDH1A1 activity and high CD44 expression, were shown to be more sensitive to chemotherapy or radiation after pre-incubation with DEABCitation84. In vitro and in vivo treatment with all-trans RA has been reported to induce differentiation and chemotherapy sensitivity of resistant and metastatic breast CSCs, which were selected as CD44+/CD24−/low/ALDEFLUOR+ cells after long-term low-dose fractionated radiation treatment in MCF7/C6 cellsCitation85. Moreover, evidence has demonstrated that NANOG signalling induces enhanced ALDH1A3 activity through activation of the Notch1 and AKT pathways, which in turn stimulates DNA double-strand break repair capacity and confers radio resistance to breast cancer cell linesCitation86. These results provide evidence for the selective suppression of ALDH1 isozymes (ALDH1A1 and ALDH1A3) as promising therapeutic targets in breast cancerCitation87.
ALDEFLUOR+ cells isolated from normal mammary epithelial cells accounted for approximately 8% of the total number of cells and displayed mammary stem cell characteristics, including sphere formation ability, colony formation ability, and multidirectional differentiationCitation24. IHC analysis further indicated that this ALDEFLUOR+ cell population mainly expressed ALDH1A1Citation24 and was distributed in luminal areas of the gland, consistent with the localisation and presumed quantity of stem cellsCitation88. In addition, ALDEFLUOR+ mammary epithelial cells participate in duct formation and can be differentiated into uncommitted, luminal epithelial, and myoepithelial cells, whereas almost all ALDEFLUOR– mammary epithelial cells produce luminal epithelial cellsCitation24. However, the results obtained by Rexer et al. indicated that ALDH1A3 determines the enzyme activity of ALDH in mouse mammary tissue and that it has a leading role in the synthesis of RACitation89. Furthermore, the normal mammary epithelial cell line MTSV1.7 expressed ALDH1A3 mRNA and did not express ALDH1A1 and ALDH1A2 mRNAsCitation89.
Viewpoints differ regarding the dominant ALDH subtype determining ALDEFLUOR activity in breast cancer. Deng et al. studied 15 breast cancer cell lines (BT-474, BT-483, BT-20, MDA-MB-468, SKBR-3, MDA-MB-231, MDA-MB-436, MDA-MB-453, MDA-MB-157, MCF7, T47D, BT-549, ZR75-1, HCC202, and HCC1428) and found that the mean ALDEFLUOR-positive rate was 3.5% and positively correlated with ALDH1A1 protein expressionCitation88. The IHC results of tissue arrays of 481 breast cancer samples indicated that 26% of the samples expressed ALDH1A1, and ALDH1A1+ breast cancer cells accounted for approximately 5% of the total cellsCitation24. IHC analysis of another group containing 69 breast cancer samples yielded similar results, showing that 20.3% (14/69) of the samples expressed ALDH1A1, and breast cancer cells strongly positive for ALDH1A1 represented only 4.3% of the 14 positive samplesCitation88. The above ALDH1A1 positivity rates are consistent with the fact that CSCs account for only a small fraction of cancer cellsCitation24. Therefore, the researchers concluded that ALDH1A1 was the dominant enzyme subtype in the ALDEFLUOR+ population. However, recent studies have suggested that in addition to ALDH1A1, other ALDH subtypes, especially ALDH1A3, might also determine the positive rate of ALDEFLUOR in breast cancerCitation66,Citation90. In breast cancer cell lines with high ALDEFLUOR activity, such as MDA-MB-468 cells, ALDH1A3 knockdown significantly decreased the ALDEFLUOR-positive rate, whereas ALDH1A1 knockdown did not have this effectCitation90. ALDH1A3 overexpression significantly increased ALDEFLUOR activity in MDA-MB-231 breast cancer cells, and the increase was greater than that caused by ALDH1A1 overexpressionCitation91. Marcato et al. analysed 4 cases of breast cancer samples and found that only 1 sample in the ALDEFLUOR+ subpopulation displayed high ALDH1A1 expressionCitation90. Correlation analysis of ALDH subtype expression with the ALDEFLUOR-positive rate in 7 breast cancer cell lines revealed higher ALDEFLUOR-positive rates in MDA-MB-468, SKBR3, and MDA-MB-435 cellsCitation90. ALDH1A1 knockdown in MAD-MB-435 cells only slightly affected the ALDEFLUOR-positive rate, whereas ALDH1A3 knockdown triggered a significant decrease (from 84.7% ± 0.95% to 36.9% ± 18.36%)Citation90. The above results indicate that the ALDEFLUOR activity of some breast cancer cell lines and specimens from breast cancer patients may be mainly determined by ALDH1A3Citation90.
To determine which ALDH isoforms play a decisive role in ALDEFLUOR activity, Zhou et al.Citation63 cloned all 19 ALDH isoforms into lentiviral vectors and then established stable breast cancer SUM159 and MDA-MB-231 overexpression cell linesCitation63. These 2 cell lines had relatively low endogenous levels of most ALDH isoforms and a low percentage of ALDH+ in the ALDEFLUOR assayCitation63. As a result, 9 of the 19 ALDH isoforms may be involved in the level of ALDEFLUOR activity, including ALDH1A1, ALDH1A2, ALDH1A3, ALDH1B1, ALDH2, ALDH3A1, ALDH3A2, ALDH3B1, and ALDH5A1Citation63. Upon stable knockdown of ALDH1A1, ALDH3, and ALDH3A2 in BT-474 cells and ALDH1A3, ALDH3A2, and ALDH3B1 in the MCF10A cell line, the ALDH+ ratio was measuredCitation63. The knockdown of ALDH2 in BT-474 cells and ALDH1A3 and ALDH3A2 in MCF10A cells significantly reduced the ALDH+ ratio, indicating that ALDH2 is a major player in the ALDEFLUOR activity of BT-474 cells, while ALDH1A3 and ALDH3A2 play an important role in the ALDEFLUOR activity of MCF10A cellsCitation63.
Digestive tract
Oral mucosa
ALDEFLUOR activity in oral mucosa keratinocytes is primarily determined by ALDH1A3 and ALDH3A1Citation92. The ALDEFLUOR-positive rate in normal oral mucosa cells is 14.1–17.6% (17.6% ± 7.4% in cells of fresh specimens and 14.1% ± 7.4% in cultured cells). IHC analysis revealed that the mucosal layer expressed ALDH1A3 and ALDH3A1 but not ALDH1A1Citation92. ALDH1A3 expression gradually increased from the lower suprabasal layer to the upper suprabasal layer. ALDH3A1 is mainly expressed in the cytoplasm in the lower suprabasal layer cells and the nuclei in basal layer cellsCitation92. In situ hybridisation of mRNA confirmed the expression patterns of ALDH1A3 and ALDH3A1 in the oral mucosaCitation92. Inhibition of ALDH1A3 or ALDH3A1 expression significantly decreased the ALDEFLUOR-positive rate, and the effect of ALDH1A3 intervention was more robustCitation92. These results indicated that the ALDEFLUOR-positive rate in oral mucosa keratinocytes was determined by both ALDH1A3 and ALDH3A1 but predominantly by ALDH1A3Citation92. Hedberg et al. reached a similar conclusion by analysing the gene expression profiles of cultured primary keratinocytes, the immortalised cell line SVpgC2a, and the buccal carcinoma cell line SqCC/Y1. The results indicated that all 3 types of cells expressed ALDH1A3 but not ALDH1A1, and ALDH3A1 was only expressed in the buccal carcinoma cell lineCitation93. Furthermore, the other enzyme subtypes ALDH3A2, ALDH4A1, ALDH7A1, and ALDH9A1 also displayed differential expression in the 3 types of cellsCitation93.
Colorectal cancer
Most researchers consider ALDH1A1 to be the dominant subtype determining ALDEFLUOR activity in colorectal cancer cells, and this enzyme subtype may be used as a marker for colorectal CSCsCitation32,Citation33. The mean ALDEFLUOR-positive rate in 3 colorectal cancer cell lines (LoVo, BE, and HT-29) was 15.5%, and ALDH1A1 displayed high expression in the ALDEFLUOR-positive subpopulationCitation88. In addition, ALDEFLUOR+ cells isolated from colorectal cancer tissues had stem cell characteristics and could develop primary tumours in immunodeficient animals, while ALDEFLUOR– cells could notCitation33. IHC analysis indicated that colorectal cancer tissues had a positive rate of 94% (63/67) for ALDH1A1 expressionCitation88. Tumour cells with strong positive ALDH1A1 expression accounted for approximately 38.8% of the total cells and were localised in crypts, showing a spatial distribution similar to that of normal stem cellsCitation88. ALDH1A1 silencing decreased the tumorigenicity of HT-29 cells in athymic miceCitation34. Therefore, these findings indicated that ALDH1A1 can be used as a colorectal cancer stem/progenitor cell marker and that it is the major enzyme subtype that determines ALDEFLUOR activity.
However, studies have demonstrated that ALDH isoforms are different in different colorectal cancer cell lines. HT-29 cells mainly express ALDH1A1 isoforms, HCT-116 cells mainly express ALDH1A3 isoforms, and LS-180 cells express ALDH1A1 and ALDH2 isoformsCitation34.
Chen et al. reported that in addition to ALDH1A1, slim columnar cells in undifferentiated normal colon tissue also express a small amount of ALDH1B1Citation65. ALDH1B1 is predominantly expressed in cells near the sides of the crypt bottom and is sporadically expressed in cells on both sides of the upper portion of the crypt, while no expression is evident in differentiated cells or stromal cellsCitation65. This distribution is highly similar to that of stem cellsCitation65. A total of 97.5% of colonic adenocarcinoma samples expressed high levels of ALDH1B1, whereas only 36.6% of samples expressed ALDH1A1, which was comparatively low. Thus, ALDH1B1 may represent another marker for colorectal CSCs and the ALDEFLUOR activity-determining enzyme subtypeCitation65.
Pancreatic cancer
ALDEFLUOR+ cells account for approximately 3% of cells in pancreatic cancer and display CSCs characteristicsCitation43–45. Two groups of researchers have reported the measurement of ALDH1A1 expression in pancreatic cancer samples and showed similar positive rates of approximately 75%Citation94. In positive samples, ALDH1A1+ cells accounted for more than 50% of the total tumour cells in 47% of the samplesCitation94. Therefore, the researchers concluded that ALDH1A1 is the major ALDH subtype that determines ALDEFLUOR activity in pancreatic cancer samples.
While the ALDEFLUOR-positive rates in the pancreatic cancer cell lines CAPAN-1, DAN-G, Panc1, and L3.6pl are 2.4–8.5%Citation44, which is equivalent to those found in pancreatic cancer tissues, a comparison of 6 pancreatic cancer cell lines (BxPC3, T3M4, PANC1, SU8686, Colo-357, and AsPC-1) and 1 noncancerous pancreatic cell line (ACBRI) revealed a differential expression pattern for ALDH1A1 mRNACitation94. Of these, BxPC3, T3M4, and PANC1 showed little or no ALDH1A1 mRNA expression, and SU8686 and Colo-357 cells exhibited relatively low expression compared with ACBRI cellsCitation94. In addition, gene expression profiling of 3 pancreatic cancer cell lines indicated that AsPC-1 and BxPC3 cells primarily expressed ALDH1A3, whereas the expression of ALDH1A1 and ALDH1A3 in MIA PaCa cells was equivalentCitation95. These results indicate that ALDEFLUOR activity in pancreatic cancer cells may not be solely determined by ALDH1A1.
Lung cancer
The ALDEFLUOR-positive subpopulation derived from non-small cell lung cancer (NSCLC) cell lines was demonstrated to be highly clonogenic, tumorigenic, and invasive and displayed resistance to chemotherapeutic drugs. Moreover, ALDH1A1 expression was associated with poor prognosis in early-stage NSCLCCitation37. Knockdown of ALDH1A1 and ALDH3A1 can inhibit clonogenicity and motilityCitation96 and induce sensitivity to 4-HC in NSCLC cell linesCitation97. ALDH1A1 expression has been reported to be positively associated with Notch transcription, and pharmacologic and genetic interference with Notch can result in decreased ALDH1A1 activity in lung CSCsCitation98,Citation99. In addition, mechanistic investigations have identified that NFATc2/SOX2 coupling can upregulate ALDH1A1 expression by binding to its 5′ enhancer, attenuate oxidative stress induced by cancer drug treatment, and lead to increased resistance to chemotherapy and targeted therapyCitation100. The embryonic transcription factor SOX9 was reported to promote stem-like properties and induce chemo resistance of NSCLC cells by trans activating ALDH1A1 expressionCitation101. ALDH1A1 knockdown was suggested to inhibit the invasive ability and in vivo tumorigenicity of ALDEFLUOR+ cellsCitation38, indicating that ALDH1A1 was the major subtype determining ALDEFLUOR activity in lung cancer. However, other studies have reported that ALDH1A3 might be the major enzyme subtype that determines ALDEFLUOR activity. The ALDEFLUOR-positive rates in Calu-1, H358, H1993, H2009, H2087, HCC44, HCC95, and HCC827 cells were 1–15%, and ALDEFLUOR+ cells exhibited relatively high ALDH1A3 expression in all cell linesCitation102. ALDH1A3 knockdown decreased the ALDEFLUOR-positive rate and suppressed the colony formation ability and tumorigenicity of ALDEFLUOR+ cellsCitation102. Furthermore, upon knockdown of ALDH3A1 and ALDH1A1 in A549 cells, ALDEFLUOR-positive cells decreased by 95%, along with attenuated colony formation and migration abilityCitation39.
Melanoma
The ALDH1A isoenzyme is expressed in melanoma. Erfani et al. detected the expression of ALDH1A1 in 12 melanoma tissue samples through IHC, of which 67% (8/12) had low expression of ALDH1A1 and 33% (4/12) had high expression of ALDH1A1Citation103. Recent studies have demonstrated that ALDH1As play essential roles in regulating the proliferation, apoptosis, and chemoresistance of melanoma CSCsCitation104 and might have therapeutic potentialCitation105. ALDEFLUOR-positive melanoma CSCs were associated with chemoresistance, and ALDH1A (ALDH1A1 and ALDH1A3) silencing could attenuate cell proliferation and induce apoptosis in ALDEFLUOR-positive melanoma CSCs in vitro and suppress melanoma tumorigenesis in vivoCitation104. Studies have found that ALDEFLUOR+ cells accounted for approximately 2% of melanoma cells (0.46–4.39%)Citation55, and ALDEFLUOR+ subpopulations were detected in 8/9 cases of fresh tumour samples and 8/10 cases of xenografted tumours, with positive rates ranging from 0.08 to 1.15%Citation104. Microarray analysis of ALDEFLUOR+ and ALDEFLUOR– cells isolated from xenografted tumours indicated that the expression of ALDH1A1 and ALDH1A3 was more than 15 times higher in the ALDEFLUOR+ subpopulationCitation104. These results suggest that ALDEFLUOR activity in primary melanoma may be mainly determined by both ALDH1A1 and ALDH1A3. However, in ALDEFLUOR+ subpopulations isolated from the 4 melanoma cell lines 1205Lu, A375, WM239A, and HS294T, ALDH1A3 mRNA expression was more than 200 times higher than that of ALDH1A1Citation104. Knockdown of ALDH1A3 expression significantly decreased the ALDEFLUOR-positive rate in 1205Lu and A375 cells, suggesting a contributing role of ALDH1A3 in determining ALDEFLUOR activityCitation104. Altogether, the reason for the differences in ALDH1A1 and ALDH1A3 expression between primary melanomas and melanoma cell lines was not clear.
Prostate cancer
Studies have shown that ALDEFLUOR-positive cells of prostatic cancer cell lines were enriched for clonogenicity, tumorigenicity, and metastatic abilityCitation106, and high ALDH1A1 expression correlates with lower overall survival, Gleason score, and pathologic stage in patients with primary prostate cancerCitation49,Citation106. The Wnt pathway was reported to directly regulate ALDH1A1 expression through β-catenin/TCF-dependent transcription, and inhibition of the Wnt/β-catenin signalling pathway was demonstrated to suppress the viability of prostate cancer cells with relatively high ALDH1A1 expressionCitation16. Work by Li et al. indicated that ALDEFLUOR+ cells in the PC3 and LNCaP cell lines possess stem cell characteristics and that ALDH1A1 was the major enzyme subtype determining ALDEFLUOR activity and may be used as a marker for prostate CSCsCitation49. IHC analysis of 18 normal tissues and 163 prostate cancer tissue sections revealed low ALDH1A1 expression in the basal cell layers of normal prostate tissues and that it coexisted with the stem cell marker CD44Citation49. In tumour tissues, ALDH1A1 was highly expressed in secretory-type cancer epithelial cells and neuroendocrine tumour cellsCitation49. Kalantari et al. studied the expression of ALDH1A1 in 105 prostate cancer tissues, 21 high-grade prostatic intraepithelial neoplasia tissues, and 31 benign prostatic hyperplasia tissues. In tumour tissues, 66% (69/105) had low expression, and 34% (36/105) had high expressionCitation107. There was a significant positive correlation between ALDH1A1 expression and the Gleason score. However, almost all BPH and HGPIN cases were not stained, and only 3% (1/31) of BPH cases showed strong stainingCitation107. However, van den Hoogen et al. found that the high ALDEFLUOR activity found in prostate cancer cells did not correlate with ALDH1A1 expression and that ALDH7A1 was the major enzyme subtype determining ALDEFLUOR activityCitation106. Analysis of 8 prostate cancer cell lines (PC-3luc, PC-3M-Pro4lucA6, PC-3M-Pro4lucBIII, LNCaP, C4, C4-2, C4-2B, and DU145) and 6 primary prostate cancer specimens indicated that the ALDEFLUOR-positive rates were approximately 0.8–31% in prostate cancer cell lines and 0.5–12.5% in prostate cancer specimensCitation106. The ALDH subtypes with higher expression levels included ALDH3A2, ALDH4A1, ALDH7A1, ALDH9A1, and ALDH18A1. High ALDH7A1 expression was observed in all primary cultured cells and in the PC-3, PC-3M-Pro4lucBIII, and DU145 cell linesCitation106. ALDH7A1 knockdown was found to decrease ALDEFLUOR activity by 21%Citation50. IHC analysis further confirmed the dominant expression of ALDH7A1 in prostate cancerCitation106. In addition, ALDH4A1 and ALDH9A1 havebeen shown to be the major enzyme subtypes accounting for ALDEFLUOR activity in prostate cancer tissues, while the dominant enzyme subtypes in nontumor tissues and high-grade prostate intraepithelial neoplasia are ALDH3A2and ALDH18A1Citation106.
Head and neck squamous cell carcinoma (HNSCC)
In HNSCC samples, the mean ALDEFLUOR-positive rate was 3.5% (n = 6, 1.0–7.8%)Citation108. ALDEFLUOR-positive HNSCC cells had stem cell characteristics with high CD44 expression and could form primary tumours in immunodeficient animalsCitation35,Citation108. The mean ALDEFLUOR-positive rate in Fanconi anemia–head and neck squamous cell carcinoma (FA–HNSCC) cells was approximately 23% (VU-1365, 31% ± 2.9%; VU-1131, 23% ± 2.7%; OHSU-974, 15% ± 2.0%), higher than that in the UMSCC-22A HNSCC cell line (10.33% ± 3.0%)Citation109. More than 10% of the tumour cells in HNSCC tumour samples expressed ALDH1A1, which was scattered as single cells or small groups of cells, whereas more than 25% of the cells expressed ALDH1A1, which was distributed as sheets or islands in FA–HNSCC tumour tissue samplesCitation109. IHC analysis of FA–HNSCC xenografted tumours yielded similar resultsCitation109. Therefore, ALDH1A1 is considered the major subtype that determines ALDEFLUOR activity in HNSCC cells.
Nevertheless, other researchers argued that ALDEFLUOR activity in HNSCC is at least partially determined by ALDH1A3Citation110. ALDH1A3 exhibited relatively high expression in FaDu and Cal33 cells compared to ALDH1A1Citation110. Analysis of xenografted tumours derived from ALDEFLUOR-positive cells also showed high ALDH1A3 expressionCitation110. In other words, ALDH1A3 might also influence ALDEFLUOR activityCitation110.
Liver cancer
ALDH1A1 has been reported to be the dominant enzyme subtype determining ALDEFLUOR activity in hepatocellular cancer (HCC) and hepatoblastoma and was a differentially expressed protein in CD133+ and CD133– cells of the Huh7 and PLC8024 HCC cell linesCitation41. The ALDEFLUOR-positive rate was 55% in Hep3B cells and 1–8% in Huh7, H2M, PLC8024, and Hep2B cells, consistent with the theory that CSCs account for only a very small percentage of tumour cells. Therefore, researchers concluded that ALDH1A1 is the dominant enzyme subtype that determines ALDEFLUOR activity in HCCCitation41. However, researchers observed no appreciable change in the HCC CSCs marker EpCAM and proliferation and sphere formation ability upon ALDH1A1 knockdown in Huh1 and Huh7 cellsCitation111. Moreover, patients with high ALDH1A1 expression had a higher degree of tumour differentiation. Thus, ALDH1A1 might otherwise be a differentiation marker with little relevance to the maintenance of stem cell characteristics of HCCCitation111. RT–PCR analysis of 60 HCC samples and 47 paired HCC and adjacent nontumorous tissues revealed that ALDH1A1 mRNA expression was not significantly different in tumour and nontumor tissues (1.36 ± 1.26 vs. 1.00 ± 0.53, p = 0.8858)Citation42. IHC analysis also indicated that ALDH1A1 was not coexpressed with stem cell markers such as BMI1, EpCAM, CD13, CD24, CD90, and CD133Citation42.
The major enzyme subtype that determines ALDEFLUOR activity was shown to be ALDH1A3 in intrahepatic CHOLCitation40. The ALDEFLUOR-positive rates in the intrahepatic CHOLcell lines HuCCT1 and SNU1079 were 9.67% and 20.32%, respectively. The ALDEFLUOR+ cell population had stem cell characteristics, with higher resistance to gemcitabine than ALDEFLUOR– cellsCitation40. Analysis of the mRNA expression of 19 ALDH subtypes in the ALDEFLUOR+ and ALDEFLUOR– subpopulations indicated higher ALDH1A3 and ALDH1L1 expression in the ALDEFLUOR+ subpopulation of HuCCT1 cells and higher expression of ALDH1A3, ALDH1B1, ALDH6A1, ALDH1A1, ALDH18A1, ALDH3B2, and ALDH3B1 in the ALDEFLUOR+ subpopulation of SUN1079 cellsCitation40. The ALDH subtype with the greatest abundance in the ALDEFLUOR+ populations in these 2 cell lines was ALDH1A3Citation40. ALDH1A3 knockdown markedly reduced not only the invasion/migration ability but also the sensitivity to gemcitabineCitation40. Altogether, these findings indicate that ALDH1A3 is the major enzyme subtype that determines ALDEFLUOR activity in CHOL.
Brain tumours
Constitutive activation of the Notch signalling pathway was reported to rescue neurosphere differentiation of glioblastoma (GBM) in an RA-dependent process, indicating a functional role of ALDH in GBM tumorigenesisCitation112. Some researchers consider ALDH1A1 to be the dominant enzyme subtype that determines ALDEFLUOR activity in glioma, and this subtype can be used as a marker of glioma stem cells (GSCs)Citation113. Flow cytometry analysis of U251 cells and 1 case of primary cultured cells revealed that the ALDEFLUOR-positive rates were 6.9% ± 2.1% and 4.2% ± 1.9%, respectively, and the ALDEFLUOR population had CSC characteristicsCitation113. RT–PCR analysis of ALDEFLUOR+ and ALDEFLUOR– subpopulations in U251 cells showed that ALDH1A1 expression was significantly higher in the ALDEFLUOR+ subpopulation and was the dominant enzyme subtype for ALDEFLUOR. IHC analysis of 237 glioma samples revealed higher ALDH1A1 expression in high-grade gliomas (WHO III-IV) than in low-grade gliomas (WHO I-II)Citation113. In addition, ALDH1A1 was the dominant subtype in the classical subtype of GBM, with 84.6% of the classical samples (n = 13) exhibiting high ALDH1A1 expression, while the ALDH1A1-positive rates were 12.5% and 21.7% in the proneural (PN) subtype (n = 36) and the other 2 subtypes (neural and mesenchymal (Mes)), respectivelyCitation113.
In contrast, the results of another study suggested that ALDH1A3 might be the dominant subtype that determines ALDEFLUOR activity in glioma. IHC analysis revealed that 15 samples of normal tissues and 7 low-grade gliomas (WHO II) did not express ALDH1A3 and that 37 of 38 GBM cases expressed ALDH1A3Citation114. In addition, Mes GSCs displayed relatively high invasiveness and intracranial tumorigenicity compared to PN GSCs, and the ALDEFLUOR activity of Mes GSCs was approximately 8 times higher than that of PN GSCsCitation114. RT–PCR analysis of 19 ALDH subtypes indicated that ALDH1A3 had significantly higher expression in Mes GSCs, with an approximately 150-fold higher expression level than that in PN GSCsCitation114, whereas the other 18 enzyme subtypes were expressed at a very low level in both groups of GSCsCitation114. Transcriptome microarray analysis also revealed 150-fold higher expression of ALDH1A3 in Mes GSCs than in PN GSCsCitation114. Moreover, inhibition of ALDH1A3 could sufficiently attenuate the proliferation, sphere formation ability, and tumorigenicity of Mes GSCsCitation114. These results indicate that ALDH1A3 is the dominant enzyme subtype that determines ALDEFLUOR activity in Mes GSCs.
The expression of different ALDH isoforms and their subcellular location in various cancer types were summarised in .
Table 1. Different ALDH isoforms and their location in various cancer types.
Articular chondrocyte stem cells
Studies of human articular chondrocyte stem cells demonstrated that ALDH1A2, ALDH1A3, and ALDH2 are strongly expressed in chondrocytes, whereas ALDH1A1 expression is low. In isolated ALDEFLUOR+ chondrocyte subpopulations, only ALDH1A2 and ALDH1A3 showed remarkably high expressionCitation115.
Possible mechanisms underlying the ALDEFLUOR-positive rate and study directions
With the completion of in-depth studies and the availability of commercial antibodies specific for ALDH enzyme subtypes, accumulative efforts have begun to clarify the characteristics of the ALDEFLUOR+ cell population. In addition to ALDH1A1, other ALDH subtypes are now understood to also influence the ALDEFLUOR-positive rate, including ALDH1A2, ALDH1A3, ALDH1B1, ALDH3A2, ALDH4A1, ALDH7A1, and ALDH9A1. The possible reasons for this phenomenon are discussed below.
BAAA is not the only substrate of ALDH1A1
In early studies, BAAA was generally considered a specific substrate of ALDH1A1, and the ALDEFLUOR-positive rate was thought to directly reflect the expression level and activity of ALDH1A1Citation116. However, all members of the ALDH family have similar physiological functions and participate in the metabolism of hydrocarbons, biogenic amines, RA, and steroids and in lipid peroxidationCitation117. With the exception of ALDH6A1, which uses CoA as a cofactor, the other enzyme subtypes all use NAD(P+) as a cofactor, and their catalytic mechanisms in the dehydrogenase and lipase functions are similar and may be associated with the highly conserved catalytic residues Cys302, Lys192, and Glu268Citation118. The substrates of each isozyme also have similar structuresCitation117. Several isozymes (1A1, 1A2, 1A3, 2, 3A1, 7A1, and 8A1) in the ALDH family are able to utilise BAAA as a substrateCitation3,Citation17,Citation66,Citation104,Citation119, suggesting that ALDEFLUOR+ cells might be a mixed population and that ALDEFLUOR activity is not simply determined by the expression level and activity of ALDH1A1Citation17. When ALDH1A2 and ALDH2 were overexpressed in Beas-2b immortalised lung epithelial cells that did not contain an ALDEFLUOR+ cell subpopulation, the resulting Beas-2BALDH1A2 or Beas-2BALDH2 cells were mixed with K562 leukaemia cells or H1299 lung cancer cells at percentages of 20–25%, and the mixed cell populations were analysed using the ALDEFLUOR systemCitation17. The results indicated that the ALDEFLUOR-positive rates increased significantly in both mixed cell populations. These results indicate that in addition to ALDH1A1, the enzyme activities of ALDH1A2 and ALDH2 also contribute to activity in the ALDEFLUOR systemCitation17. However, although researchers now recognise that BAAA may be a common substrate of various ALDH subtypes, progress in identifying specific substrates for ALDH subtypes has been limited, and this topic should become a key research topic in the future.
DEAB does not specifically inhibit ALDH1A1 activity and is a substrate for various ALDH subtypes
DEAB was previously considered a selective inhibitor of ALDH1A1Citation39,Citation120 and was often used as a negative control in ALDEFLUOR analysis (). However, recent studies have found that DEAB is not a specific inhibitor of ALDH1A1 and that it has a stronger inhibitory effect (IC50 0.057–15 μM) on other enzyme subtypes, including ALDH1A2, ALDH1A3, ALDH1B1, ALDH2, and ALDH5A1Citation17,Citation63,Citation64,Citation90. The order of sensitivity of ALDH subtypes to DEAB was ALDH1A1 < ALDH2 < ALDH1A2 < ALDH1B1 < ALDH1A3 < ALDH3A1 < ALDH5A1Citation63,Citation64. The order of the catalytic efficiency of different ALDH subtypes on DEAB, from strong to weak, was shown to be ALDH1A1 > ALDH1B1, ALDH1A3, ALDH5A1 > ALDH2Citation64. Furthermore, DEAB had inhibitory effects on ALDH3A1 in oral mucosal epithelial cellsCitation92, and the Vmax/Km of ALDH3A1 on DEAB was even higher than that of its conventional substrate benzaldehydeCitation64. These findings at least partially explain why increased DEAB concentration and increased incubation time may both influence the ALDEFLUOR-positive rate observed during practical application of the method. For example, in cell lines with higher ALDEFLUOR-positive rates, such as MDA-MB-468, SKBR3, and MDA-MB-435 cells, a reduction in the DEAB concentration can significantly increase the ALDEFLUOR-positive rateCitation66. However, in cell lines with moderate ALDEFLUOR-positive rates, such as BT-20 cells, and in those with lower ALDEFLUOR-positive rates, such as MCF7, T47D, and MDA-MB-231 cells, the effects of using different concentrations of DEAB (15 μM and 100 μM) on ALDEFLUOR-positive rates are not as obviousCitation66. Similarly, inhibition of ALDH1A2 and ALDH2 by DEAB showed an obvious concentration dependence in K562 leukaemia cells and H1299 lung cancer cellsCitation17. Therefore, exploration and verification of the optimal incubation time and the optimal concentration of DEAB for use in different cells is another desirable area for future research.
Ibrahim et al.Citation121 investigated the inhibitory effect of DEAB analogs on different isoforms of ALDH, which were synthesised using either aliphatic or aromatic nucleophilic substitution in a one-step reaction. Analogs 18 (4-(dipropylamino)-3-nitrobenzaldehyde) and 19 (4-(diethylamino)-3-nitrobenzaldehyde) showed potent inhibitory activity against ALDH3A1Citation121. Analogs 14 (3-bromo-4-(dipropylamino)benzaldehyde), 15 (3-methyl-4-(piperidin-1-yl)benzaldehyde), and 16 (4-isopropoxybenzaldehyde) had potent inhibitory activity against ALDH1A3Citation121. Analog 14 showed the highest antiproliferative activity in the prostate cancer cell lines DU145 and PC3, with IC50 values of 61 and 47 μM, respectively, and analog 14 also showed very low activity as a substrate for ALDH1A1 and ALDH3A1 isoforms, demonstrating the advantage of its higher selectivityCitation121. The specificity of the ALDEFLUOR assay was improved by using compounds 14 and 15 to label ALDH1A3-expressing cells or 18 and 19 for ALDH3A1-expressing cells and compared to the results from DEABCitation121 ().
To date, specific inhibitors of most ALDH isozymes, including ALDH1A1, have not been systematically identifiedCitation118. Nevertheless, natural and synthesised compounds have been developed, including disulphiram (), which can irreversibly inhibit ALDH1A1 and ALDH2 at the same time and is clinically used to treat alcoholismCitation118 and cocaine addictionCitation122. In addition, the H2 receptor antagonist cimetidine, which is used in the treatment of peptic ulcers, has weaker non-competitive inhibitory effects on ALDH1A1 enzyme activity in the liver, small intestine, and stomach at therapeutic concentrations (0.015 mM) and competitive inhibitory effects on ALDH2 and ALDH3A1Citation123. Therefore, the identification of specific inhibitors of each individual ALDH enzyme subtype should also be a key focus of future researchCitation119. Notably, the identification of specific inhibitors of individual ALDH subtypes will not only help confirm the specific isozyme determining ALDEFLUOR activity in cells but is also necessary for accurate intervention in all ALDH subtypes with different functions.
ALDH isozymes have a high degree of sequence homology, with ALDH1A1 sharing more than 70% sequence homology with ALDH1A2 and ALDH1A3Citation124–127. The development of selective inhibitors for each ALDH isoform has been hampered by a high degree of sequence and structural homology. Li et al. used the structure of human ALDH1A combined with in silico modelling to identify a selective, active-site inhibitor of ALDH1A3, MCI-INI-3Citation124 (). The compound MCI-INI-3 is a potent selective inhibitor of recombinant human ALDH1A3, with greater than 140-fold selectivity for ALDH1A3 compared to the closely related isoform ALDH1A1Citation124. NR6 is a new selective ALDH1A3 inhibitorCitation128 (). The crystal structure determined by X-ray analysis reveals that NR6 binds to a nonconserved tyrosine residue of ALDH1A3, which drives the selectivity for this isoform, which is supported by computational binding simulationsCitation128. NR6 has cytotoxic activity against glioblastoma and colorectal cancer cellsCitation128.
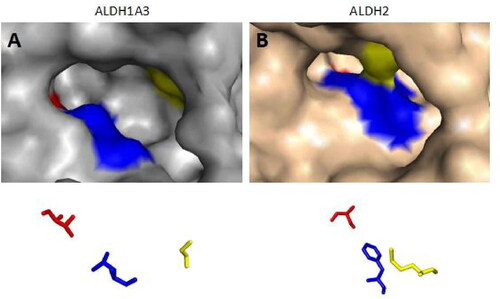
Because the Rossmann fold structure at the NAD(P)+ cofactor-binding site of other dehydrogenase families, such as lactate dehydrogenase and alcohol dehydrogenase, is different from that of the ALDH familyCitation129, this structure is an important target for developing inhibitors for the ALDH family. Recently, Feng et al. discovered a selective imidazolium-based inhibitor of ALDH1B1 (half-maximum inhibitory concentration (IC50) = 57 nM)Citation130. This inhibitor acted in a non-competitive manner with respect to the aldehyde substrate and exhibited an uncompetitive relationship with NAD+Citation130. However, the Rossmann fold structures at the NAD(P)+ binding sites of all ALDH enzyme subtypes display high similarity. Further studies indicated that different aldehyde group binding sites have evolved in different subtypes in the ALDH familyCitation117,Citation131. Therefore, Morgan et al. considered that targeting substrate binding sites or allosteric sites other than the active sites might be suitable for screening specific inhibitorsCitation132. Specifically, 3 topologically conserved residues located within the ligand-binding pocket control the substrate specificity of different ALDH subtypes, such as ALDH1A3 and ALDH2Citation133 (). Of course, the direct use of the high-throughput screening method offers another strategy for acquiring inhibitors for all ALDH subtypes. However, how to evaluate their inhibitory efficiency is also worth exploring. In the past, an ultraviolet light spectrophotometer was used to evaluate the inhibitory efficiency of candidate molecules on ALDH activity by measuring the intensity of the absorption peak of NADH at 340 nm. However, compounds in the library to be screened might have the same ultraviolet light absorption wavelength as NADH, which would reduce the accuracy of the results. ALDH family members also have esterase activity and can hydrolyse p-nitrophenyl acetate into nitrophenol, producing an absorption peak at 405 nmCitation132. Therefore, in recent years, some researchers have also used the intensity of the absorption peak at 405 nm to evaluate the inhibition of ALDH by candidate molecules. For example, Morgan et al. adopted this strategy to identify a small molecule among 6,400 compounds, CM037 (), that specifically inhibits ALDH1A1 with an IC50 of 4.6 ± 0.8 μMCitation132. This compound significantly influenced the sphere formation ability of ovarian cancer cellsCitation134. Parajuli et al. combined these 2 strategies using a method that involved measuring the intensity of the absorption peaks at 340 nm and 405 nm to identify 14 small-molecule inhibitors that could better inhibit the dehydrogenase and esterase activities of ALDH3A1Citation135. ABMM-15 and ABMM-16 are selective ALDH1A3 inhibitors designed based on the similarity of physiological substrates and are a class of compounds with a benzyloxybenzaldehyde scaffoldCitation136 (). ALDH-positive A549 and ALDH-negative H1299 cells were used to validate the selectivity and cytotoxicity of the compounds against ALDH1A1, ALDH1A3, and ALDH3A1Citation136. The 2 compounds were found to be the most potent and selective inhibitors of ALDH1A3Citation136.
Figure 2. The substrate binding positions of ALDH1A3 and ALDH2. Surface representation of ALDH1A3 (A, in gray); the conserved ligand-binding residues are occupied by G136, L471, and T315 (PDB:5FHZ). Surface representation of human mitochondrial ALDH2 (B, in pink), with positions 124, 459, and 303 shown (PDB:1O01). The binding sites from the two monomers are shown as sticks and coloured red, blue, and yellow according to their positions.
Tumour type and tissue/cell origin are important influencing factors
As mentioned above, BAAA can be catalysed by various ALDH subtypes. One important question is whether differences in the expression of ALDH subtypes in various normal tissues and tumours result in differences in the dominant ALDH subtypes in cells isolated and acquired using the ALDEFLUOR system.
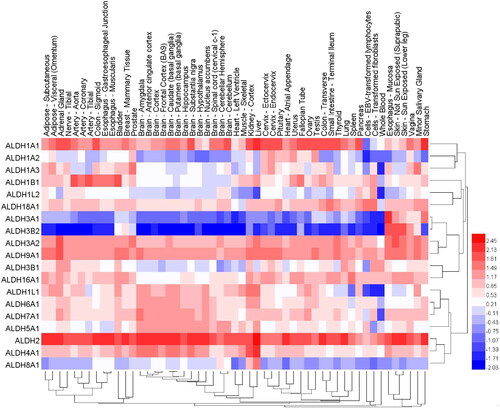
We used public databases to analyse the mRNA expression of different ALDH subtypes in 30 normal tissues (). The results indicated that except for blood, which contained ALDH3B1 as the dominant subtype, the normal oesophageal mucosa, which contained ALDH3A1 as the dominant subtype, and prostate and salivary glands, which contained ALDH1A3 as the dominant subtype, most tissues all had ALDH2 and ALDH1A1 as the dominant subtypes. These findings are consistent with the results showing that ALDH1A1 is predominantly expressed in the testis, brain, eyeCitation137, liverCitation138, kidney epithelium, neural stem cells, and HSCsCitation139,Citation140, while ALDH1A3 is mainly expressed in the kidney, salivary glands, stomachCitation141, foetal nasal mucosaCitation142, and mammary glandsCitation89. Studies of buccal carcinomaCitation93, HNSCCCitation110, and breast cancerCitation66 tissue also indicated that ALDH1A3 was the dominant subtype and that tumour cells with high ALDH1A3 expression had CSCs characteristics; this result is consistent with the high expression of ALDH1A3 in normal tissues of the same origin.
Figure 3. The expression levels of different ALDH family isoforms in 30 normal tissues. The data were derived from the GTEx (Genotype-Tissue Expression Project). Dendrograms and heat maps of ALDH mRNA expression in various normal tissues are shown.
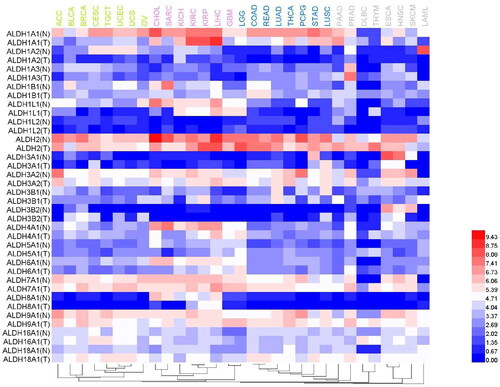
Despite the foregoing information, researchers have reached inconsistent conclusions regarding some tissues and tumours. First, analysis of data in public databases indicated that the expression of ALDH3B1 in normal lung tissues was higher than that reported above. Some researchers demonstrated that the expression of ALDH3B1 in lung cancer was upregulatedCitation143, which is consistent with the trend in which some subtypes with higher expression in normal tissues were also highly expressed in tumours. However, some studies found that ALDH1A3 was the major subtype in NSCLCCitation102. Second, ALDH1A1 was reported to be the dominant ALDH subtype in liver and brain tissues. Most researchers consider that ALDH1A1 is also the dominant ALDH subtype expressed in liver cancerCitation41 and brain cancerCitation113. However, some researchers reported that ALDH1A3 was the dominant subtype and that cells expressing ALDH1A3 had CSCs characteristicsCitation40. Other studies found that the dominant subtype in normal tissues was not dominant in tumours derived from those tissues. For example, the dominant subtype in blood was ALDH3B1; however, the expression of ALDH1A1, ALDH1A2, and ALDH1B1 in the K562 chronic myeloid leukaemia cell line was higher than that of ALDH3B1 and ALDH3A1Citation17. Marchitti et al. reported that ALDH3B1 expression was upregulated in various tumours, including breast cancer, ovarian cancer, and colorectal cancerCitation143. However, data in public databases indicate that the expression of ALDH3B1 in normal mammary, ovarian, and colonic tissues occurs at a lower level and that the dominant enzyme subtype in these tissues is ALDH1A1. Similarly, we also used public data resources to analyse the expression of different ALDH subtypes in various tumour tissues and corresponding paracancerous tissues (). Hepatocellular carcinoma is a special case because the expression of many ALDH subtypes, including ALDH1A1, ALDH1L1, ALDH2, ALDH3A2, ALDH4A1, ALDH7A1, and ALDH9A1, is relatively high in this cancer. High expression of ALDH1L2, ALDH5A1, and ALDH6A1 can be observed in low-grade glioma. The results also revealed that ALDH1A2 expression is highest in mesothelioma (data not shown) and that the tumour type with the highest ALDH1A3 expression level is prostate cancer. In addition, the expression of ALDH3A2 in adenocarcinoma, ALDH7A1 in prostate cancer and glioma, and ALDH9A1 in breast cancer and thyroid cancer is also high. In general, the expression of ALDHs in tumour tissues is higher than that in corresponding paracancerous tissues. For example, ALDH1L1, ALDH4A1, ALDH6A1, and ALDH8A1 expression in sarcoma is lower than that in corresponding mesenchymal tissues. However, this trend is sometimes reversed in thymoma: ALDH1A1, ALDH1B1, ALDH3A2, ALDH5A1, ALDH6A1, ALDH7A1, ALDH9A1, and ALDH18A1 expression is higher in thymoma than in corresponding paracancerous tissues.
Figure 4. Comparison of the expression levels of different ALDH subtypes in various tumour tissues and corresponding normal tissues. A heat map was generated based on a TCGA dataset. The dendrogram of tumours (top) was divided into 4 parts using different colours.
Interestingly, when tumours, primary specimens, and cell lines derived from the same tissues were compared, the dominant ALDH subtypes that determined the ALDEFLUOR-positive rate were found not to be the same. As previously mentioned, ALDEFLUOR activity in primary melanoma may be mainly determined by ALDH1A1 and ALDH1A3. However, in melanoma cell lines (1205Lu, A375, WM239A, and HS294T), ALDEFLUOR activity may be mainly determined by ALDH1A3. Determination of the reason for this discrepancy will require further in-depth studies.
Clinical significance of ALDH subtypes in different cancers
The catalytic activity of ALDH can be used as a marker for the identification and isolation of tumour stem cells and is associated with prognosis in patients with a variety of tumour typesCitation24,Citation144,Citation145.
ALDH1A1, a marker of tumour stem cells in breast, prostate, colon, and lung cancers, is a cytoplasmic enzyme that is upregulated in tumour cells; ALDH1A1 is associated with poor prognosis of many tumours (breast cancer, melanoma, etc.) and has an important role in promoting tumour angiogenesis and metastasis and in acquiring resistance to anticancer drugsCitation146–148.
Low ALDH1A2 expression is associated with poor prognosis and shorter disease-free and overall survival for ovarian cancer patients. ALDH1A2 suppresses the proliferation and migration of epithelial ovarian cancer cells by downregulating STAT3, and enhanced ALDH1A2-related signalling may provide new opportunities for therapeutic intervention in ovarian cancerCitation149.
ALDH1A3 activates the PI3K/AKT/mTOR signalling pathway and its downstream target PPARγ, leading to increased expression of HK2, which in turn increases glycolysis in pancreatic ductal carcinoma cells, thereby promoting tumour metastasisCitation150. CHOL cells with high ALDH1A3 levels not only migrated more quickly but were more resistant to gemcitabineCitation40. In addition, knockdown of ALDH1A3 expression in HuCCT1 cells markedly reduced not only their sensitivity to gemcitabine, which might be attributed to a decreased expression of ribonucleotide reductase M1, but also their migrationCitation40. ALDH1A3 plays an important role in enhancing the malignant behaviour of CHOL and serves as a new therapeutic targetCitation40.
Our previous studies in colorectal cancer have demonstrated that ALDH1A3 can promote the invasion and metastasis of colorectal cancer cells through the miR-200-ZEB1/SANI2 axis and that inhibition of ALDH1A3 with the identified compound YD1701 may be an effective therapeutic approach to prevent colorectal cancer metastasisCitation151. Other investigators have reported that knockdown of ALDH1B1 in colorectal cancer SW480 cells downregulates Wnt, Notch, and PI3K/Akt signalling and that the growth of 5-fluorouracil-resistant colorectal cancer spheroids and patient-derived organoids can be inhibited using small-molecule inhibitors of ALDH1B1Citation130,Citation152.
Studies have demonstrated that high expression of ALDH3B2 in perihilar CHOL is associated with higher tumour T stage and M stage and a higher incidence of neuroinvasion. High ALDH3B2 expression was an independent prognostic factor for intrahepatic CHOL and distal CHOLCitation153. Inhibition of ALDH3B2 expression may inhibit the proliferation and clonogenic ability of CHOL by inducing G1 phase arrest. ALDH3B2 may also promote CHOL progression by promoting epithelial–mesenchymal transitionCitation153. Targeted inhibition of ALDH3B2 function is expected to inhibit the metastasis of CHOL cells and prolong the survival of patientsCitation153.
The expression of ALDH18A1 was significantly associated with the overall survival of neuroblastoma patientsCitation154. On the one hand, ALDH18A1 positively regulates MYCN gene expression at the transcriptional and posttranscriptional levels through the miR-29b/SP1 self-regulatory loop and miRNA regulatory network, respectively, which in turn affects the division and proliferation, self-renewal, and tumorigenic capacity of neuroblastoma cellsCitation154. On the other hand, the transcription factor N-Myc can directly target the ALDH18A1 promoter through specific binding sites to activate ALDH18A1 transcription and increase ALDH18A1 expression levelsCitation154. Administration of the ALDH18A1-specific inhibitor YG1702 inhibits N-Myc expression and attenuates the growth of neuroblastoma cellsCitation154.
Clinical trials on ALDH in cancers
Targeting CSCs has great prospects for preventing metastasis and reducing the risk of drug resistance and recurrence. The vast majority of patients with AML achieve complete remission after standard induction chemotherapy. However, most patients die from tumour recurrence. LSCs are more resistant to chemotherapy and are a major cause of relapseCitation79. DIMATE is an active enzyme-dependent, competitive, irreversible, broad spectrum inhibitor of ALDHs 1s and 3sCitation155. A clinical study was proposed to test the in vitro sensitivity of leukaemia-derived CD4/CD38 cells to the DIMATE compound and to analyse the effect of DIMATE on the parameters of proliferation, apoptosis, secretory cytokines, and transcriptomic changesto better define the therapeutic index of the compound. HSC from healthy donors were used as controlsCitation155. The study did not provide resultsCitation155 (). Venton et al. reported that in cultured cells, DIMATE was a potent inhibitor of ALDH 1 s and ALDH 3 s and had major cytotoxic activity against human AML cell linesCitation79. In addition, DIMATE was selectively cytotoxic in a leukemic population enriched in LSCs, but unlike conventional chemotherapy, DIMATE was not toxic to healthy HSCCitation79.
Table 2. Clinical trials on ALDH in cancers.
A forthcoming case–control study of OSCC patients and healthy volunteers will collect saliva samples from both groups and submit them for sequencing analysis to evaluate the frequency of ALDH1b1 and ALDH2 polymorphisms in the Brazilian population and correlate OSCC risk to alcohol consumption or smoking through applied questionnaires. These data will help characterise genetic variants of ALDH1B1 and ALDH2 in the Brazilian population and support the development of future public policies to reduce the main risk factors for OSCC, especially in those with these genetic variants (NCT04270201) ().
In addition, a few clinical trials are evaluating the effectiveness of treatment on the expression of CSC markers, mainly measuring the activity of ALDH, including breast cancer (clinical trial numbers: NCT00949013, NCT01190345, NCT01688609, NCT04142892, NCT01077453, NCT01426880), lung cancer (NCT04634630), lymphoma (NCT00369681), gynecological tumours (NCT02903771), and prostate cancer (NCT03410030) ().
Conclusions
In this review, we summarise the predominant enzymes and groups of enzymes among the 19 members of the ALDH family that determine the positive rate of ALDEFLUOR in different normal tissues and tumours. In addition to ALDH1A1, other ALDH isoforms, such as ALDH1A3, ALDH2, and ALDH3A1, should also be considered in ALDEFLUOR analyses.
Although the ALDEFLUOR system provides a powerful tool for in-depth study of the characteristics of stem cells and cancers, this system has some limitations due to the specificity of the fluorescent substrate BAAA and the control inhibitor DEAB. Future research should focus on the development of new systems based on cell-type-specific substrates and inhibitors.
Ethics approval and consent to participate
This article does not contain any experimental studies on human participants or animals performed by the authors.
Author contributions
JJD and JC collected the related literature and drafted the manuscript. SCY and LG participated in the design of the review and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Prof. Xindong Liu for his constructive suggestions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability and materials
The materials that support the conclusions of this review have been included within the article.
Additional information
Funding
References
- Black WJ, Stagos D, Marchitti SA, Nebert DW, Tipton KF, Bairoch A, Vasiliou V. Human aldehyde dehydrogenase genes: alternatively spliced transcriptional variants and their suggested nomenclature. Pharmacogenet Genom. 2009;19(11):893–902.
- Ma I, Allan AL. The role of human aldehyde dehydrogenase in normal and cancer stem cells. Stem Cell Rev Rep. 2011;7(2):292–306.
- Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-p450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4(6):697–720.
- Jackson B, Brocker C, Thompson DC, Black W, Vasiliou K, Nebert DW, Vasiliou V. Update on the aldehyde dehydrogenase gene (aldh) superfamily. Hum Genomics. 2011;5(4):283–303.
- Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev. 2004;36(2):279–299.
- Dockham PA, Lee MO, Sladek NE. Identification of human liver aldehyde dehydrogenases that catalyze the oxidation of aldophosphamide and retinaldehyde. Biochem Pharmacol. 1992;43(11):2453–2469.
- el Akawi Z, Napoli JL. Rat liver cytosolic retinal dehydrogenase: comparison of 13-cis-, 9-cis-, and all-trans-retinal as substrates and effects of cellular retinoid-binding proteins and retinoic acid on activity. Biochemistry. 1994;33(7):1938–1943.
- Grun F, Hirose Y, Kawauchi S, Ogura T, Umesono K. Aldehyde dehydrogenase 6, a cytosolic retinaldehyde dehydrogenase prominently expressed in sensory neuroepithelia during development. J Biol Chem. 2000;275(52):41210–41218.
- Wang X, Penzes P, Napoli JL. Cloning of a cdna encoding an aldehyde dehydrogenase and its expression in escherichia coli. Recognition of retinal as substrate. J Biol Chem. 1996;271(27):16288–16293.
- Lin M, Napoli JL. Cdna cloning and expression of a human aldehyde dehydrogenase (aldh) active with 9-cis-retinal and identification of a rat ortholog, aldh12. J Biol Chem. 2000;275(51):40106–40112.
- Lin M, Zhang M, Abraham M, Smith SM, Napoli JL. Mouse retinal dehydrogenase 4 (raldh4), molecular cloning, cellular expression, and activity in 9-cis-retinoic acid biosynthesis in intact cells. J Biol Chem. 2003;278(11):9856–9861.
- Sophos NA, Pappa A, Ziegler TL, Vasiliou V. Aldehyde dehydrogenase gene superfamily: the 2000 update. Chem Biol Interact. 2001;130–132(1–3):323–337.
- Keysar SB, Jimeno A. More than markers: biological significance of cancer stem cell-defining molecules. Mol Cancer Ther. 2010;9(9):2450–2457.
- Visus C, Ito D, Amoscato A, Maciejewska-Franczak M, Abdelsalem A, Dhir R, Shin DM, Donnenberg VS, Whiteside TL, DeLeo AB. Identification of human aldehyde dehydrogenase 1 family member a1 as a novel cd8+ t-cell-defined tumor antigen in squamous cell carcinoma of the head and neck. Cancer Res. 2007;67(21):10538–10545.
- Yoon KA, Nakamura Y, Arakawa H. Identification of aldh4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49(3):134–140.
- Clark DW, Palle K. Aldehyde dehydrogenases in cancer stem cells: potential as therapeutic targets. Ann Transl Med. 2016;4(24):518–518.
- Moreb JS, Ucar D, Han S, Amory JK, Goldstein AS, Ostmark B, Chang LJ. The enzymatic activity of human aldehyde dehydrogenases 1a2 and 2 (aldh1a2 and aldh2) is detected by aldefluor, inhibited by diethylaminobenzaldehyde and has significant effects on cell proliferation and drug resistance. Chem Biol Interact. 2012;195(1):52–60.
- Giorgianni F, Bridson PK, Sorrentino BP, Pohl J, Blakley RL. Inactivation of aldophosphamide by human aldehyde dehydrogenase isozyme 3. Biochem Pharmacol. 2000;60(3):325–338.
- Magni M, Shammah S, Schiro R, Mellado W, Dalla-Favera R, Gianni AM. Induction of cyclophosphamide-resistance by aldehyde-dehydrogenase gene transfer. Blood. 1996;87(3):1097–1103.
- Storms RW, Trujillo AP, Springer JB, Shah L, Colvin OM, Ludeman SM, Smith C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc Natl Acad Sci USA. 1999;96(16):9118–9123.
- Kastan MB, Schlaffer E, Russo JE, Colvin OM, Civin CI, Hilton J. Direct demonstration of elevated aldehyde dehydrogenase in human hematopoietic progenitor cells. Blood. 1990;75(10):1947–1950.
- Estes BT, Wu AW, Storms RW, Guilak F. Extended passaging, but not aldehyde dehydrogenase activity, increases the chondrogenic potential of human adipose-derived adult stem cells. J Cell Physiol. 2006;209(3):987–995.
- Jean E, Laoudj-Chenivesse D, Notarnicola C, Rouger K, Serratrice N, Bonnieu A, Gay S, Bacou F, Duret C, Carnac G. Aldehyde dehydrogenase activity promotes survival of human muscle precursor cells. J Cell Mol Med. 2011;15(1):119–133.
- Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG, Liu S, et al. Aldh1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567.
- Dalley AJ, AbdulMajeed AA, Upton Z, Farah CS. Organotypic culture of normal, dysplastic and squamous cell carcinoma-derived oral cell lines reveals loss of spatial regulation of cd44 and p75 ntr in malignancy. J Oral Pathol Med. 2013;42(1):37–46.
- Lohberger B, Rinner B, Stuendl N, Absenger M, Liegl-Atzwanger B, Walzer SM, Windhager R, Leithner A. Aldehyde dehydrogenase 1, a potential marker for cancer stem cells in human sarcoma. PLoS One. 2012;7(8):e43664.
- Minato T, Yamamoto Y, Seike J, Yoshida T, Yamai H, Takechi H, Yuasa Y, Furukita Y, Goto M, Bando Y, et al. Aldehyde dehydrogenase 1 expression is associated with poor prognosis in patients with esophageal squamous cell carcinoma. Ann Surg Oncol. 2013;20(1):209–217.
- Wang Y, Zhe H, Gao P, Zhang N, Li G, Qin J. Cancer stem cell marker aldh1 expression is associated with lymph node metastasis and poor survival in esophageal squamous cell carcinoma: a study from high incidence area of northern china. Dis Esophagus. 2012;25(6):560–565.
- Rassouli FB, Matin MM, Saeinasab M. Cancer stem cells in human digestive tract malignancies. Tumour Biol. 2016;37(1):7–21.
- Katsuno Y, Ehata S, Yashiro M, Yanagihara K, Hirakawa K, Miyazono K. Coordinated expression of reg4 and aldehyde dehydrogenase 1 regulating tumourigenic capacity of diffuse-type gastric carcinoma-initiating cells is inhibited by tgf-beta. J Pathol. 2012;228(3):391–404.
- Wakamatsu Y, Sakamoto N, Oo HZ, Naito Y, Uraoka N, Anami K, Sentani K, Oue N, Yasui W. Expression of cancer stem cell markers aldh1, cd44 and cd133 in primary tumor and lymph node metastasis of gastric cancer. Pathol Int. 2012;62(2):112–119.
- Carpentino JE, Hynes MJ, Appelman HD, Zheng T, Steindler DA, Scott EW, Huang EH. Aldehyde dehydrogenase-expressing colon stem cells contribute to tumorigenesis in the transition from colitis to cancer. Cancer Res. 2009;69(20):8208–8215.
- Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, Fields JZ, Wicha MS, Boman BM. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (sc) and tracks sc overpopulation during colon tumorigenesis. Cancer Res. 2009;69(8):3382–3389.
- Kozovska Z, Patsalias A, Bajzik V, Durinikova E, Demkova L, Jargasova S, Smolkova B, Plava J, Kucerova L, Matuskova M. Aldh1a inhibition sensitizes colon cancer cells to chemotherapy. BMC Cancer. 2018;18(1):656.
- Chen YC, Chen YW, Hsu HS, Tseng LM, Huang PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385(3):307–313.
- Kurashige T, Shimamura M, Yasui K, Mitsutake N, Matsuse M, Nakashima M, Minami S, Eguchi S, Nagayama Y. Studies on expression of aldehyde dehydrogenase in normal and cancerous tissues of thyroids. Horm Metab Res. 2015;47(3):194–199.
- Jiang F, Qiu Q, Khanna A, Todd NW, Deepak J, Xing L, Wang H, Liu Z, Su Y, Stass SA, et al. Aldehyde dehydrogenase 1 is a tumor stem cell-associated marker in lung cancer. Mol Cancer Res. 2009;7(3):330–338.
- Li X, Wan L, Geng J, Wu CL, Bai X. Aldehyde dehydrogenase 1a1 possesses stem-like properties and predicts lung cancer patient outcome. J Thorac Oncol. 2012;7(8):1235–1245.
- Moreb JS, Baker HV, Chang LJ, Amaya M, Lopez MC, Ostmark B, Chou W. Aldh isozymes downregulation affects cell growth, cell motility and gene expression in lung cancer cells. Mol Cancer. 2008;7:87.
- Chen MH, Weng JJ, Cheng CT, Wu RC, Huang SC, Wu CE, Chung YH, Liu CY, Chang MH, Chen MH, et al. Aldh1a3, the major aldehyde dehydrogenase isoform in human cholangiocarcinoma cells, affects prognosis and gemcitabine resistance in cholangiocarcinoma patients. Clin Cancer Res. 2016;22(16):4225–4235.
- Ma S, Chan KW, Lee TK, Tang KH, Wo JY, Zheng BJ, Guan XY. Aldehyde dehydrogenase discriminates the cd133 liver cancer stem cell populations. Mol Cancer Res. 2008;6(7):1146–1153.
- Tanaka K, Tomita H, Hisamatsu K, Nakashima T, Hatano Y, Sasaki Y, Osada S, Tanaka T, Miyazaki T, Yoshida K, et al. Aldh1a1-overexpressing cells are differentiated cells but not cancer stem or progenitor cells in human hepatocellular carcinoma. Oncotarget. 2015;6(28):24722–24732.
- Rasheed Z, Wang Q, Matsui W. Isolation of stem cells from human pancreatic cancer xenografts. J Vis Exp. 2010;43:2169.
- Rasheed ZA, Yang J, Wang Q, Kowalski J, Freed I, Murter C, Hong SM, Koorstra JB, Rajeshkumar NV, He X, et al. Prognostic significance of tumorigenic cells with mesenchymal features in pancreatic adenocarcinoma. J Natl Cancer Inst. 2010;102(5):340–351.
- Ishizawa K, Rasheed ZA, Karisch R, Wang Q, Kowalski J, Susky E, Pereira K, Karamboulas C, Moghal N, Rajeshkumar NV, et al. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell. 2010;7(3):279–282.
- Honoki K, Fujii H, Kubo A, Kido A, Mori T, Tanaka Y, Tsujiuchi T. Possible involvement of stem-like populations with elevated aldh1 in sarcomas for chemotherapeutic drug resistance. Oncol Rep. 2010;24(2):501–505.
- Martins-Neves SR, Corver WE, Paiva-Oliveira DI, van den Akker BE, Briaire-de-Bruijn IH, Bovee JV, Gomes CM, Cleton-Jansen AM. Osteosarcoma stem cells have active Wnt/β-catenin and overexpress SOX2 and KLF4. J Cell Physiol. 2016;231(4):876–886.
- Martins-Neves SR, Paiva-Oliveira DI, Wijers-Koster PM, Abrunhosa AJ, Fontes-Ribeiro C, Bovee JV, Cleton-Jansen AM, Gomes CM. Chemotherapy induces stemness in osteosarcoma cells through activation of Wnt/β-catenin signaling. Cancer Lett. 2016;370(2):286–295.
- Li T, Su Y, Mei Y, Leng Q, Leng B, Liu Z, Stass SA, Jiang F. Aldh1a1 is a marker for malignant prostate stem cells and predictor of prostate cancer patients’ outcome. Lab Invest. 2010;90(2):234–244.
- van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Pelger RC, van der Pluijm G. The aldehyde dehydrogenase enzyme 7a1 is functionally involved in prostate cancer bone metastasis. Clin Exp Metastasis. 2011;28(7):615–625.
- Falso MJ, Buchholz BA, White RW. Stem-like cells in bladder cancer cell lines with differential sensitivity to cisplatin. Anticancer Res. 2012;32(3):733–738.
- Ferreira-Teixeira M, Parada B, Rodrigues-Santos P, Alves V, Ramalho JS, Caramelo F, Sousa V, Reis F, Gomes CM. Functional and molecular characterization of cancer stem-like cells in bladder cancer: a potential signature for muscle-invasive tumors. Oncotarget. 2015;6(34):36185–36201.
- Su Y, Qiu Q, Zhang X, Jiang Z, Leng Q, Liu Z, Stass SA, Jiang F. Aldehyde dehydrogenase 1 a1-positive cell population is enriched in tumor-initiating cells and associated with progression of bladder cancer. Cancer Epidemiol Biomarkers Prev. 2010;19(2):327–337.
- Rasper M, Schafer A, Piontek G, Teufel J, Brockhoff G, Ringel F, Heindl S, Zimmer C, Schlegel J. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro Oncol. 2010;12(10):1024–1033.
- Boonyaratanakornkit JB, Yue L, Strachan LR, Scalapino KJ, LeBoit PE, Lu Y, Leong SP, Smith JE, Ghadially R. Selection of tumorigenic melanoma cells using aldh. J Invest Dermatol. 2010;130(12):2799–2808.
- Chang B, Liu G, Xue F, Rosen DG, Xiao L, Wang X, Liu J. Aldh1 expression correlates with favorable prognosis in ovarian cancers. Mod Pathol. 2009;22(6):817–823.
- Januchowski R, Wojtowicz K, Sterzyſska K, Sosiſska P, Andrzejewska M, Zawierucha P, Nowicki M, Zabel M. Inhibition of aldh1a1 activity decreases expression of drug transporters and reduces chemotherapy resistance in ovarian cancer cell lines. Int J Biochem Cell Biol. 2016;78:248–259.
- Huddle BC, Grimley E, Buchman CD, Chtcherbinine M, Debnath B, Mehta P, Yang K, Morgan CA, Li S, Felton J, et al. Structure-based optimization of a novel class of aldehyde dehydrogenase 1a (aldh1a) subfamily-selective inhibitors as potential adjuncts to ovarian cancer chemotherapy. J Med Chem. 2018;61(19):8754–8773.
- Rao Q-X, Yao T-T, Zhang B-Z, Lin R-C, Chen Z-L, Zhou H, Wang L-J, Lu H-W, Chen Q, Di N, et al. Expression and functional role of aldh1 in cervical carcinoma cells. Asian Pac J Cancer Prev. 2012;13(4):1325–1331.
- Yao T, Chen Q, Zhang B, Zhou H, Lin Z. The expression of aldh1 in cervical carcinoma. Med Sci Monit. 2011;17(8):HY21–26.
- Jin N, Zhu X, Cheng F, Zhang L. Disulfiram/copper targets stem cell-like aldh(+) population of multiple myeloma by inhibition of aldh1a1 and hedgehog pathway. J Cell Biochem. 2018;119(8):6882–6893.
- Levi BP, Yilmaz OH, Duester G, Morrison SJ. Aldehyde dehydrogenase 1a1 is dispensable for stem cell function in the mouse hematopoietic and nervous systems. Blood. 2009;113(8):1670–1680.
- Zhou L, Sheng D, Wang D, Ma W, Deng Q, Deng L, Liu S. Identification of cancer-type specific expression patterns for active aldehyde dehydrogenase (aldh) isoforms in aldefluor assay. Cell Biol Toxicol. 2019;35(2):161–177.
- Morgan CA, Parajuli B, Buchman CD, Dria K, Hurley TD. N,n-diethylaminobenzaldehyde (deab) as a substrate and mechanism-based inhibitor for human aldh isoenzymes. Chem Biol Interact. 2015;234:18–28.
- Chen Y, Orlicky DJ, Matsumoto A, Singh S, Thompson DC, Vasiliou V. Aldehyde dehydrogenase 1b1 (aldh1b1) is a potential biomarker for human colon cancer. Biochem Biophys Res Commun. 2011;405(2):173–179.
- Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10(9):1378–1384.
- Yang X, Yao R, Wang H. Update of aldh as a potential biomarker and therapeutic target for aml. Biomed Res Int. 2018;2018:9192104.
- Smith C, Gasparetto M, Humphries K, Pollyea DA, Vasiliou V, Jordan CT. Aldehyde dehydrogenases in acute myeloid leukemia. Ann NY Acad Sci. 2014;1310:58–68.
- Gasparetto M, Sekulovic S, Brocker C, Tang P, Zakaryan A, Xiang P, Kuchenbauer F, Wen M, Kasaian K, Witty MF, et al. Aldehyde dehydrogenases are regulators of hematopoietic stem cell numbers and b-cell development. Exp Hematol. 2012;40(4):318–329 e312.
- Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, Matsumoto A, Thompson DC, Vasiliou V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic Biol Med. 2013;56:89–101.
- Gasparetto M, Smith CA. Aldhs in normal and malignant hematopoietic cells: potential new avenues for treatment of aml and other blood cancers. Chem Biol Interact. 2017;276:46–51.
- Pearce DJ, Taussig D, Simpson C, Allen K, Rohatiner AZ, Lister TA, Bonnet D. Characterization of cells with a high aldehyde dehydrogenase activity from cord blood and acute myeloid leukemia samples. Stem Cells. 2005;23(6):752–760.
- Moreb JS, Turner C, Sreerama L, Zucali JR, Sladek NE, Schweder M. Interleukin-1 and tumor necrosis factor alpha induce class 1 aldehyde dehydrogenase mrna and protein in bone marrow cells. Leuk Lymphoma. 1995;20(1–2):77–84.
- Moreb JS, Maccow C, Schweder M, Hecomovich J. Expression of antisense rna to aldehyde dehydrogenase class-1 sensitizes tumor cells to 4-hydroperoxycyclophosphamide in vitro. J Pharmacol Exp Ther. 2000;293(2):390–396.
- Su M, Alonso S, Jones JW, Yu J, Kane MA, Jones RJ, Ghiaur G. All-trans retinoic acid activity in acute myeloid leukemia: role of cytochrome p450 enzyme expression by the microenvironment. PLoS One. 2015;10(6):e0127790.
- Xu B, Wang S, Li R, Chen K, He L, Deng M, Kannappan V, Zha J, Dong H, Wang W. Disulfiram/copper selectively eradicates aml leukemia stem cells in vitro and in vivo by simultaneous induction of ros-jnk and inhibition of nf-kappab and nrf2. Cell Death Dis. 2017;8(5):e2797.
- Conticello C, Martinetti D, Adamo L, Buccheri S, Giuffrida R, Parrinello N, Lombardo L, Anastasi G, Amato G, Cavalli M, et al. Disulfiram, an old drug with new potential therapeutic uses for human hematological malignancies. Int J Cancer. 2012;131(9):2197–2203.
- Bista R, Lee DW, Pepper OB, Azorsa DO, Arceci RJ, Aleem E. Disulfiram overcomes bortezomib and cytarabine resistance in down-syndrome-associated acute myeloid leukemia cells. J Exp Clin Cancer Res. 2017;36(1):22.
- Venton G, Perez-Alea M, Baier C, Fournet G, Quash G, Labiad Y, Martin G, Sanderson F, Poullin P, Suchon P, et al. Aldehyde dehydrogenases inhibition eradicates leukemia stem cells while sparing normal progenitors. Blood Cancer J. 2016;6(9):e469.
- Ratajczak J, Zuba-Surma E, Klich I, Liu R, Wysoczynski M, Greco N, Kucia M, Laughlin MJ, Ratajczak MZ. Hematopoietic differentiation of umbilical cord blood-derived very small embryonic/epiblast-like stem cells. Leukemia. 2011;25(8):1278–1285.
- He X, Gonzalez V, Tsang A, Thompson J, Tsang TC, Harris DT. Differential gene expression profiling of cd34+ cd133+ umbilical cord blood hematopoietic stem progenitor cells. Stem Cells Dev. 2005;14(2):188–198.
- Lassen N, Bateman JB, Estey T, Kuszak JR, Nees DW, Piatigorsky J, Duester G, Day BJ, Huang J, Hines LM, et al. Multiple and additive functions of aldh3a1 and aldh1a1: cataract phenotype and ocular oxidative damage in aldh3a1(–/–)/aldh1a1(–/–) knock-out mice. J Biol Chem. 2007;282(35):25668–25676.
- Zhao D, Mo Y, Li MT, Zou SW, Cheng ZL, Sun YP, Xiong Y, Guan KL, Lei QY. Notch-induced aldehyde dehydrogenase 1a1 deacetylation promotes breast cancer stem cells. J Clin Invest. 2014;124(12):5453–5465.
- Croker AK, Allan AL. Inhibition of aldehyde dehydrogenase (aldh) activity reduces chemotherapy and radiation resistance of stem-like aldhhicd44(+) human breast cancer cells. Breast Cancer Res Treat. 2012;133(1):75–87.
- Yan Y, Li Z, Xu X, Chen C, Wei W, Fan M, Chen X, Li JJ, Wang Y, Huang J. All-trans retinoic acids induce differentiation and sensitize a radioresistant breast cancer cells to chemotherapy. BMC Complement Altern Med. 2016;16(113):113.
- Dehghan Harati M, Rodemann HP, Toulany M. Nanog signaling mediates radioresistance in aldh-positive breast cancer cells. Int J Mol Sci. 2019;20(5):1151.
- Moreb JS, Ucar-Bilyeu DA, Khan A. Use of retinoic acid/aldehyde dehydrogenase pathway as potential targeted therapy against cancer stem cells. Cancer Chemother Pharmacol. 2017;79(2):295–301.
- Deng S, Yang X, Lassus H, Liang S, Kaur S, Ye Q, Li C, Wang LP, Roby KF, Orsulic S, et al. Distinct expression levels and patterns of stem cell marker, aldehyde dehydrogenase isoform 1 (aldh1), in human epithelial cancers. PLoS One. 2010;5(4):e10277.
- Rexer BN, Zheng WL, Ong DE. Retinoic acid biosynthesis by normal human breast epithelium is via aldehyde dehydrogenase 6, absent in mcf-7 cells. Cancer Res. 2001;61(19):7065–7070.
- Marcato P, Dean CA, Pan D, Araslanova R, Gillis M, Joshi M, Helyer L, Pan L, Leidal A, Gujar S, et al. Aldehyde dehydrogenase activity of breast cancer stem cells is primarily due to isoform aldh1a3 and its expression is predictive of metastasis. Stem Cells. 2011;29(1):32–45.
- Marcato P, Dean CA, Liu RZ, Coyle KM, Bydoun M, Wallace M, Clements D, Turner C, Mathenge EG, Gujar SA, et al. Aldehyde dehydrogenase 1a3 influences breast cancer progression via differential retinoic acid signaling. Mol Oncol. 2015;9(1):17–31.
- Kato H, Izumi K, Saito T, Ohnuki H, Terada M, Kawano Y, Nozawa-Inoue K, Saito C, Maeda T. Distinct expression patterns and roles of aldehyde dehydrogenases in normal oral mucosa keratinocytes: differential inhibitory effects of a pharmacological inhibitor and RNAI-mediated knockdown on cellular phenotype and epithelial morphology. Histochem Cell Biol. 2013;139(6):847–862.
- Hedberg JJ, Grafstrom RC, Vondracek M, Sarang Z, Warngard L, Hoog JO. Micro-array chip analysis of carbonyl-metabolising enzymes in normal, immortalised and malignant human oral keratinocytes. Cell Mol Life Sci. 2001;58(11):1719–1726.
- Kahlert C, Bergmann F, Beck J, Welsch T, Mogler C, Herpel E, Dutta S, Niemietz T, Koch M, Weitz J. Low expression of aldehyde dehydrogenase 1a1 (aldh1a1) is a prognostic marker for poor survival in pancreatic cancer. BMC Cancer. 2011;11:275.
- Kim IG, Lee JH, Kim SY, Kim JY, Cho EW. Fibulin-3 negatively regulates aldh1 via c-met suppression and increases gamma-radiation-induced sensitivity in some pancreatic cancer cell lines. Biochem Biophys Res Commun. 2014;454(3):369–375.
- Moreb JS, Muhoczy D, Ostmark B, Zucali JR. Rnai-mediated knockdown of aldehyde dehydrogenase class-1a1 and class-3a1 is specific and reveals that each contributes equally to the resistance against 4-hydroperoxycyclophosphamide. Cancer Chemother Pharmacol. 2007;59(1):127–136.
- Moreb JS, Gabr A, Vartikar GR, Gowda S, Zucali JR, Mohuczy D. Retinoic acid down-regulates aldehyde dehydrogenase and increases cytotoxicity of 4-hydroperoxycyclophosphamide and acetaldehyde. J Pharmacol Exp Ther. 2005;312(1):339–345.
- Sullivan JP, Spinola M, Dodge M, Raso MG, Behrens C, Gao B, Schuster K, Shao C, Larsen JE, Sullivan LA, et al. Aldehyde dehydrogenase activity selects for lung adenocarcinoma stem cells dependent on notch signaling. Cancer Res. 2010;70(23):9937–9948.
- Zhang Q, Lu C, Fang T, Wang Y, Hu W, Qiao J, Liu B, Liu J, Chen N, Li M, et al. Notch3 functions as a regulator of cell self-renewal by interacting with the beta-catenin pathway in hepatocellular carcinoma. Oncotarget. 2015;6(6):3669–3679.
- Xiao ZJ, Liu J, Wang SQ, Zhu Y, Gao XY, Tin VP, Qin J, Wang JW, Wong MP. Nfatc2 enhances tumor-initiating phenotypes through the nfatc2/sox2/aldh axis in lung adenocarcinoma. Elife. 2017;6:e26733.
- Voronkova MA, Rojanasakul LW, Kiratipaiboon C, Rojanasakul Y. Sox9-aldh axis determines resistance to chemotherapy in non-small cell lung cancer. Mol Cell Biol. 2019;40(2):e00307-19.
- Shao C, Sullivan JP, Girard L, Augustyn A, Yenerall P, Rodriguez-Canales J, Liu H, Behrens C, Shay JW, Wistuba II, et al. Essential role of aldehyde dehydrogenase 1a3 for the maintenance of non-small cell lung cancer stem cells is associated with the stat3 pathway. Clin Cancer Res. 2014;20(15):4154–4166.
- Erfani E, Roudi R, Rakhshan A, Sabet MN, Shariftabrizi A, Madjd Z. Comparative expression analysis of putative cancer stem cell markers cd44 and aldh1a1 in various skin cancer subtypes. Int J Biol Markers. 2016;31(1):e53-61–e61.
- Luo Y, Dallaglio K, Chen Y, Robinson WA, Robinson SE, McCarter MD, Wang J, Gonzalez R, Thompson DC, Norris DA, et al. Aldh1a isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells. 2012;30(10):2100–2113.
- Vasiliou V, Thompson DC, Smith C, Fujita M, Chen Y. Aldehyde dehydrogenases: from eye crystallins to metabolic disease and cancer stem cells. Chem Biol Interact. 2013;202(1–3):2–10.
- van den Hoogen C, van der Horst G, Cheung H, Buijs JT, Lippitt JM, Guzman-Ramirez N, Hamdy FC, Eaton CL, Thalmann GN, Cecchini MG, et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70(12):5163–5173.
- Kalantari E, Saadi FH, Asgari M, Shariftabrizi A, Roudi R, Madjd Z. Increased expression of aldh1a1 in prostate cancer is correlated with tumor aggressiveness: a tissue microarray study of Iranian patients. Appl Immunohistochem Mol Morphol. 2017;25(8):592–598.
- Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, Wicha MS, Prince ME. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32(9):1195–1201.
- Wu J, Mu Q, Thiviyanathan V, Annapragada A, Vigneswaran N. Cancer stem cells are enriched in fanconi anemia head and neck squamous cell carcinomas. Int J Oncol. 2014;45(6):2365–2372.
- Kurth I, Hein L, Mabert K, Peitzsch C, Koi L, Cojoc M, Kunz-Schughart L, Baumann M, Dubrovska A. Cancer stem cell related markers of radioresistance in head and neck squamous cell carcinoma. Oncotarget. 2015;6(33):34494–34509.
- Suzuki E, Chiba T, Zen Y, Miyagi S, Tada M, Kanai F, Imazeki F, Miyazaki M, Iwama A, Yokosuka O. Aldehyde dehydrogenase 1 is associated with recurrence-free survival but not stem cell-like properties in hepatocellular carcinoma. Hepatol Res. 2012;42(11):1100–1111.
- Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, Guerrero-Cazares H, Quinones-Hinojosa A, Laterra J, Xia S. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30(31):3454–3467.
- Xu SL, Liu S, Cui W, Shi Y, Liu Q, Duan JJ, Yu SC, Zhang X, Cui YH, Kung HF, et al. Aldehyde dehydrogenase 1a1 circumscribes high invasive glioma cells and predicts poor prognosis. Am J Cancer Res. 2015;5(4):1471–1483.
- Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1a3. Proc Natl Acad Sci USA. 2013;110(21):8644–8649.
- Unguryte A, Bernotiene E, Bagdonas E, Garberyte S, Porvaneckas N, Jorgensen C. Human articular chondrocytes with higher aldehyde dehydrogenase activity have stronger expression of col2a1 and sox9. Osteoarthritis Cartilage. 2016;24(5):873–882.
- Russo JE, Hauguitz D, Hilton J. Inhibition of mouse cytosolic aldehyde dehydrogenase by 4-(diethylamino)benzaldehyde. Biochem Pharmacol. 1988;37(8):1639–1642.
- Wang MF, Han CL, Yin SJ. Substrate specificity of human and yeast aldehyde dehydrogenases. Chem Biol Interact. 2009;178(1–3):36–39.
- Koppaka V, Thompson DC, Chen Y, Ellermann M, Nicolaou KC, Juvonen RO, Petersen D, Deitrich RA, Hurley TD, Vasiliou V. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64(3):520–539.
- Pors K, Moreb JS. Aldehyde dehydrogenases in cancer: an opportunity for biomarker and drug development? Drug Discov Today. 2014;19(12):1953–1963.
- Russo JE, Hilton J. Characterization of cytosolic aldehyde dehydrogenase from cyclophosphamide resistant l1210 cells. Cancer Res. 1988;48(11):2963–2968.
- Ali IM, Ibrahim EB, Sneha S, Jiménez R, Pequerul R, Parés X, Rüngeler T, Jha V, Tuccinardi T, Sadiq M, et al. Klaus Pors Expansion of the 4-(diethylamino)benzaldehyde scaffold to explore the impact on aldehyde dehydrogenase activity and antiproliferative activity in prostate cancer. J Med Chem. 2022;65(5):16.
- Yao L, Fan P, Arolfo M, Jiang Z, Olive MF, Zablocki J, Sun HL, Chu N, Lee J, Kim HY, et al. Inhibition of aldehyde dehydrogenase-2 suppresses cocaine seeking by generating thp, a cocaine use-dependent inhibitor of dopamine synthesis. Nat Med. 2010;16(9):1024–1028.
- Lai CL, Li YP, Liu CM, Hsieh HS, Yin SJ. Inhibition of human alcohol and aldehyde dehydrogenases by cimetidine and assessment of its effects on ethanol metabolism. Chem Biol Interact. 2013;202(1–3):275–282.
- Jianfeng Li SG, Ye Z, Moretti A, Belyaeva OV, Beiser A, Ibrahim M, Wilk A, McClellan S, Klyuyeva AV, Goggans KR, et al. A specific inhibitor of aldh1a3 regulates retinoic acid biosynthesis in glioma stem cells. Commun Biol. 2021;4(1):1420.
- Kumar LLS, Trainor PA, Koentgen F, Duester G. Alcohol and aldehyde dehydrogenases: retinoid metabolic effects in mouse knockout models. Biochim Biophys Acta. 2012;1821(1):8.
- Karen Niederreither VF, Garnier J-M, Chambon P, Dollé P. Differential expression of retinoic acid-synthesizing (raldh) enzymes during fetal development and organ differentiation in the mouse. Mech Dev. 2002;110(1–2):5.
- Bingyan Li Kang Yang DL, Jiang C, Ma Z. Discovery and development of selective aldehyde dehydrogenase 1a1 (aldh1a1) inhibitors. Eur J Med Chem. 2021;2021:209.
- Gelardi ELM, Colombo G, Picarazzi F, Ferraris DM, Mangione A, Petrarolo G, Aronica E, Rizzi M, Mori M, La Motta C, et al. A selective competitive inhibitor of aldehyde dehydrogenase 1a3 hinders cancer cell growth, invasiveness and stemness in vitro. Cancers . 2021;2021(2):13.
- Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J, et al. The first structure of an aldehyde dehydrogenase reveals novel interactions between nad and the rossmann fold. Nat Struct Biol. 1997;4(4):317–326.
- Feng Z, Hom ME, Bearrood TE, Rosenthal ZC, Fernandez D, Ondrus AE, Gu Y, McCormick AK, Tomaske MG, Marshall CR, et al. Targeting colorectal cancer with small-molecule inhibitors of aldh1b1. Nat Chem Biol. 2022;18(10):1065–1075.
- Li Y, Zhang T, Korkaya H, Liu S, Lee HF, Newman B, Yu Y, Clouthier SG, Schwartz SJ, Wicha MS, et al. Sulforaphane, a dietary component of broccoli/broccoli sprouts, inhibits breast cancer stem cells. Clin Cancer Res. 2010;16(9):2580–2590.
- Morgan CA, Hurley TD. Development of a high-throughput in vitro assay to identify selective inhibitors for human aldh1a1. Chem Biol Interact. 2015;234:29–37.
- Moretti A, Li J, Donini S, Sobol RW, Rizzi M, Garavaglia S. Crystal structure of human aldehyde dehydrogenase 1a3 complexed with nad(+) and retinoic acid. Sci Rep. 2016;6:35710.
- Condello S, Morgan CA, Nagdas S, Cao L, Turek J, Hurley TD, Matei D. Beta-catenin-regulated aldh1a1 is a target in ovarian cancer spheroids. Oncogene. 2015;34(18):2297–2308.
- Parajuli B, Kimble-Hill AC, Khanna M, Ivanova Y, Meroueh S, Hurley TD. Discovery of novel regulators of aldehyde dehydrogenase isoenzymes. Chem Biol Interact. 2011;191(1–3):153–158.
- Ibrahim AIM, Ikhmais B, Batlle E, AbuHarb WK, Jha V, Jaradat KT, Jimenez R, Pequerul R, Pares X, Farres J, et al. Design, synthesis, biological evaluation and in silico study of benzyloxybenzaldehyde derivatives as selective aldh1a3 inhibitors. Molecules. 2021;26(19):5770.
- King G, Hirst L, Holmes R. Human corneal and lens aldehyde dehydrogenases. Localization and function(s) of ocular aldh1 and aldh3 isozymes. Adv Exp Med Biol. 1999;463:189–198.
- Zhai Y, Sperkova Z, Napoli JL. Cellular expression of retinal dehydrogenase types 1 and 2: effects of vitamin a status on testis MRNA. J Cell Physiol. 2001;186(2):220–232.
- Armstrong L, Stojkovic M, Dimmick I, Ahmad S, Stojkovic P, Hole N, Lako M. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells. 2004;22(7):1142–1151.
- Chute JP, Muramoto GG, Whitesides J, Colvin M, Safi R, Chao NJ, McDonnell DP. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc Natl Acad Sci USA. 2006;103(31):11707–11712.
- Hsu LC, Chang WC, Hiraoka L, Hsieh CL. Molecular cloning, genomic organization, and chromosomal localization of an additional human aldehyde dehydrogenase gene, aldh6. Genomics. 1994;24(2):333–341.
- Zhang X, Zhang QY, Liu D, Su T, Weng Y, Ling G, Chen Y, Gu J, Schilling B, Ding X. Expression of cytochrome p450 and other biotransformation genes in fetal and adult human nasal mucosa. Drug Metab Dispos. 2005;33(10):1423–1428.
- Marchitti SA, Orlicky DJ, Brocker C, Vasiliou V. Aldehyde dehydrogenase 3b1 (aldh3b1): immunohistochemical tissue distribution and cellular-specific localization in normal and cancerous human tissues. J Histochem Cytochem. 2010;58(9):765–783.
- Singh S, Arcaroli J, Thompson DC, Messersmith W, Vasiliou V. Acetaldehyde and retinaldehyde-metabolizing enzymes in colon and pancreatic cancers. Adv Exp Med Biol. 2015;815:281–294.
- Cheung AM, Wan TS, Leung JC, Chan LY, Huang H, Kwong YL, Liang R, Leung AY. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior nod/scid engrafting potential. Leukemia. 2007;21(7):1423–1430.
- Liu C, Qiang J, Deng Q, Xia J, Deng L, Zhou L, Wang D, He X, Liu Y, Zhao B, et al. Aldh1a1 activity in tumor-initiating cells remodels myeloid-derived suppressor cells to promote breast cancer progression. Cancer Res. 2021;81(23):5919–5934.
- Ciccone V, Terzuoli E, Ristori E, Filippelli A, Ziche M, Morbidelli L, Donnini S. ALDH1A1 overexpression in melanoma cells promotes tumor angiogenesis by activating the IL-8/Notch signaling cascade. Int J Mol Med. 2022;50(1):5155.
- Choi SA, Lee JY, Phi JH, Wang KC, Park CK, Park SH, Kim SK. Identification of brain tumour initiating cells using the stem cell marker aldehyde dehydrogenase. Eur J Cancer. 2014;50(1):137–149.
- Wang Y, Shao F, Chen L. L C. Aldh1a2 suppresses epithelial ovarian cancer cell proliferation and migration by downregulating stat3. Onco Targets Ther. 2018;11:599–608.
- Nie S, Qian X, Shi M, Li H, Peng C, Ding X, Zhang S, Zhang B, Xu G, Lv Y, et al. Aldh1a3 accelerates pancreatic cancer metastasis by promoting glucose metabolism. Front Oncol. 2020;10:915.
- Duan JJ, Wang D, Cai J, Chen JJ, Zheng XX, Chen TQ, Wang J, Zhang X, Yang QK, Yu SC. An aldehyde dehydrogenase 1a3 inhibitor attenuates the metastasis of human colorectal cancer. Cancer Lett. 2022;536:215662.
- Crunkhorn S. Targeting aldh1b1 in colorectal cancer. Nat Rev Drug Discov. 2022;21(9):636.
- Wang Y, Li K, Zhao W, Liu Z, Liu J, Shi A, Chen T, Mu W, Xu Y, Pan C, et al. Aldehyde dehydrogenase 3b2 promotes the proliferation and invasion of cholangiocarcinoma by increasing integrin beta 1 expression. Cell Death Dis. 2021;12(12):1158.
- Guo YF, Duan JJ, Wang J, Li L, Wang D, Liu XZ, Yang J, Zhang HR, Lv J, Yang YJ, et al. Inhibition of the aldh18a1-mycn positive feedback loop attenuates mycn-amplified neuroblastoma growth. Sci Transl Med. 2020;12:531.
- Canuto RA, Muzio G, Salvo RA, Maggiora M, Trombetta A, Chantepie J, Fournet G, Reichert U, Quash G. The effect of a novel irreversible inhibitor of aldehyde dehydrogenases 1 and 3 on tumour cell growth and death. Chem Biol Interact. 2001;130–132(1–3):209–218.